Abstract
Constitutive expression of the chimeric NPM/ALK fusion protein encoded by the t(2;5)(p32;q35) is a key oncogenic event in the pathogenesis of most anaplastic large cell lymphomas (ALCLs). The proteomic network alterations produced by this aberration remain largely uncharacterized. Using a mass spectrometry (MS)–driven approach to identify changes in protein expression caused by the NPM/ALK fusion, we identified diverse NPM/ALK-induced changes affecting cell proliferation, ribosome synthesis, survival, apoptosis evasion, angiogenesis, and cytoarchitectural organization. MS-based findings were confirmed using Western blotting and/or immunostaining of NPM/ALK-transfected cells and ALK-deregulated lymphomas. A subset of the proteins distinguished NPM/ALK-positive ALCLs from NPM/ALK-negative ALCLs and Hodgkin lymphoma. The multiple NPM/ALK-deregulated pathways identified by MS analysis also predicted novel biologic effects of NPM/ALK expression. In this regard, we showed loss of cell adhesion as a consequence of NPM/ALK expression in a kinase-dependent manner, and sensitivity of NPM/ALK-positive ALCLs to inhibition of the RAS, p42/44ERK, and FRAP/mTOR signaling pathways. These findings reveal that the NPM/ALK alteration affects diverse cellular pathways, and provide novel insights into NPM/ALK-positive ALCL pathobiology. Our studies carry important implications for the use of MS-driven approaches for the elucidation of neoplastic pathobiology, the identification of novel diagnostic biomarkers, and pathogenetically relevant therapeutic targets.
Introduction
Chromosomal translocations are among the most frequent genetic alterations identified in cancer1,2 and constitute a common mechanism of oncogenic activation. A frequent consequence of chromosomal translocation is the generation of a chimeric transcript that encodes a fusion protein,2 leading to constitutive activation of tyrosine kinases that are central to the development and progression of many types of cancer.3
Anaplastic large cell lymphoma (ALCL) is the most common form of peripheral T-cell lymphoma in children.4 Most ALCLs harbor chromosomal translocations involving the anaplastic lymphoma kinase (ALK) gene, of which the t(2;5)(p23;q35) is by far the most frequent.5,6 The t(2;5) results in overexpression of a chimeric oncoprotein, nucleophosmin-anaplastic lymphoma kinase (NPM/ALK). In vitro and in vivo studies have shown that the constitutively active tyrosine kinase function of NPM/ALK is a key oncogenic event in the pathogenesis of t(2;5)-positive ALCLs.7,8
NPM/ALK modulates signaling pathways including PI3-K/AKT9 and JAK/STAT,9-11 however, the global impact of constitutive NPM/ALK expression in tumor cells is unknown. We reasoned that an unbiased approach to comprehensively assess the proteomic consequences of NPM/ALK expression would lead to novel insights with respect to the signaling pathways and functional alterations induced by the oncogene. In this regard, we performed quantitative proteomic analysis of NPM/ALK-positive and NPM/ALK-negative lymphoid cells using cleavable isotope-coded affinity tags (cICATs) and electrospray-ionization tandem mass spectrometry.12,13 Our studies reveal that NPM/ALK deregulates the expression of numerous proteins within previously described and novel signaling modules important for cell proliferation, survival, apoptosis evasion, and tumor dissemination. Knowledge of the proteomic changes produced by the singular NPM/ALK molecular alteration provides better understanding of ALCL pathobiology, and enables the identification of novel biomarkers and therapeutic targets.
Methods
Cell lines and drugs
Jurkat cells (ATCC TIB 152) were derived from acute T-lymphoblastic leukemia. The SUDHL-1 (DSMZ ACC 356) and Karpas 299 (DSMZ ACC 31) cell lines were derived from ALCLs, and carry a t(2;5)(p23;q35) aberration producing the NPM/ALK fusion gene. The Mac2A cell line was derived from a peripheral T-cell lymphoma and does not harbor the t(2;5)(p23;q35). All cells were cultures as described previously.14 U0126 (Upstate Biotechnology), rapamycin (Calbiochem), and FTI-277 (Calbiochem) were dissolved in dimethyl sulfoxide (DMSO).
Transfections
The NPM/ALK gene was obtained from Dr Stephan Morris (Department of Experimental Oncology, St Jude's Children's Research Hospital). Jurkat cells were transfected as outlined in supplemental information (available on the Blood website; see the Supplemental Materials link at the top of the online article). After G418 selection for 2 to 3 weeks, the expression of the fusion gene was enhanced by culturing transfected cells in medium containing 30% fetal bovine serum. Transfectants expressing NPM/ALK were screened by reverse-transcription–polymerase chain reaction (RT-PCR) and immunoblotting. The levels of NPM/ALK mRNA and protein were comparable with those seen in the ALCL-derived cell lines Karpas 299 and SUDHL-1 (as shown in supplemental Figure 1).
RT-PCR
Total RNA was extracted using TRIzol reagent (Gibco BRL), and Superscript II RT-PCR kit (Gibco BRL) was used to generate cDNA. The presence of NPM/ALK transcripts was detected with a primer pair spanning the NPM/ALK translocation breakpoint.15
Cell lysis and Western blot analysis
Cell lysis and Western blot analysis were performed according to protocol in Crockett.14 The antibodies are listed in supplemental Table 2.
