Abstract
Forced expression of MN1 in primitive mouse hematopoietic cells causes acute myeloid leukemia and impairs all-trans retinoic acid-induced granulocytic differentiation. Here, we studied the effects of MN1 on myeloid differentiation and proliferation using primary human CD34+ hematopoietic cells, lineage-depleted mouse bone marrow cells, and bipotential (granulocytic/monocytic) human acute myeloid leukemia cell lines. We show that exogenous MN1 stimulated the growth of CD34+ cells, which was accompanied by enhanced survival and increased cell cycle traverse in cultures supporting progenitor cell growth. Forced MN1 expression impaired both granulocytic and monocytic differentiation in vitro in primary hematopoietic cells and acute myeloid leukemia cell lines. Endogenous MN1 expression was higher in human CD34+ cells compared with both primary and in vitro–differentiated monocytes and granulocytes. Microarray and real-time reverse-transcribed polymerase chain reaction analysis of MN1-overexpressing CD34+ cells showed down-regulation of CEBPA and its downstream target genes. Reintroduction of conditional and constitutive CEBPA overcame the effects of MN1 on myeloid differentiation and inhibited MN1-induced proliferation in vitro. These results indicate that down-regulation of CEBPA activity contributes to MN1-modulated proliferation and impaired myeloid differentiation of hematopoietic cells.
Introduction
The decision between self-renewal and lineage commitment of hematopoietic stem cells is mainly regulated by extracellular signals from the hematopoietic microenvironment that induce differential expression of key transcription factors. Aberrant expression or function of these factors contributes significantly to leukemogenesis.1,2 Clonal expansion of immature hematopoietic cells resulting from alterations of differentiation-inducing transcription factors is the hallmark of acute myeloid leukemia (AML).2,3
MN1, meningioma 1, was identified in a patient with meningioma, as a target of a chromosome translocation t(4;22).4,5 Subsequent studies showed that MN1 is also involved in AML either as a partner of a recurrent translocation t(12;22)(p12;q12), encoding an MN1-TEL fusion protein,6 or as an overexpressed gene in 2 subsets of AML specified by the presence of inv(16) or by overexpression of ecotropic viral integration site 1.7,8 Our recent studies showed that ectopic expression of MN1 or MN1-TEL in hematopoietic cells causes myeloid disease in mice.9,10 Moreover, MN1 cooperates with CBFβ-MYH11, the product of inv(16), in a murine model of inv(16)-AML and accelerates leukemogenesis.9 MN1 also prevents granulocytic differentiation and abrogates all-trans retinoic acid (ATRA)–mediated growth inhibition in primary mouse hematopoietic cells.11 In addition, MN1 overexpression eliminated the beneficial effects of the addition of ATRA to the maintenance chemotherapy of nonacute promyelocytic leukemia–AML patients; whereas in AML patients with low MN1 expression, ATRA administration increased survival.11
The transcription factor CEBPA, CCAAT enhancer binding protein-α (C/EBPα), plays an essential, nonredundant role in the decision of hematopoietic stem cells to self-renew or differentiate into a myeloid direction12 and it is activated by ATRA.13 CEBPA regulates expression of several myeloid-specific genes, such as granulocyte-colony stimulating factor receptor (G-CSFR), monocyte-colony stimulating factor receptor (M-CSFR),14 and miR-22313 via direct binding to sites in their promoters. Cebpa knockout mice showed a profound defect in the generation of granulocyte-monocyte progenitors from common myeloid progenitors, resulting in a lack of granulocytes and impaired macrophage development.12,14,15 Concordantly, the absence of Cebpa enhances the repopulating and self-renewal capacity of hematopoietic stem/progenitor cells (HSPCs),12,15 whereas its forced expression in human CD34+ primary cells and AML cells induced myeloid differentiation.16,17 It is therefore logical that the loss of CEBPA function is frequently observed in AML.18-21
Although MN1 is a novel player in AML pathophysiology, the molecular mechanisms via which MN1 inhibits differentiation and stimulates proliferation of hematopoietic cells remain elusive. Here, we expanded on previous studies and addressed the following questions: (1) Is monocytic differentiation protected in MN1 cells at the expense of granulocytic differentiation? (2) Are the effects of MN1 on primary hematopoietic cells similar to those in human AML cell lines? (3) How does the transcriptome of primary human CD34+-MN1 cells differ from that of parental CD34+ cells? To answer these questions, we used cell culture experiments to analyze the effects of MN1 overexpression on the proliferation and myeloid differentiation of human and mouse primary HSPC and AML cell lines. To gain insight into molecular mechanisms orchestrating the MN1-induced phenotype, we performed microarray analysis using RNA from control and MN1-overexpressing CD34+ hematopoietic cells. Furthermore, we determined the effects of reintroduction of conditional and constitutive CEBPA on the MN1-mediated inhibition of differentiation and stimulation of proliferation because CEBPA expression was repressed in CD34+-MN1 cells.
Methods
Plasmids and viral packaging
Human CEBPA (Origene) and MN1 cDNAs22 were cloned into MSCV-IRES-GFP (MIG) or MSCV-IRES-YFP (MIY) retroviral vectors as Xho/EcoRI and EcoRI fragments, respectively. A tamoxifen inducible MIGR1-CEBPA-ER retroviral vector was kindly provided by Dr Alan D. Friedman (Johns Hopkins University, Baltimore, MD). We generated VSVg-pseudotyped retrovirus by transient transfection of 293T cells maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS; Hyclone; Thermo Fisher Scientific) Glutamax (2 mM) and penicillin/streptomycin (100 U/mL each, Invitrogen; referred to as complete medium). For virus production, we used complete medium supplemented with 10% heat inactivated FBS.