MTT cell proliferation assay
Proliferation assay was performed using MTT reagent (3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide; ATCC) as described in Lin et al.16
Cell cycle analysis
Cells were stained with propidium iodide and analyzed by FACScan flow cytometry (Becton Dickinson) using Cell Quest software (Becton Dickinson), and G1, G2, + S phase values were recorded.16
Caspase-3 activity assay
Caspase-3 activity (CaspACE Assay System; Promega) measurements were conducted on cells grown under stated conditions. U0126 (Calbiochem) was added to SUDHL-1 cells to create a positive (induced apoptosis) control. Z-VAD-FMK inhibitor was used at a final concentration of 50 μM. A negative control was prepared using untreated cells.
ICAT mass spectrometry and statistical analysis
Tandem mass spectrometry (MS/MS) analysis was performed on the LCQ Deca XP ion trap mass spectrometer (ThermoFisher), using the Surveyor (Thermo) autosampler and MS pumps with Xcalibur software Version 1.4 (ThermoScientific) installed. ICAT labeling and 3-dimensional liquid chromatography were performed as described previously.12,17 We have provided the details of our mass spectrometry–drive proteomic analyses listed according to the stipulations of the “Proposed Guidelines for the Analysis and Documentation of Peptide and Protein Identifications”18 as supplemental information.
Analysis of NPM/ALK-deregulated pathways by knowledge-based structured network analysis tool (Ingenuity Pathway Analysis)
Differentially expressed proteins were further analyzed using Ingenuity Pathways Analysis. Files were submitted online for analysis and comparison with the Ingenuity gene/protein interaction knowledge base (http://www.ingenuity.com).
Immunofluorescence and confocal microscopy
Cells were fixed with 3.7% formaldehyde, and incubated with primary antibodies (1:100 or 1:200). Primary antibodies include ALK-1 (Cell Signaling), phospho-ALK (Cell Signaling), phospho-Tyr (Upstate 4G10), RhoA (Santa Cruz Biotechnology), and paxillin (Santa Cruz Biotechnology). After additional PBS washes, cells were incubated with 7.5 μg/mL FITC goat anti–rabbit IgG (Molecular Probes) and a 1:25 dilution of Texas Red-X Phalloidin (Molecular Probes). Confocal images were acquired using an Olympus FluoView (FVX200) laser scanning microscope (Olympus) with a 60 × oil inversion objective on a Olympus confocal microscope.
Adhesion and invasion assay
Cells were plated onto human plasma fibronectin (Chemicon International) and allowed to adhere for 24 hours. Media were removed and adherent cells were stained with crystal violet and manually counted. For invasion assays, cells were plated on BD Biosciences BioCoat membranes with Matrigel. Vector- and NPM/ALK-transfected NIH3T3 cells were scraped off the dish after 22 to 24 hours and plated onto BD BioCoat (BD Biosciences) membranes (1.25 × 105 cells/0.25 mL) and incubated for an additional 24 hours. Noninvading cells were swabbed from the top surface of the membranes. Cells remaining were washed one time in 1 × PBS and then fixed for 4 minutes in methanol. Cells were immersed in a crystal violet stain (20% ethanol, 0.05% crystal violet) for 30 minutes to 16 hours. Membranes were cut from inserts and embedded in gelvatol and sandwiched between microscope slides; cells were scored by light microscopy under 20 × magnification.
Tissue microarray construction and immunohistochemistry
Tumor specimens were obtained from the surgical pathology files of the Department of Pathology, University of Utah School of Medicine, and the Department of Hematopathology, The University of Texas M. D. Anderson Cancer Center. This study was approved by the Institutional Review Boards of the University of Utah and M. D. Anderson Cancer Center. Malignant lymphoma cases were classified according to the World Health Organization classification of lymphoid neoplasms4 and reviewed by 4 hematopathologists (M.S.L. and K.E.J. at the University of Michigan and G.R. and L.J.M. at M. D. Anderson Cancer Center). Immunohistochemistry was performed using protocol outlined in Lim et al.19
Results
Comparative proteomic analysis of NPM/ALK-positive and NPM/ALK-negative cells
To evaluate the proteomic consequences of NPM/ALK overexpression, we performed quantitative analysis using cICAT labeling followed by electrospray-ionization tandem mass spectrometry. See Figure 1 for overall experimental strategy. We ectopically expressed NPM/ALK in Jurkat T cells (supplemental Figure 1) to ensure the expression of NPM/ALK transcript and protein (both native ALK and phosphorylated ALK). We performed quantitative proteomic profiling of NPM/ALK-transfected cells compared with vector-transfected cells by cICAT. One-hundred ten proteins showed a 1.5-fold or greater change in the NPM/ALK-positive cells compared with the vector control cells (supplemental Table 1). Of these, 73 proteins were up-regulated, whereas 37 proteins were down-regulated. Figure 2 illustrates the relative overexpression (1.7-fold) and MS/MS identification of ribosomal S6 kinase in NPM/ALK-positive cells.
Strategy for determining proteomic consequences of NPM/ALK expression. Jurkat cells transfected with NPM/ALK cDNA were compared with those that were transfected with control LacZ (vector). One milligram from each cell lysate was labeled with either the light cICAT reagent (Lac Z Jurkat) or the heavy cICAT reagent (NPM/ALK-transfected Jurkat). The labeled proteins were combined; digested; subjected to cation-exchange chromatography, avidin chromatography, and reversed-phase chromatography; and analyzed by MS/MS. Selected proteins were verified by Western blot analysis, immunofluorescence microscopy, and immunohistochemistry and cellular pathways were functionally validated.
Strategy for determining proteomic consequences of NPM/ALK expression. Jurkat cells transfected with NPM/ALK cDNA were compared with those that were transfected with control LacZ (vector). One milligram from each cell lysate was labeled with either the light cICAT reagent (Lac Z Jurkat) or the heavy cICAT reagent (NPM/ALK-transfected Jurkat). The labeled proteins were combined; digested; subjected to cation-exchange chromatography, avidin chromatography, and reversed-phase chromatography; and analyzed by MS/MS. Selected proteins were verified by Western blot analysis, immunofluorescence microscopy, and immunohistochemistry and cellular pathways were functionally validated.