Isolation and culture of human CD34+ cells and AML patient cells
After approval by the Institutional Review Board of St Jude Children's Research Hospital, bone marrow (BM) or RNA samples were obtained from AML patients and mononuclear cells were separated via Ficoll-gradient centrifugation of apheresis samples of G-CSF–mobilized transplantation donors. Informed consent was provided according to the Declaration of Helsinki. Monocytes, granulocytes, and CD34+ cells were isolated from mononuclear cells, expanded, and differentiated using existing protocols.23 Purity of isolated cells was confirmed by fluorescence-activated cell sorting (FACS) analysis. CD34+ cells (2-4 × 105 cells/1 mL per well of retronectin-coated 24-well plates) were transduced 3 times during a 2-day interval with MIG or MIG-MN1 virus prepared in growth medium (serum-free X-VIVO15, BioWhittaker, 04-744Q; supplemented with Glutamax and penicillin/streptomycin), diluted 1:1, and supplemented with progenitor cytokines (human stem cell factor [SCF], fms-related tyrosine kinase 3 ligand, and thrombopoietin; 50 ng/mL each).23 For in vitro differentiation assays, 2- to 3-day expanded CD34+ cells were plated (2-3 × 104 cells/mL) in growth medium containing monocyte cytokines: macrophage colony-stimulating factor (M-CSF; 100 ng/mL), SCF (20 ng/mL), interleukin-6 (IL-6; 20 ng/mL), fms-related tyrosine kinase 3 ligand (50 ng/mL), ± vitamin D3 (60 nM; Sigma-Aldrich) to induce monocytic differentiation; or granulocyte cytokines [G-CSF (100 ng/mL) and SCF (20 ng/mL; all cytokines from PeproTech) to induce granulocytic differentiation.23 Half of the medium was replaced every 3 days, and differentiation was assessed on days 10 to 14 of culture using FACS analysis.
Human BM-derived CD34+ cells (StemCell Technologies) were expanded and transduced with mock or MN1 retrovirus. One day later, RNA was isolated from GFP+/CD34+ FACS-sorted cells using Trizol (Sigma-Aldrich) and was subjected to microarray analysis following Affymetrix protocols (Affymetrix) using the GeneChip Human U133 Plus 2.0 array. The microarray data for this study have been deposited into GEO under accession no. GSE16745.24
Methyl-thiazol-tetrazolium (MTT) assay was performed following the manufacturer's instructions (Promega). Methylcellulose assays were performed by plating FACS-sorted, engineered cells into MethoCult GF H4444 (StemCell Technologies). Primary AML cells were expanded for 1 day and transduced using the progenitor cytokines and the same conditions as those used for primary CD34+ cultures.
Generation of MN1 and CEBPA cell lines
HL60 and U937 cells (2.5 × 105/well, 12-well plate) were transduced with 2 mL of the corresponding MIG or MIY retrovirus 1:1 diluted in complete RPMI containing 10% heat-inactivated FBS in the presence of 8 μg/mL polybrene (Chemicon) for 2 days (total 2 or 3 times). GFP+ or YFP+ cells were sorted using FACS and induced to differentiate with different doses of vitamin D3 (Sigma-Aldrich, D1530), ATRA (Sigma-Aldrich, R2625), or the same volume of vehicle for 2 to 3 days. Differentiation was assessed using FACS analysis and May-Grünwald-Giemsa staining of cytospin preparations. CEBPA-ER-expressing cells were additionally treated with 4-hydroxytamoxifen (Sigma-Aldrich, H7904) or vehicle for the indicated times and doses.
Quantitative real-time RT-PCR
Total RNA was extracted using Trizol or the mirVana miRNA isolation kit (Ambion) following the manufacturer's protocol. cDNA was synthesized using hexamer primers and Superscript II reverse transcriptase (Invitrogen). Primer and probes for human HPRT, GAPDH, RARβ, CEBPA, p21, G-CSFR, M-CSFR, and CALR were from Applied Biosystems (assay ID: 4326321E, 4310884E, Hs00977137-m1, Hs00269972-s1, Hs99999142-m1, Hs00167918-m1, Hs00911250-m1, Hs00189032-m1, respectively). The MN1 primer and probe sets were as described.9 Expression levels were obtained using the standard curve method (after normalization with a housekeeping gene). The TaqMan microRNA assay kit for RNU6B (used as endogenous control and normalizer) and miR-223 were from Applied Biosystems (part no. 4373381 and 4373075, respectively). Reverse-transcribed polymerase chain reaction (RT-PCR) reactions were performed according to the manufacturer's instructions.
FACS analysis
Cells were washed and taken up in blocking solution (phosphate-buffered saline containing 5% fetal bovine serum, 2 mM sodium azide, and 0.1 mg/mL human gamma globulin) for 30 minutes on ice. After washing, cells were stained with the indicated directly conjugated antibodies for 30 minutes on ice, washed, and analyzed using an LSR II flow cytometer (BD Biosciences). Anti–human CD11b-allophycocyanin (APC), CD14-phycoerythrin (PE), CD15-APC, CD45-APC, CD34-PE, and CD38-APC were from Miltenyi Biotec, and anti–mouse antibodies (Mac1-PE, Gr1-APC, Sca1-PerCP-Cy5-5) were from BD Biosciences, except for ckit-APC-Alexa750 (eBioscience). In some experiments, cells engineered to express either GFP or YFP or both were counterstained with propidium iodide to mark dead cells and were analyzed using FACS Vantage-SE-DiVa cell sorters (BD Biosciences); subpopulations of viable cells expressing these fluorescent proteins were collected by FACS. Cell cycle and apoptosis assays were performed using FACS as described.25
Western blots and immunocytochemistry
Total cell lysates were prepared by sonication for 15 seconds after incubating the cells on ice in radio immunoprecipitation assay buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate) containing protease and phosphate inhibitors (Sigma-Aldrich). Lysates were subjected to Western blot analysis to detect CEBPA (39306; active motif) and GAPDH (MAB374; Millipore) expression following the manufacturer's instructions. Immunocytochemistry of HL60-MN1 and U937-MN1 cells using anti-MN1 antibody was performed as described.9 Images of cytospins were captured with an Olympus BX-50 microscope (equipped with UPlanFL 40×/0.75 and UPlanFL 60×/1.25 numerical aperatures with a SPOT camera and SPOT Advanced Imaging software (Diagnostic Instruments). The original magnification for cytospins was ×400 or ×600.
Mouse BM culture
Statistical analysis
Unpaired t test analysis (2-tailed) was performed using the GraphPad Prism, version 4.0c for Mac (GraphPad Software; www.graphpad.com).