Identification and quantitation of ribosomal protein S6 kinase in NPM/ALK-transfected cells by ICAT-MS/MS. Data-dependent MS full scan and MS/MS sequencing scans for one ICAT pair identified ribosomal protein S6 kinase as 1.7-fold overexpressed (inset) in the NPM/ALK-positive cell lysate in comparison with NPM/ALK-negative cells. The b ion series (blue) and the y ion series (red) match the tryptic peptide AYSFCGTVEYMAPEVVNR. The b ion series represent peptide fragment ions wherein the positive charge is maintained on the N-terminus (blue m/z peaks and numbers). The y ion series represent the fragment ions where the positive charge is retained on C-terminus (red m/z peaks and numbers). The differences between successive b or y ions in a series indicate the m/z of the individual amino acid residues. In the bottom panel, column 1 (AA) represents amino acid residue. Column 2 represents the position of the amino acid residue in the peptide identified read in the N-to-C-terminal direction. Column 3 represents the m/z of the peptide at a specific amino acid position in the identified peptide read from an N-to-C-terminal direction. Column 4 represents the m/z of the peptide at a specific amino acid position C-to-N-terminal direction. Column 5 represents the position of the amino acid residue in the peptide read from the C-to-N-terminal direction.
Identification and quantitation of ribosomal protein S6 kinase in NPM/ALK-transfected cells by ICAT-MS/MS. Data-dependent MS full scan and MS/MS sequencing scans for one ICAT pair identified ribosomal protein S6 kinase as 1.7-fold overexpressed (inset) in the NPM/ALK-positive cell lysate in comparison with NPM/ALK-negative cells. The b ion series (blue) and the y ion series (red) match the tryptic peptide AYSFCGTVEYMAPEVVNR. The b ion series represent peptide fragment ions wherein the positive charge is maintained on the N-terminus (blue m/z peaks and numbers). The y ion series represent the fragment ions where the positive charge is retained on C-terminus (red m/z peaks and numbers). The differences between successive b or y ions in a series indicate the m/z of the individual amino acid residues. In the bottom panel, column 1 (AA) represents amino acid residue. Column 2 represents the position of the amino acid residue in the peptide identified read in the N-to-C-terminal direction. Column 3 represents the m/z of the peptide at a specific amino acid position in the identified peptide read from an N-to-C-terminal direction. Column 4 represents the m/z of the peptide at a specific amino acid position C-to-N-terminal direction. Column 5 represents the position of the amino acid residue in the peptide read from the C-to-N-terminal direction.
Functional groups of differentially expressed proteins in NPM/ALK-positive cells
We used the bioinformatics tool GoMiner (National Cancer Institute)20 to classify proteins deregulated by NPM/ALK expression into functional categories based on their gene ontology annotations.21 The proteins identified are from nuclear, cytoplasmic, membrane, and extracellular compartments (supplemental Figure 2A), representing multiple functional categories (supplemental Figure 2B), and include high and low abundance proteins such as myosin heavy polypeptide and transcription factors, respectively.
Identification of signaling proteins modulated by NPM/ALK
The proteins modulated by NPM/ALK expression are summarized in supplemental Table 1. We identified up-regulated expression of several proteins previously implicated in NPM/ALK signaling including PI3-K,9,10,22 JAK1,10 and SRC.23 We also identified MAPK pathway components that were up-regulated as a consequence of constitutive NPM/ALK expression. These included MEK kinase 1 (2.4-fold), which activates ERK1/2 and JNK1-3 MAPKs, and MAPK-APK (1.9-fold), a substrate of ERK1/2 and p38 MAPK. MAP/ERK kinase kinase 3 (MKK3) and MAPK4 (p63 MAPK) were also overexpressed by 5.9- and 4.5-fold, respectively.
Significantly, we identified several proteins that have not been previously recognized to be affected by constitutive NPM/ALK expression. We identified up-regulation of receptor tyrosine kinases including TEK (9.1-fold) and the Eph family receptor protein tyrosine kinase members, Eph tyrosine kinase 2 (5-fold), EphA3 (1.6-fold), EphB1 (3.4-fold), and HEK5 (2.8-fold).
Notably, our quantitative proteomic studies revealed modulation of protein phosphatases previously unknown to be affected by NPM/ALK expression. These include protein tyrosine phosphatase receptor type (CD148 or DEP1; 1.9-fold), protein tyrosine phosphatase 1B (PTP1B; 3.6-fold), and tyrosine phosphatase 1A-2β (3.0-fold), which are up-regulated, whereas phosphoprotein phosphatase (PP2AC) is down-regulated (2.5-fold).
Another previously unrecognized and therapeutically relevant finding is the identification of downstream targets of the FRAP/mTOR pathway that are up-regulated by NPM/ALK. These include ribosomal S6 kinase (1.6-fold), translational initiation factor eIF4 (4.8-fold), ribosomal protein L11 (4.8-fold), eukaryotic translation initiation factor 3 (3.2-fold), translation initiation factor IF-2 homolog (4.3-fold), and translation initiation factor eIF-2alpha kinase (3.4-fold). Figure 2 illustrates the data-dependent MS full scan and MS/MS sequencing scans for one ICAT pair that led to the identification of ribosomal protein S6 kinase as 1.7-fold overexpressed in the NPM/ALK-positive cell lysate in comparison with NPM/ALK-negative cells. The b ion series (blue) and the y ion series (red) match the tryptic peptide AYSFCGTVEYMAPEVVNR.
Western blot analysis corroborates MS-based quantitative proteomics
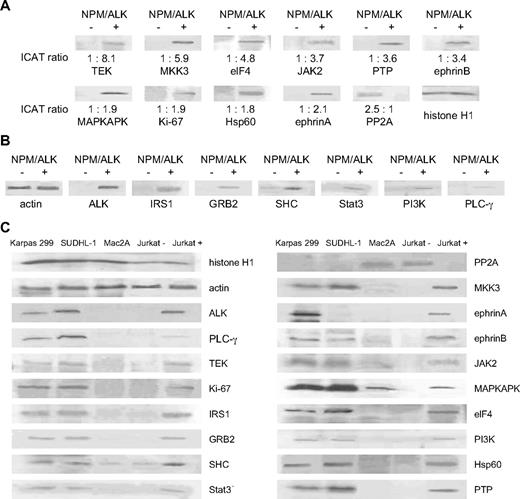
We performed Western blot analysis to validate the results obtained by cICAT-MS/MS. We observed complete concordance in the trends of differential expression of the proteins, despite differences in the quantitative ranges of the 2 approaches. Figure 3A demonstrates the differential expression of 11 selected proteins (10 up-regulated: TEK, MKK3, eIF4, Jak2, PTP, ephrinB, MAPKAPK, Ki-67, HSP60, ephrinA) and (1 down-regulated: PP2A) in the NPM/ALK-transfected cells compared with vector control cells by Western blot analysis with the corresponding ICAT ratios. Western blot analyses for an additional 6 proteins (IRS1, GRB2, SHC, Stat3, PI3-K, PLC-γ) also confirm the overexpression of proteins that have been previously reported to be up-regulated in NPM/ALK-positive ALCLs24 (Figure 3B).
Western blot analyses corroborate cICAT liquid chromatography–MS/MS studies and show parallel expression in NPM/LK-positive cell lines. (A) The differential expression identified by cICAT liquid chromatography–MS/MS was validated by Western blot analysis using control LacZ (NPM/ALK-negative) and NPM/ALK-transfected Jurkat cells (NPM/ALK positive). Expression of TEK, MKK3, eIF4, Jak2, PTP1B, ephrinB, MAPKAPK, Ki-67, Hsp60, ephrinA, and PP2A was compared with the cICAT ratios shown at the bottom of the Western blots. Histone H1 was used as protein loading control. (B) Up-regulated expression of ALK, and proteins previously reported to be mediators of NPM/ALK (IRS1, GRB2, SHC, Stat3, PI3-K, and PLCg), is corroborated by Western blot analyses. Actin was used as protein loading control. (C) Cell lines derived from NPM/ALK-positive ALCLs (SUDHL-1 and Karpas 299) were used to evaluate the expression of proteins found to be differentially expressed by the cICAT analysis as well as proteins previously known to be regulated by ALK. An ALK-negative ALCL cell line (Mac2A), and NPM/ALK-positive (Jurkat+) and NPM/ALK-negative (Jurkat−) cells are evaluated in comparison.
Western blot analyses corroborate cICAT liquid chromatography–MS/MS studies and show parallel expression in NPM/LK-positive cell lines. (A) The differential expression identified by cICAT liquid chromatography–MS/MS was validated by Western blot analysis using control LacZ (NPM/ALK-negative) and NPM/ALK-transfected Jurkat cells (NPM/ALK positive). Expression of TEK, MKK3, eIF4, Jak2, PTP1B, ephrinB, MAPKAPK, Ki-67, Hsp60, ephrinA, and PP2A was compared with the cICAT ratios shown at the bottom of the Western blots. Histone H1 was used as protein loading control. (B) Up-regulated expression of ALK, and proteins previously reported to be mediators of NPM/ALK (IRS1, GRB2, SHC, Stat3, PI3-K, and PLCg), is corroborated by Western blot analyses. Actin was used as protein loading control. (C) Cell lines derived from NPM/ALK-positive ALCLs (SUDHL-1 and Karpas 299) were used to evaluate the expression of proteins found to be differentially expressed by the cICAT analysis as well as proteins previously known to be regulated by ALK. An ALK-negative ALCL cell line (Mac2A), and NPM/ALK-positive (Jurkat+) and NPM/ALK-negative (Jurkat−) cells are evaluated in comparison.
Further, we evaluated the expression of these proteins in 2 cell lines derived from NPM/ALK-positive ALCLs (SUDHL-1 and Karpas 299), and compared them with a cell line derived from a ALK-negative ALCL (Mac2A) as well as the NPM/ALK-transfected Jurkat cells and the vector control cells (Figure 3C). Proteins up-regulated by NPM/ALK were also highly expressed in the SUDHL-1 and Karpas 299 cell lines. The down-regulation of PP2A seen in NPM/ALK-transfected Jurkat cells was reflected by relatively low levels of PP2A in the SUDHL-1 and Karpas 299 cell lines.
NPM/ALK regulates proteins associated with cytoskeletal architecture
Our proteomic profiling demonstrates differential expression of numerous proteins associated with cytoskeletal function, adhesion, motility, and invasion in NPM/ALK-positive cells. Up-regulated proteins include nephrocystin (4.5-fold), a component of FAK complex, TIAM-1 (1.5-fold), myosin (4.3-fold), Nck (5.3-fold), symplekin (4.0-fold), and fibrillin (2.8-fold). Proteins associated with adhesion such as alpha-5 β-1 integrin (2.1-fold), FAT gene product (cadherin; 2.3-fold), L-type amino acid transporter (11.1-fold), and Hic-5 (12.5-fold) are down-regulated.