Results
MN1 overexpression inhibits ATRA- or vitamin D3-induced myeloid differentiation of AML cell lines
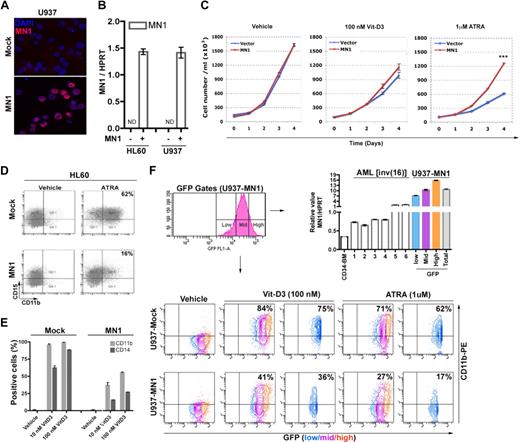
Forced expression of MN1 in mouse hematopoietic cells prevented granulocytic differentiation, abrogated ATRA-mediated growth inhibition, and caused myeloid disease in mouse models.9,11 To further determine the effects of MN1 on myeloid differentiation of human AML cell lines, HL60 and U937 cells were transduced with MIG or MIG-MN1 retroviruses, respectively. Immunocytochemistry and real-time RT-PCR analysis of GFP+ FACS-sorted cells showed MN1 expression in the nucleus (Figure 1A bottom panel) and similar levels of MN1 expression in the MN1-transduced cell lines, whereas mock-transduced HL60 and U937 cells did not express endogenous MN1 (Figure 1B).
Exogenous MN1 inhibits the differentiation of AML cell lines toward monocytes and granulocytes. (A) Cytospin preprations of FACS-sorted GFP+ U937 and HL60 cell lines transduced with control GFP (mock) or MN1-expressing retroviruses (MN1), stained with anti-MN1 antibody (red). Blue represents nuclear staining with 4,6-diamidino-2-phenylindole. (B) MN1 expression in transduced HL60 and U937 cells was analyzed using real-time RT-PCR. Each sample was analyzed in triplicate, and the signals were normalized for endogenous HPRT expression. Histogram shows mean ± SEM of duplicates. MN1(−) indicates mock; MN1(+), MN1-transduced cells; and ND, not determined. (C) Sorted GFP+ HL60 cells were seeded at day 0 in duplicates in 6-well plates (105 cells/mL, 4 mL/well) with indicated amounts of vitamin D3, ATRA, or vehicle, and cells were counted at consecutive days (mean ± SEM of duplicates). ***P = .001. (D) Cells were treated with 1 μM ATRA or (E) with the indicated amount of vitamin D3 or the same volume of vehicle in duplicates for 72 hours. Expression of CD11b and CD15 was analyzed by FACS. The result from 1 representative example is shown. (F) Low, medium (mid), or high GFP-expressing U937-MN1 cells were FACS-sorted (top left panel; GFP gates) and MN1 expression (normalized to HPRT) was compared with that of human CD34-BM cells and AML patients (1-6) using real-time RT-PCR (top right histogram). CD11b expression of cells treated for 48 hours with vehicle or the indicated amounts of vitamin D3 or ATRA was determined using FACS in all GFP+ cells (bottom panels, blue, magenta and orange together; U937-mock and U937-MN1) or in GFP-low fractions (blue panels). All experiments were repeated a minimum of 3 times.
Exogenous MN1 inhibits the differentiation of AML cell lines toward monocytes and granulocytes. (A) Cytospin preprations of FACS-sorted GFP+ U937 and HL60 cell lines transduced with control GFP (mock) or MN1-expressing retroviruses (MN1), stained with anti-MN1 antibody (red). Blue represents nuclear staining with 4,6-diamidino-2-phenylindole. (B) MN1 expression in transduced HL60 and U937 cells was analyzed using real-time RT-PCR. Each sample was analyzed in triplicate, and the signals were normalized for endogenous HPRT expression. Histogram shows mean ± SEM of duplicates. MN1(−) indicates mock; MN1(+), MN1-transduced cells; and ND, not determined. (C) Sorted GFP+ HL60 cells were seeded at day 0 in duplicates in 6-well plates (105 cells/mL, 4 mL/well) with indicated amounts of vitamin D3, ATRA, or vehicle, and cells were counted at consecutive days (mean ± SEM of duplicates). ***P = .001. (D) Cells were treated with 1 μM ATRA or (E) with the indicated amount of vitamin D3 or the same volume of vehicle in duplicates for 72 hours. Expression of CD11b and CD15 was analyzed by FACS. The result from 1 representative example is shown. (F) Low, medium (mid), or high GFP-expressing U937-MN1 cells were FACS-sorted (top left panel; GFP gates) and MN1 expression (normalized to HPRT) was compared with that of human CD34-BM cells and AML patients (1-6) using real-time RT-PCR (top right histogram). CD11b expression of cells treated for 48 hours with vehicle or the indicated amounts of vitamin D3 or ATRA was determined using FACS in all GFP+ cells (bottom panels, blue, magenta and orange together; U937-mock and U937-MN1) or in GFP-low fractions (blue panels). All experiments were repeated a minimum of 3 times.
Compared with mock-transduced cells, exogenous MN1 inhibited ATRA-induced growth arrest of HL60 cells by 2-fold (Figure 1C right panel) and differentiation, as monitored by the induction of granulocytic markers (CD11b+/CD15+ or CD11b) in both HL60-MN1 (from 62% to 16%) and U937-MN1 cells (from 71% to 27%; Figure 1D,F). Although vitamin D3-mediated growth arrest was not inhibited significantly (Figure 1C middle panel), vitamin D3-induced monocytic differentiation was also inhibited in both MN1-transduced cell lines (Figure 1E-F). FACS analysis indicated that vitamin D3-induced expression of CD11b was decreased by 2.6-fold (P = .005) and 1.8-fold (P = .001), and CD14 expression by 4-fold (P = .004) and 3.2-fold (P < .001) in HL60-MN1 cells treated with 10 nM and 100 nM vitamin D3, respectively (mock and MN1, Figure 1E). Similar vitamin D3 effects were seen in U937-MN1 cells with a 2-fold reduction in CD11b expression (vitamin D3, U937-mock, and U937-MN1, Figure 1F). Thus, MN1 overexpression impaired not only granulocytic but also monocytic differentiation of AML cell lines in response to ATRA and vitamin D3. To determine the relevance to AML patients who overexpress MN1, we sorted U937-MN1 cells into a low, a medium, and a high GFP-expressing fraction and compared the amount of MN1 RNA in these fractions with that in CD34+ BM cells and in 6 different inv(16) AML patient samples that overexpress MN1. Expression in the low fraction of U937-MN1 cells was 3.4-fold higher than in the highest expressing patient sample (Figure 1F top right panel), whereas the medium and high fractions are 5-fold and 7.4-fold higher, respectively. Curiously, the inhibition of CD11b expression by MN1 is the strongest in the low fraction (vitamin D3, 2.1-fold; ATRA, 3.6-fold, Figure 1F) slightly less effective in the medium (vitamin D3, 2.4-fold; ATRA, 3.3-fold) and least effective in the high fraction (vitamin D3, 1.3-fold; ATRA, 1.4-fold; blue, magenta, and orange in Figure 1F bottom panel, U937-mock and U937-MN1). This suggests that disproportionate overexpression of MN1 actually impairs its differentiation-inhibiting effect, which bodes well for the effects mediated by the lower MN1 overexpression levels in patient samples.
Endogenous MN1 expression is down-regulated in primary human granulocytes and monocytes
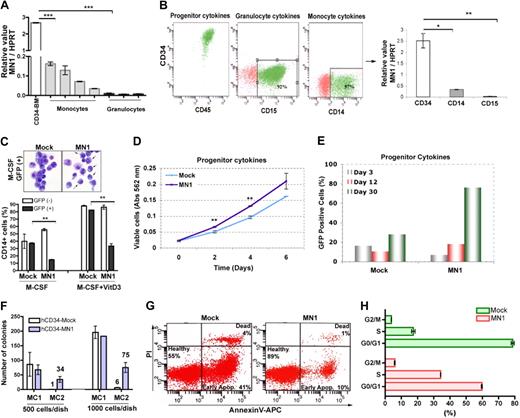
We next analyzed endogenous MN1 expression in RNA samples obtained from human BM-derived CD34+ cells as well as from primary peripheral blood monocytes and granulocytes of G-CSF-mobilized healthy donors. Compared with CD34+ cells, the level of endogenous MN1 mRNA was down-regulated in both monocytes (17- to 86-fold) and granulocytes (216- to 370-fold; Figure 2A). In an independent experiment, CD34+ cells were maintained in cultures supporting HSPC growth for 5 days followed by differentiation for 14 days in the presence of monocyte or granulocyte cytokine cocktails.23 FACS analysis confirmed that CD34+ cells maintained their identity in the presence of progenitor cytokines (Figure 2B), whereas the presence of G-CSF or M-CSF induced differentiation and expression of CD15 (92%) and CD14 (57%), respectively (Figure 2B). Similar to uncultured peripheral blood monocytes and granulocytes, endogenous MN1 expression was down-regulated in both in vitro-differentiated monocytes (8.3-fold; CD14 graph, Figure 2B) and granulocytes compared with cultured CD34+ cells (92-fold; CD15 graph, Figure 2B). Collectively, these results suggested that the endogenous MN1 expression must have been switched off during the differentiation process.
MN1 overexpression inhibits M-CSF or vitamin D3-induced differentiation and enhances growth of CD34+ primary human hematopoietic cells. (A) Real-time RT-PCR analysis of endogenous MN1 expression in BM-derived human CD34+ cells (CD34-BM), peripheral blood monocytes, and granulocytes, normalized for endogenous HPRT expression (mean ± SEM of triplicates). ***P < .001. (B) Primary human CD34+ cells from peripheral blood of G-CSF mobilized healthy donors maintained in cultures containing progenitor, granulocyte, or monocyte cytokine cocktails, respectively. Marker analysis was performed using FACS of day 5 CD34+ stem cell/progenitors (progenitor cytokines) cultures and day 14 in vitro differentiated granulocytes (granulocyte cytokines; CD15) or monocytes (monocyte cytokines; CD14) cultures. Endogenous MN1 expression in these cells was analyzed using real-time RT-PCR. The graph shows MN1 expression normalized for endogenous HPRT expression (mean ± SEM of duplicates). *P = .01; **P = .008. (C) CD34+ cells were transduced with MIG (mock) or MIG-MN1 (MN1) retroviruses, and unsorted cells were cultured with monocyte-cytokines (M-CSF) in the absence or presence of vitamin D3 (M-CSF + vitamin D3). GFP+ and GFP− cells in mock or MN1 cultures were analyzed for CD14 expression by FACS at day 14 (mean ± SEM of duplicates). **P < .002. In an independent experiment, GFP+ cells from M-CSF cultures were FACS-sorted and cytospin preparations were stained with May-Grünwald-Giemsa (top panel; original magnification ×600). (D) Human CD34+ cells were expanded 1 day and transduced 3 times in 2 consecutive days in media containing progenitor cytokines. One day after the last transduction, CD34+/GFP+ mock or MN1 cells were sorted into 96-well plates containing 100 μL of media with progenitor cytokines (2000 cells/well; triplicate). Cell proliferation was determined at indicated times using the MTT assay (mean ± SEM). **P < .007. (E) In an independent experiment, the percentage of GFP+ cells in unsorted CD34-mock and CD34-MN1 cultures were analyzed at 3, 12, and 30 days after transduction. Data represent one of 2 independent experiments. (F) Human CD34+ cells were transduced and sorted as in panel D, and 500 or 1000 cells were plated in 1 mL of methylcellulose medium in 35-mm dishes in duplicate. Eight days later, colonies were counted and GFP+ cells were replated following the same procedure (mean ± SEM). (G) GFP+ CD34-mock and MN1 cells were sorted 3 days after transduction. After 3-day expansion, cells were seeded at the same density (day 0; 1.2 × 105 cells/well in 1 mL media, 24-well plate) in medium supplemented with progenitor cytokines and followed for 7 days. Media were refreshed every 3 days, and survival (Annexin V) and cell-cycle analysis (H) was performed in duplicate using FACS at day 7. Panel H represents the mean of duplicate ± SEM.