To further investigate the extent to which NPM/ALK influences components of the cytoskeletal architecture, we analyzed the expression of paxillin and RhoA25,26 in NPM/ALK-transfected HEK293T cells. As shown in Figure 4A, RhoA and paxillin are overexpressed in NPM/ALK-transfected HEK293T cells compared with vector control cells. Furthermore, the induction of RhoA and paxillin is abrogated in cells transfected with the K210R kinase–defective mutant of NPM/ALK (Figure 4A). Similarly, the expression of tyrosine-phosphorylated proteins as detected by 4G10 antibody is increased in NPM/ALK-transfected cells and decreased in the K210R-transfected cells (Figure 4A), indicating that phosphorylation of RhoA and paxillin may be regulated by the kinase activity of NPM/ALK.
NPM/ALK expression induces changes in proteins associated with cytoskeleton, and affects cell adhesion and invasion. (A) HEK293T cells transfected with vector, wild-type (Wt), and kinase-defective mutant (K210R) NPM/ALK were evaluated by confocal immunofluorescence microscopy for the expression of the cytoskeletal proteins RhoA and paxillin. ALK (green) is expressed in Wt NPM/ALK-transfected HEK293T cells but not in vector-transfected HEK293T cells. The level of ALK expression is slightly decreased in the K210R-transfected cells. The Texas Red–phalloidin (red) staining of the vector-, Wt-, and K210R mutant–transfected cells does not show significant differences in actin expression. The expression of RhoA, paxillin, and phospho-Tyr proteins is significantly reduced in the K210R compared with Wt. (B) HEK293T cells transfected with vector, Wt, or the K210R NPM/ALK were assayed for adhesion to fibronectin. Data represent the mean of 2 experiments with error bars representing SD of triplicate mean. The unpaired Student t test was used (Wt vs K210R, P = .017). (C) The effect of NPM/ALK on invasive properties was evaluated using NIH3T3 cells transfected with vector or Wt NPM/ALK plated onto BioCoat Matrigel invasion chambers, and cells present at the opposite side of the coated membranes were stained with crystal violet. Cells per high-power field (400× magnification) were counted. The graphs represent the results of replicate independent assays.
NPM/ALK expression induces changes in proteins associated with cytoskeleton, and affects cell adhesion and invasion. (A) HEK293T cells transfected with vector, wild-type (Wt), and kinase-defective mutant (K210R) NPM/ALK were evaluated by confocal immunofluorescence microscopy for the expression of the cytoskeletal proteins RhoA and paxillin. ALK (green) is expressed in Wt NPM/ALK-transfected HEK293T cells but not in vector-transfected HEK293T cells. The level of ALK expression is slightly decreased in the K210R-transfected cells. The Texas Red–phalloidin (red) staining of the vector-, Wt-, and K210R mutant–transfected cells does not show significant differences in actin expression. The expression of RhoA, paxillin, and phospho-Tyr proteins is significantly reduced in the K210R compared with Wt. (B) HEK293T cells transfected with vector, Wt, or the K210R NPM/ALK were assayed for adhesion to fibronectin. Data represent the mean of 2 experiments with error bars representing SD of triplicate mean. The unpaired Student t test was used (Wt vs K210R, P = .017). (C) The effect of NPM/ALK on invasive properties was evaluated using NIH3T3 cells transfected with vector or Wt NPM/ALK plated onto BioCoat Matrigel invasion chambers, and cells present at the opposite side of the coated membranes were stained with crystal violet. Cells per high-power field (400× magnification) were counted. The graphs represent the results of replicate independent assays.
NPM/ALK reduces cell adhesion and enhances invasion
Based on the differential expression of numerous proteins associated with cell adhesion and integrin signaling (supplemental Table 1), we performed functional validation by directly comparing the adhesive properties of NPM/ALK-positive and NPM/ALK-negative cells. Alpha 5 β1 integrin, which was down-regulated in NPM/ALK-positive cells (supplemental Table 1), is the major receptor for fibronectin.27 Thus, we examined the adhesive property of HEK293T cells transfected with NPM/ALK with that of vector-transfected cells on fibronectin substratum. Figure 4B demonstrates that expression of NPM/ALK is associated with a significant decrease in cell adhesion to fibronectin (15.6 vs 6.67; P = .08). Furthermore, the expression of the kinase-defective mutant of NPM/ALK (K210R) abrogated the reduction in adhesion (16.6 vs 6.67; P = .017), suggesting that the kinase activity of NPM/ALK is required for this phenotype. Furthermore, NIH3T3 cells that express NPM/ALK demonstrate significantly enhanced invasion through Matrigel compared with vector-expressing cells (39.5 vs 22.2; P = .004; Figure 4C). Our data suggest that NPM/ALK expression leads to decreased cell adhesion via down-regulation of adhesion proteins.