MN1 overexpression inhibits M-CSF or vitamin D3-induced differentiation and enhances growth of CD34+ primary human hematopoietic cells. (A) Real-time RT-PCR analysis of endogenous MN1 expression in BM-derived human CD34+ cells (CD34-BM), peripheral blood monocytes, and granulocytes, normalized for endogenous HPRT expression (mean ± SEM of triplicates). ***P < .001. (B) Primary human CD34+ cells from peripheral blood of G-CSF mobilized healthy donors maintained in cultures containing progenitor, granulocyte, or monocyte cytokine cocktails, respectively. Marker analysis was performed using FACS of day 5 CD34+ stem cell/progenitors (progenitor cytokines) cultures and day 14 in vitro differentiated granulocytes (granulocyte cytokines; CD15) or monocytes (monocyte cytokines; CD14) cultures. Endogenous MN1 expression in these cells was analyzed using real-time RT-PCR. The graph shows MN1 expression normalized for endogenous HPRT expression (mean ± SEM of duplicates). *P = .01; **P = .008. (C) CD34+ cells were transduced with MIG (mock) or MIG-MN1 (MN1) retroviruses, and unsorted cells were cultured with monocyte-cytokines (M-CSF) in the absence or presence of vitamin D3 (M-CSF + vitamin D3). GFP+ and GFP− cells in mock or MN1 cultures were analyzed for CD14 expression by FACS at day 14 (mean ± SEM of duplicates). **P < .002. In an independent experiment, GFP+ cells from M-CSF cultures were FACS-sorted and cytospin preparations were stained with May-Grünwald-Giemsa (top panel; original magnification ×600). (D) Human CD34+ cells were expanded 1 day and transduced 3 times in 2 consecutive days in media containing progenitor cytokines. One day after the last transduction, CD34+/GFP+ mock or MN1 cells were sorted into 96-well plates containing 100 μL of media with progenitor cytokines (2000 cells/well; triplicate). Cell proliferation was determined at indicated times using the MTT assay (mean ± SEM). **P < .007. (E) In an independent experiment, the percentage of GFP+ cells in unsorted CD34-mock and CD34-MN1 cultures were analyzed at 3, 12, and 30 days after transduction. Data represent one of 2 independent experiments. (F) Human CD34+ cells were transduced and sorted as in panel D, and 500 or 1000 cells were plated in 1 mL of methylcellulose medium in 35-mm dishes in duplicate. Eight days later, colonies were counted and GFP+ cells were replated following the same procedure (mean ± SEM). (G) GFP+ CD34-mock and MN1 cells were sorted 3 days after transduction. After 3-day expansion, cells were seeded at the same density (day 0; 1.2 × 105 cells/well in 1 mL media, 24-well plate) in medium supplemented with progenitor cytokines and followed for 7 days. Media were refreshed every 3 days, and survival (Annexin V) and cell-cycle analysis (H) was performed in duplicate using FACS at day 7. Panel H represents the mean of duplicate ± SEM.
Ectopic MN1 enhances proliferation and survival of human CD34+ cells and inhibits their in vitro monocytic differentiation
We next analyzed whether MN1 expression also promoted proliferation and inhibited differentiation of primary human CD34+ cells. We transduced CD34+ cells with mock or MN1-expressing MIG retroviruses and 2 days later induced differentiation of unsorted cells in liquid cultures containing a monocyte-cytokine cocktail in the presence or absence of vitamin D3.23 Coculturing of normal and transduced cells in this experiment provided an internal control for the culture conditions, as well as a test for possible autocrine/paracrine effects between untransduced and transduced cells. FACS analysis showed that, in the mock-transduced sample, M-CSF treatment resulted in a similar percentage of CD14-expressing cells in both the GFP− (39%) and GFP+ (37%) cells, and this percentage was increased 2.2-fold in the presence of vitamin D3 (Figure 2C). Although M-CSF and M-CSF + vitamin D3 treatment of the MN1-transduced culture resulted in a similar percentage of CD14-expressing GFP− cells, the MN1-expressing (GFP+) cells displayed a 3.6-fold and 2.5-fold drop in CD14 expressing cells, respectively (Figure 2C). Indeed, cytospin preparations of GFP+ FACS-sorted mock cells from M-CSF cultures displayed different levels of monocytic differentiation, whereas MN1 cells showed mostly blast morphology (Figure 2C top panel). Reduced sensitivity to M-CSF in CD34+-MN1 cells correlated with decreased expression of M-CSFR (Figure 3A). Similar results were obtained in an independent experiment in which we cocultured FACS-sorted GFP+ and GFP− cells at a 1:1 ratio in a monocytic differentiation assay (data not shown). Therefore, forced MN1 expression impaired the monocytic differentiation of human CD34+ cells in vitro through cell-autonomous mechanisms.