Immunohistochemical validation using tissue microarrays of primary ALCL biopsy specimens
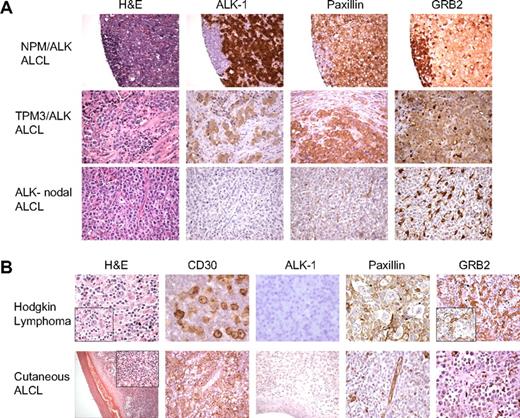
To further establish the relevance of the NPM/ALK-associated proteomic changes involving proteins mediating cellular cytoarchitecture identified by cICAT-MS/MS, we assessed the expression of the focal adhesion protein paxillin and the adaptor scaffold GRB2 using tissue microarrays containing cases of primary human ALK-positive and nodal ALK-negative ALCLs. Figure 5A illustrates a representative H&E, ALK-1, paxillin, and GRB2 immunoreactivity in a tissue biopsy specimen of an NPM/ALK-positive ALCL (top row), an ALCL with a TPM3-ALK translocation (middle row), and an ALK-negative nodal ALCL (bottom row). Notably, paxillin expression is observed in 14 of 14 ALK-positive lymphomas but is absent in 16 ALK-negative nodal ALCLs. Similarly, GRB2 expression is observed in 8 of 11 ALK-positive lymphomas (Figure 5A top and middle row), whereas 0 of 6 ALK-negative nodal ALCLs expressed GRB2 (Figure 5A bottom row). We analyzed 2 other types of malignant lymphoma that morphologically mimic NPM/ALK-positive ALCL and express the lymphocyte activation marker CD30, namely cutaneous ALCL and Hodgkin lymphoma.4 The malignant cells of either Hodgkin lymphoma (n = 24 cases) or cutaneous ALCL (n = 5 cases) do not express either paxillin or GRB2 (Figure 5B).
Immunohistochemical validation of paxillin and GRB2 expression in human malignant lymphomas. (A) Tissue microarrays composed of ALCLs (ALK positive and ALK negative) were used to perform immunohistochemical analyses for paxillin and GRB2. The morphologic features evaluated by H&E stains and expression of ALK-1, paxillin, and GRB2 in representative cases of NPM/ALK-positive ALCL, a variant ALK-positive ALCL (TPM3/ALK), and an NPM/ALK-negative nodal ALCL are shown. Paxillin and GRB2 are selectively overexpressed in the ALK-positive lymphoma cells in contrast to the ALK-negative nodal lymphoma cells. (B) The top panel demonstrates the features of a classical Hodgkin lymphoma with strong expression of CD30 in the neoplastic Reed-Sternberg cells that do not express ALK, paxillin, or GRB2. The bottom panel illustrates the morphologic features of a cutaneous ALCL with strong expression of CD30 but without expression of ALK-1, paxillin, or GRB2 within the neoplastic T cells.
Immunohistochemical validation of paxillin and GRB2 expression in human malignant lymphomas. (A) Tissue microarrays composed of ALCLs (ALK positive and ALK negative) were used to perform immunohistochemical analyses for paxillin and GRB2. The morphologic features evaluated by H&E stains and expression of ALK-1, paxillin, and GRB2 in representative cases of NPM/ALK-positive ALCL, a variant ALK-positive ALCL (TPM3/ALK), and an NPM/ALK-negative nodal ALCL are shown. Paxillin and GRB2 are selectively overexpressed in the ALK-positive lymphoma cells in contrast to the ALK-negative nodal lymphoma cells. (B) The top panel demonstrates the features of a classical Hodgkin lymphoma with strong expression of CD30 in the neoplastic Reed-Sternberg cells that do not express ALK, paxillin, or GRB2. The bottom panel illustrates the morphologic features of a cutaneous ALCL with strong expression of CD30 but without expression of ALK-1, paxillin, or GRB2 within the neoplastic T cells.
Analysis of cellular pathways deregulated by NPM/ALK
Interaction networks generated with respect to putative canonical pathway components modulated by NPM/ALK are highlighted using a knowledge-based structured-network analysis tool (Ingenuity Pathway Analysis; http://www.ingenuity.com). Supplemental Figure 1C depicts the diverse signaling pathways affected by NPM/ALK expression. They represent proteins involved in survival, cell cycle regulation, proliferation, growth factor/cytokine signaling, antiapoptosis, neoangiogenesis, adhesion, and migration, and include PI3-K/AKT, JAK/STAT, IGF1, IL4, IL6, NF-κB, PPAR, p38/MAPK, ERK/MAPK, G-protein, PDGF, Wnt/β-catenin, and integrin signaling pathways.
Ras/ERK/MAPK and FRAP/mTOR signaling pathways mediate the growth and survival signals of NPM/ALK
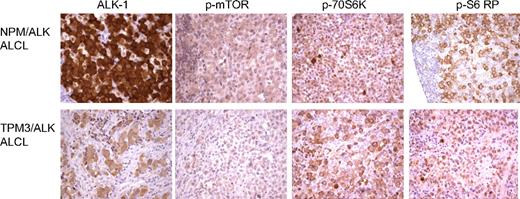
Pathway analysis of our proteomic data highlights deregulation of several signaling cascades including the ras/ERK/MAPK and FRAP/mTOR pathways (supplemental Table 1). Downstream targets of the FRAP/mTOR pathway were also identified by cICAT-MS/MS (supplemental Table 1) to be up-regulated by NPM/ALK expression. The FRAP/mTOR pathway plays a key role in the regulation of cell growth and proliferation and positively regulates translation, ribosome biogenesis, cell size, and proliferation.28 mTOR phosphorylates 2 key translational regulators, ribosomal S6K and 4EBP-1. Ribosomal S6K phosphorylates the 40S ribosomal protein S6 and initiates the translation of 5′ terminal oligopyrimidine tract-containing mRNAs that encode components of the protein synthesis machinery.29 Immunohistochemical studies on tissue biopsies of ALK-positive lymphomas show increased expression of phospho-mTOR, phospho-p70S6 kinase, and phospho-S6 ribosomal protein (Figure 6) relative to reactive lymphocytes.
Expression of FRAP/mTOR pathway proteins in ALK-positive ALCLs. Tissue microarrays of ALK-positive ALCLs were analyzed for the expression of proteins in the FRAP/mTOR pathway. Both the NPM/ALK-positive (top panel) and TPM3/ALK-postive (bottom panel) ALCLs demonstrate expression of phospho-mTOR, phospho-p70S6K, and phospho-S6 ribosomal proteins.
Expression of FRAP/mTOR pathway proteins in ALK-positive ALCLs. Tissue microarrays of ALK-positive ALCLs were analyzed for the expression of proteins in the FRAP/mTOR pathway. Both the NPM/ALK-positive (top panel) and TPM3/ALK-postive (bottom panel) ALCLs demonstrate expression of phospho-mTOR, phospho-p70S6K, and phospho-S6 ribosomal proteins.
To determine the functional role of the ras/ERK/MAPK and the FRAP/mTOR pathways in the survival of NPM/ALK-positive ALCL, we evaluated the effect of target-selective small molecule inhibitors on the cell viability of SUDHL-1 cells. Figure 7A demonstrates that exposure of SUDHL-1 cells to the Ras inhibitor (FTI-277) results in a dose- and time-dependent decrease in cell viability (40% reduction by 5 μM FTI-277 at 72 hours). Cell cycle analysis demonstrates a significant increase of cells in G2/M phase (38.09% compared with 7.18% control) without induction of caspase-3 (Figure 7A), consistent with cell cycle arrest in G2/M. Western blot analysis of cell lysates treated with FTI-277 (Figure 7D) demonstrates the down-regulation of 2 proteins downstream of the mTOR pathway, phospho-p70S6K1 and 4EBP-1. Interestingly, the levels of NPM/ALK also decrease after rapamycin and FTI-277. This may indicate that the steady-state levels of NPM/ALK may be regulated by proteins affected by these small molecules.
Ras/mitogen-activated protein kinase (MAPK) and FRAP/mTOR signaling pathways are required for the survival of NPM/ALK-positive cells. (A) NPM/ALK-positive SUDHL-1 cells were incubated with various concentrations of FTI-277 (ras inhibitor), (B) U0126 (p44/42 inhibitor), and (C) rapamycin (mTOR inhibitor) for variable time periods. Cell viability was evaluated by the MTT assay and compared with DMSO-treated control cells. The data represent mean percentage of viability relative to DMSO control obtained from triplicate experiments. Cell cycle analysis was performed as outlined in “Methods.” Representative data obtained from 3 independent experiments are shown. Caspase-3 activity assay was performed as described in “Methods.” U0126 (Calbiochem) was added to SUDHL-1 cells to create a positive (induced apoptosis) control. Z-VAD-FMK inhibitor was used at a final concentration of 50 μM. A negative control was prepared using untreated cells. The data represent the mean values of triplicate measurements. (D) Western blot analyses of drug-treated cells.
Ras/mitogen-activated protein kinase (MAPK) and FRAP/mTOR signaling pathways are required for the survival of NPM/ALK-positive cells. (A) NPM/ALK-positive SUDHL-1 cells were incubated with various concentrations of FTI-277 (ras inhibitor), (B) U0126 (p44/42 inhibitor), and (C) rapamycin (mTOR inhibitor) for variable time periods. Cell viability was evaluated by the MTT assay and compared with DMSO-treated control cells. The data represent mean percentage of viability relative to DMSO control obtained from triplicate experiments. Cell cycle analysis was performed as outlined in “Methods.” Representative data obtained from 3 independent experiments are shown. Caspase-3 activity assay was performed as described in “Methods.” U0126 (Calbiochem) was added to SUDHL-1 cells to create a positive (induced apoptosis) control. Z-VAD-FMK inhibitor was used at a final concentration of 50 μM. A negative control was prepared using untreated cells. The data represent the mean values of triplicate measurements. (D) Western blot analyses of drug-treated cells.
Treatment of SUDHL-1 cells with a p44/42 MAPK inhibitor (U0126) also results in a significant reduction of cell viability (40% reduction at 36 hours). The decrease in cell viability was associated with a G1 arrest (55.7% relative to 40.2% in control), with significant induction of caspase-3 (Figure 7B) consistent with G1 arrest followed by apoptosis. Western blot analysis demonstrates substantially reduced levels of phospho-p44/42 levels in U0126-treated cells (Figure 7D), whereas the levels of total p44/42 remained unchanged. Consistent with caspase-3–mediated apoptosis, enhanced cleavage of the 116-kDa form of PARP is also observed (Figure 7D). Our data indicate that specific activation of ERK1/2 may be critical for the survival of NPM/ALK-positive cells.
As shown in Figure 7C, rapamycin, an inhibitor of the FRAP/mTOR pathway, potently decreased the viability of SUDHL-1 cells (30% reduction by 10 nM at 48 hours), and resulted in G1 arrest (62.08% compared with 46.94%) without induction of caspase-3 activity. Western blot analysis demonstrated a reduction in the level of NPM/ALK, phospho-p70S6K, as well as 4EBP-1 levels (Figure 7D). These data are consistent with the critical role of mTOR activation in survival of NPM/ALK-positive ALCL.