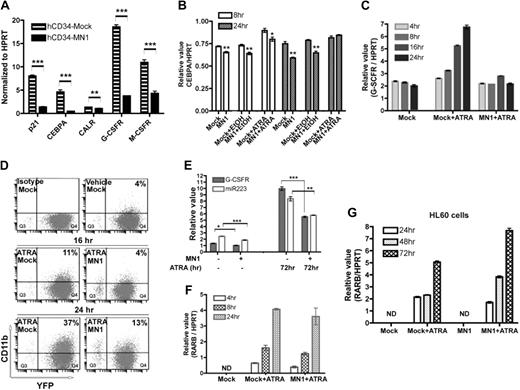
Exogenous MN1 inhibits expression of CEBPA and its downstream targets in CD34+ cells and in ATRA-treated U937 cells. (A) GFP-expressing human CD34+-mock and MN1 cells were sorted 3 days after the last transduction and expanded for 3 more days in media supplemented with progenitor cytokines. Real-time RT-PCR analysis was used to determine the expression of the indicated genes. Results show the expression of each gene relative to endogenous HPRT (mean of triplicates ± SEM). **P = .009; ***P < .001. (B) U937-mock or MN1 cells were seeded (105 cells/mL in 4 mL medium) 24 hours before addition of vehicle or 1 μM ATRA, and endogenous CEBPA expression of untreated (mock or MN1) as well as 8- and 24-hour ATRA-treated cells was determined using real-time RT-PCR (normalized to HPRT; mean of triplicates ± SEM). (C) Mock or MN1-U937 cells were treated with 1 μM ATRA for the indicated times, and expression of G-CSFR relative to endogenous HPRT was determined by real-time RT-PCR of RNA of untreated (mock) and treated cells (mock + ATRA; MN1 + ATRA; mean of triplicates ± SEM). (D) Expression of CD11b was detected using FACS of 16- or 24-hour 1 μM ATRA-treated U937-mock or MN1 cells. Controls are mock cells vehicle-treated for 24 hours. (E) Real-time RT-PCR analysis was performed to determine G-CSFR and miR-223 expression in 72-hour 1-μM ATRA-treated U937-mock and MN1 cells (endogenous HPRT or RNU6B expression was used to normalize G-CSFR or miR-223 expression, respectively; mean of triplicates ± SEM). *P = .012; **P = .009; ***P < .001. (F) Similarly, expression of the RAR/RXR downstream target RARβ was determined using real-time RT-PCR in the same RNA samples that were used in panel B (mean of triplicates ± SEM). (G) Real-time RT-PCR was performed to analyze RARβ and HPRT expression in HL60-mock and MN1 cells treated with 1 μM ATRA for the indicated times (mean of triplicates ± SEM).
Exogenous MN1 inhibits expression of CEBPA and its downstream targets in CD34+ cells and in ATRA-treated U937 cells. (A) GFP-expressing human CD34+-mock and MN1 cells were sorted 3 days after the last transduction and expanded for 3 more days in media supplemented with progenitor cytokines. Real-time RT-PCR analysis was used to determine the expression of the indicated genes. Results show the expression of each gene relative to endogenous HPRT (mean of triplicates ± SEM). **P = .009; ***P < .001. (B) U937-mock or MN1 cells were seeded (105 cells/mL in 4 mL medium) 24 hours before addition of vehicle or 1 μM ATRA, and endogenous CEBPA expression of untreated (mock or MN1) as well as 8- and 24-hour ATRA-treated cells was determined using real-time RT-PCR (normalized to HPRT; mean of triplicates ± SEM). (C) Mock or MN1-U937 cells were treated with 1 μM ATRA for the indicated times, and expression of G-CSFR relative to endogenous HPRT was determined by real-time RT-PCR of RNA of untreated (mock) and treated cells (mock + ATRA; MN1 + ATRA; mean of triplicates ± SEM). (D) Expression of CD11b was detected using FACS of 16- or 24-hour 1 μM ATRA-treated U937-mock or MN1 cells. Controls are mock cells vehicle-treated for 24 hours. (E) Real-time RT-PCR analysis was performed to determine G-CSFR and miR-223 expression in 72-hour 1-μM ATRA-treated U937-mock and MN1 cells (endogenous HPRT or RNU6B expression was used to normalize G-CSFR or miR-223 expression, respectively; mean of triplicates ± SEM). *P = .012; **P = .009; ***P < .001. (F) Similarly, expression of the RAR/RXR downstream target RARβ was determined using real-time RT-PCR in the same RNA samples that were used in panel B (mean of triplicates ± SEM). (G) Real-time RT-PCR was performed to analyze RARβ and HPRT expression in HL60-mock and MN1 cells treated with 1 μM ATRA for the indicated times (mean of triplicates ± SEM).
To determine whether the inhibitory effect of MN1 on differentiation was accompanied by increased proliferation, sorted (GFP+) or unsorted mock and MN1-transduced human CD34+ cells were maintained in progenitor media, and the proliferation capacity of the cells was assessed using the MTT assay (at day 0, 2, 4, and 6; Figure 2D) or by performing periodic GFP analysis at 3, 12, and 30 days after transduction, respectively. Sorted (CD34+/GFP+) cells were also subjected to serial replating in methylcellulose assays to determine the effect of MN1 on self-renewal. As in MN1-transduced mouse HSPCs,9 ectopic MN1 expression increased the self-renewal activity (Figure 2F) and proliferation rate of CD34+ cells as the percentage of GFP+ cells increased from 7% at day 3 to 76% at day 30, whereas it remained stable (16%-28%) in the mock culture (Figure 2E). In a separate experiment, FACS-sorted mock and MN1 cells were cultured in the presence of progenitor cytokines for 10 days and subjected to FACS analysis to determine the cell cycle status and survival of the cells. Annexin-V analysis showed 55% viability in mock cells, whereas it was increased to 89% in MN1 cells (Figure 2G). MN1 cells also showed decreased G0/G1 (MN1, 60%; mock, 79%, P = .002) and an increased S/G2M ratio (MN1, 40%; mock, 21%, P < .01; Figure 2H). Together, these data suggested that ectopic MN1 enhanced the self-renewal and proliferation of human CD34+ cells via increased cell cycle traverse and survival in vitro, which was accompanied by impaired differentiation.
MN1 overexpression down-regulates CEBPA and its downstream target genes in human CD34+ cells
To gain insight into molecular mechanisms orchestrating the MN1-induced effects, we performed microarray analysis of RNA samples of human BM-derived FACS-sorted CD34+ cells transduced with mock or MN1-expressing retroviruses (CD34+/GFP+). Among the many differentially expressed genes, MN1 cells showed decreased expression of CEBPA (2.1-fold) and of 2 of its downstream targets: miR-22313 (3.5-fold) and p2128 (1.9-fold; data not shown). Real-time RT-PCR analysis using RNA from G-CSF mobilized peripheral blood CD34+ cells showed similarly decreased levels of CEBPA (10-fold), and its downstream targets p21 (5.3-fold), G-CSFR (4.8-fold), and M-CSFR (2.6-fold) in MN1 cells, whereas expression of calreticulin (CALR), which regulates CEBPA expression at the translational level,29 was moderately (1.3-fold) but significantly (P = .009) affected (Figure 3A).