Discussion
The t(2;5)(p23;q35) that results in the constitutive activation of NPM/ALK tyrosine kinase is a key oncogenic event in the development of ALCL. In this study, we have used quantitative proteomic techniques to explore the global proteomic consequences of this singular molecular aberration leading to aberrant expression of the chimeric protein NPM/ALK. Our studies reveal that the cellular ramifications of expression of oncogenic tyrosine kinase such as NPM/ALK are diverse and affect many different cellular pathways and functions that support the neoplastic phenotype. The cICAT approach used in the current study may reveal both direct and indirect changes that occur in response to NPM/ALK expression. In addition, the cICAT strategy selectively labels peptides that contain cysteine residues and will not identify all of the protein changes that occur as a consequence of NPM/ALK expression. In addition to several of the known downstream targets of NPM/ALK, our quantitative proteomic analysis reveals effects on several proteins and pathways previously not recognized to be associated with NPM/ALK expression.
Our proteomic profiling demonstrates NPM/ALK induced differential expression of numerous proteins associated with modification of cytoskeletal architecture (supplemental Table 1). Our data suggest that NPM/ALK deregulates the expression of cytoskeletal proteins, such as RhoA and paxillin, in a kinase-dependent manner to induce changes in cytoskeletal architecture. Furthermore, NPM/ALK leads to decreased cell adhesion via down-regulation of adhesion proteins and provide a biologic mechanism for the enhanced invasive properties. These observations support the typical sinusoidal growth pattern of ALCLs in lymph nodes30 and their high propensity for extranodal dissemination. Recent studies have shown that NPM/ALK can mediate cell shape changes and actin filament depolymerization via its interactions with p130Cas; this is mediated by the adaptor protein growth factor receptor-bound protein 2 (GRB2).31,32 A limitation of the current study may be the use of nonhematopoietic cells, and it is possible that the functional alterations in cytoskeletal organization may not be completely applicable to ALCLs. Furthermore, adhesion to other substrates in addition to fibronectin is likely to occur in vivo. However, collectively, these data indicate that NPM/ALK, in addition to enhancing cell survival via proliferation and apoptosis evasion, has a role in the adhesive and invasive properties of tumor cells. Furthermore, immunohistochemical analysis of primary clinical tissue biopsy samples demonstrated the selective expression of paxillin and GRB2 in ALK-positive ALCLs but not in other ALK-negative CD30+ lymphoid malignancies that do not express ALK supporting the use of these proteins as novel biomarkers in immunohistologic panels aimed at distinguishing ALK-positive lymphomas from their morphologic mimics.
We also identified several centrosome and microtubule-associated proteins including CEP250 (2.5-fold), mitotic kinesin-like protein (2.4-fold), and microtubule associated protein 4 (2.0-fold), which were up-regulated in NPM/ALK-positive cells. Recent studies have demonstrated that abnormal centrosome size and number are common in ALCLs,33 but the biologic basis for this observation has not been provided. Our findings suggest the characteristic nuclear morphology including large size and multinucleation in ALCLs is linked to the effects of NPM/ALK based on the expression of centrosomal and microtubule-associated proteins.
Similarly, our proteomic studies show that proteins that carry out chaperone function, and are associated with the ubiquitin-proteasome machinery, are modulated by NPM/ALK. Specifically, we identified RanBP2 SUMO E3 ligase (up-regulated by 2-fold) and ubiquitin carboxy-terminal hydrolase 24 (down-regulated by 3.3-fold). In addition, proteins associated with the stress response including DnaJ (HSP40) homolog (2.4-fold) and HSP90/HSP90-organizing protein (2.1-fold)34 are up-regulated. Stress response proteins are important or survival and prevent stress-induced apoptosis in cancer cells. These findings have important implications for a potential role for heat shock protein 90 and proteasome inhibitors in the treatment of ALK-deregulated malignancies.
In addition to many protein kinases, we also identified the up-regulation of PTP1B and down-regulation of PP2A. Interestingly, PTP1B expression is enhanced by overexpression of the oncogenic kinase p210 BCR/ABL. The up-regulation of PTP1B in NPM/ALK-expressing cells suggests a similar role in ALCLs. Functional inactivation of PP2A, a serine/threonine phosphatase, is associated with cell transformation,35 and favors disease progression in BCR/ABL-positive leukemias.36
Pathway analysis highlighted the deregulation of several signaling cascades including the ras/ERK/MAPK and FRAP/mTOR pathways, which were functionally tested by targeting with selective small molecule inhibitors. Cell viability assays, cell cycle analysis, and apoptosis assays indicate that ras/ERK/MAPK and FRAP/mTOR signaling pathways mediate the growth and survival signals of NPM/ALK. Notably, 2 other groups have reported the activation of components of these pathways in ALCLs.37-39 Importantly, the selective targeting of one or combination of these pathways with small molecule inhibitors should be considered as novel therapeutic approaches in patients with NPM/ALK-positive ALCL.
In conclusion, our study demonstrates the global cellular consequences of the t(2;5)(p23;q35) translocation. Our improved understanding of ALCL pathobiology by elucidation of global NPM/ALK effects has important implications for the development of novel diagnostic markers and rational therapeutic targets in patients with ALCL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The work was supported by funds from the Children's Oncology Group Translational Research Award (M.S.L.) and the ARUP Institute for Experimental Pathology (M.S.L., K.S.J.E.-J.).
Authorship
Contribution: M.S.L. and K.S.J.E.-J. designed research, analyzed data, and wrote the paper; M.L.C., D.K.C., G.C.F., D.R.A., O.F.E.-J., S.R.T. and P.S. performed research; and G.Z.R. and L.J.M. contributed specimens.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Megan S. Lim or Kojo S. J. Elenitoba-Johnson, M5242D, Med Sci I, University of Michigan, 1301 Catherine Rd, Ann Arbor, MI 48109; e-mail: meganlim@umich.edu or kojoelen@umich.edu.