Expression of CEBPA downstream targets are not induced by ATRA in U937-MN1 cells
Our microarray data suggested that the down-regulation of CEBPA might contribute to the observed lack of differentiation and increased proliferative effects of MN1 in hematopoietic cells. To test this hypothesis, we first analyzed the expression of CEBPA and its direct targets in response to ATRA, which normally induces the expression of CEBPA targets.13 U937-mock or MN1 cells were treated with 1 μM ATRA for 4, 8, 16, and 24 hours, and expression of CEBPA or G-CSFR was determined using real-time RT-PCR. CEBPA was slightly but significantly down-regulated at 8 and 24 hours in untreated or vehicle-treated MN1 cells compared with mock cells (Figure 3B). Addition of ATRA slightly but significantly (P = .003) increased CEBPA expression at 8 hours in mock cells compared with vehicle-treated mock cells. Although a similar increase was observed after 8 hours of ATRA treatment of MN1 cells compared with vehicle-treated cells, the increase was still significantly lower (P = .042) than in ATRA-treated mock cells at the same time points. ATRA treatment (24 hours) also induced a gradual 3.5-fold increase in G-CSFR expression in U937-mock cells (Figure 3C), which was accompanied by increased expression of the differentiation marker CD11b (Figure 3D). In contrast, G-CSFR expression in MN1 cells was maintained at the level of that in untreated mock cells during ATRA treatment (Figure 3C), and CD11b expression was 2.8-fold lower in MN1 cells treated for 24 hours compared with treated mock cells (Figure 3D). Up-regulation of miR-223, another direct target of CEBPA that is critical for granulocytic differentiation,13 was also partially blocked in ATRA-treated U937-MN1 cells (Figure 3E).
We also analyzed the expression of RARβ, an ATRA-activated RAR/RXR target, in the same RNA samples in Figure 3C. As expected, untreated U937-mock cells did not express RARβ, whereas mock and MN1 cells showed similar levels of RARβ induction after 4, 8, and 24 hours of ATRA treatment (Figure 3F). In a separate experiment, the RARβ level was also determined in 24-, 48-, and 72-hour ATRA-treated HL60-mock or MN1 cells and similar results were obtained (Figure 3G). We concluded that MN1 overexpression moderately down-regulates CEBPA expression in U937 cells; and in response to ATRA, activation of the CEBPA target genes, G-CSFR and miR-223, was impaired in MN1 cells, whereas RAR/RXR activity was preserved.
Ectopic CEBPA expression overrides the MN1-mediated effects on hematopoietic cells
Given the effects of forced MN1 expression on CEBPA expression and its target genes in CD34+ cells, we reasoned that reintroduction of CEBPA might bypass the effects of MN1 on differentiation and proliferation. Therefore, YFP+-sorted U937-mock and MN1 cells were transduced with MIG retrovirus expressing human CEBPA, and the proliferation of YFP+/GFP+-sorted cells was analyzed. CEBPA overexpressing mock and MN1 cells showed a 2.7-fold decrease in growth rate at day 4 of this analysis (Figure 4A), which was accompanied by differentiation as May-Grünwald-Giemsa–stained cytospin preparations displayed morphologic features of differentiated cells, such as segmented nuclei, in mock + CEBPA and MN1 + CEBPA cells but not in the respective parental cell lines (Figure 4B). Moreover, FACS analysis showed that compared with the parental cells, CD11b was induced in 55% of CEBPA-overexpressing mock and MN1 cells (Figure 4C). Additional real-time RT-PCR analysis of RNA samples obtained from day 4 cells showed a 2-fold increase in G-CSFR expression in both mock + CEBPA and MN1 + CEBPA cells compared with controls (Figure 4D).
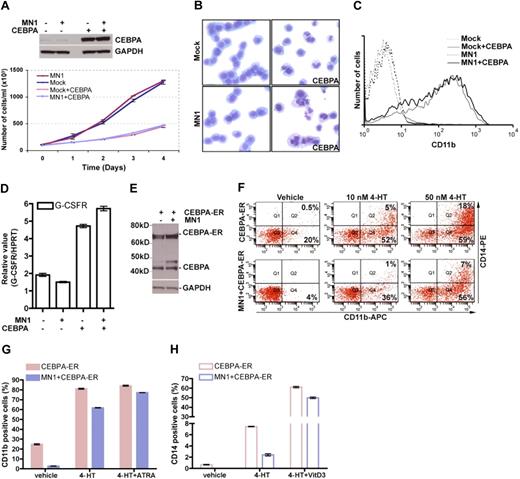
Overexpression of CEBPA overrides MN1's inhibitory effects on differentiation and sensitizes MN1 cells to ATRA and vitamin D3. (A) YFP+-sorted U937-mock and MN1 cells were transduced with GFP-expressing CEBPA-retrovirus. YFP+/GFP+ cells were sorted and used for Western blot analysis to determine CEBPA expression (top panel). Sorted cells were seeded in duplicate at the same density (105/mL) at day 0, and cell numbers were counted during 4 consecutive days of culture. (B) Day 4 cytospin preparations were stained with May-Grünwald-Giemsa (original magnification ×400). (C) CD11b expression was analyzed using FACS. (D) RNA was isolated and subjected to real-time RT-PCR to determine G-CSFR and HPRT expression (mean of triplicates ± SEM). (E) Similarly, YFP+ FACS-sorted U937-mock and MN1 cells were transduced with GFP-expressing CEBPA-ER retrovirus, and YFP+/GFP+ FACS-sorted cells were used for Western blot analysis using anti-CEBPA and GAPDH antibodies. (F) Unsorted cells from panel E were treated for 48 hours with vehicle or the indicated amount of tamoxifen or with the same volume of vehicle (ethanol), and expression of CD11b and CD14 was detected using FACS by gating on GFP+/YFP+ cells (figure represents one of duplicates). (G-H) Unsorted cells from panel E were treated with 100 nM 4-hydroxytamoxifen (4-HT) in the presence or absence of 1 μM ATRA or 100 nM vitamin D3, respectively. Expression of CD11b (G, filled bars) and CD14 (H, open bars) was determined using FACS by gating on GFP+/YFP+ cells. CEBPA-ER indicates mock + CEBPA-ER.
Overexpression of CEBPA overrides MN1's inhibitory effects on differentiation and sensitizes MN1 cells to ATRA and vitamin D3. (A) YFP+-sorted U937-mock and MN1 cells were transduced with GFP-expressing CEBPA-retrovirus. YFP+/GFP+ cells were sorted and used for Western blot analysis to determine CEBPA expression (top panel). Sorted cells were seeded in duplicate at the same density (105/mL) at day 0, and cell numbers were counted during 4 consecutive days of culture. (B) Day 4 cytospin preparations were stained with May-Grünwald-Giemsa (original magnification ×400). (C) CD11b expression was analyzed using FACS. (D) RNA was isolated and subjected to real-time RT-PCR to determine G-CSFR and HPRT expression (mean of triplicates ± SEM). (E) Similarly, YFP+ FACS-sorted U937-mock and MN1 cells were transduced with GFP-expressing CEBPA-ER retrovirus, and YFP+/GFP+ FACS-sorted cells were used for Western blot analysis using anti-CEBPA and GAPDH antibodies. (F) Unsorted cells from panel E were treated for 48 hours with vehicle or the indicated amount of tamoxifen or with the same volume of vehicle (ethanol), and expression of CD11b and CD14 was detected using FACS by gating on GFP+/YFP+ cells (figure represents one of duplicates). (G-H) Unsorted cells from panel E were treated with 100 nM 4-hydroxytamoxifen (4-HT) in the presence or absence of 1 μM ATRA or 100 nM vitamin D3, respectively. Expression of CD11b (G, filled bars) and CD14 (H, open bars) was determined using FACS by gating on GFP+/YFP+ cells. CEBPA-ER indicates mock + CEBPA-ER.
To determine whether ATRA or vitamin D3 treatment could further enhance the differentiation of MN1 + CEBPA cells, YFP-expressing U937-mock or MN1 cells were transduced with tamoxifen-inducible MIGR1-CEBPA-ER retrovirus.26 Treatment of mock and MN1 cells with 10 or 50 nM tamoxifen for 2 days showed a dose-dependent induction of differentiation markers (Figure 4F). In a separate experiment, cells were treated with vehicle or tamoxifen (100 nM) for 2 days in the presence or absence of ATRA and vitamin D3, respectively. FACS analysis of GFP+/YFP+ cells showed that expression of CD11b was induced to a similar degree (81% vs 84%) in tamoxifen and tamoxifen + ATRA-treated CEBPA-ER cells (mock + CEBPA-ER), and cotreatment with tamoxifen + ATRA further enhanced the expression level of CD11b in MN1 + CEBPA-ER cells compared with cells treated with tamoxifen alone (77% and 62%, respectively; Figure 4G). Tamoxifen + vitamin D3 treatment synergistically increased the expression of the monocytic marker CD14 compared with tamoxifen alone treated cells in MN1 + CEBPA-ER (20-fold increase) as well as in mock + CEBPA + ER cells (8-fold increase; Figure 4H).
CEBPA restores myeloid differentiation of MN1-expressing mouse HSPC and inhibits colony formation
To study the differentiation and proliferation capacity of primary mouse HSPC-expressing MN1 or MN1 + CEBPA-ER, lin− BM cells were transduced sequentially with YFP-expressing mock or MN1 retroviruses and GFP-expressing CEBPA-ER retroviruses (Figure 5A diagram). FACS profiles of transduced cells after a second lineage depletion (this time point referred as day 0), which eliminated the cells that committed during the transduction procedure,26 showed that the majority of the untreated day 0 cells were c-Kit+ or Sca1+/c-Kit+ positive progenitor/stem cells (Figure 5B). These cells were cultured with vehicle or tamoxifen for 2 days in the presence of SCF, IL-3, and IL-6, and myeloid differentiation during this culture period was monitored using FACS. At day 2, the percentage of Mac1+/Gr+ or Gr1+ positive granulocytes had increased from 22% (Figure 5C) to 69% (Figure 5D) in vehicle-treated YFP+/GFP−-mock cells. In contrast, granulocytic differentiation was inhibited in untreated day 0 (Figure 5C) and vehicle treated day 2 MN1 cells (Figure 5D) by 2.6- and 4.3-fold, respectively. The percentage of Mac1+/Gr1− monocytic cells was also reduced by ectopic MN1 expression at untreated day 0 and vehicle-treated day 2 cells, by 3-fold and 1.7-fold, respectively (Figure 5C-D). The MN1 effect on myeloid differentiation was eliminated when CEBPA-ER was reintroduced and activated in MN1 cells, as judged by increased numbers of Mac1+/Gr1+ granulocytes and Mac1+ monocytes (Figure 5E).
CEBPA overrides the MN1-induced inhibition of differentiation of primary mouse lin−BM cells. (A) The protocol used for generation of mock (YFP+/GFP−), MN1 (YFP+/GFP−), mock + CEBA-ER (YFP+/GFP+), and MN1 + CEBPA-ER (YFP+/GFP+) transduced primary mouse BM cell populations. (B) Aliquots of cells were analyzed using FACS after a second lineage depletion at the day 0 time point (before 4-HT treatment). Expression of Sca1 and c-Kit surface markers was determined of untreated YFP+ (top panels: mock; bottom panels: MN1) or YFP+/GFP+ (top panels: mock + CEBPA-ER; bottom panels: MN1 + CEBPA-ER) populations. (C) Similarly, Mac1/Gr1 expression was analyzed using the indicated gates at day 0 (before 4-HT treatment). The percentage of Mac1−/Gr1+(1) or Mac1+/Gr1+(2) granulocytes and Mac1+/Gr1−(3) monocytes is shown in mock and MN1 cells (without CEBPA-ER), and in mock + CEBPA-ER, MN1 + CEBPA-ER, or CEBPA-ER populations. (D) Day 0 cells were seeded (2 × 105/mL) in duplicate with vehicle or 4-HT (100 nM) in the presence of SCF, IL-3, and IL-6. Two days later, Mac1/Gr1 expression was determined using FACS of YFP+/GFP− gated cells in mock and MN1 cells treated with vehicle [4-HT (−)] or 4-HT (mean of duplicates ± SEM). *P < .04; **P = .007; ***P < .001. (E) Similarly, Mac1/Gr1 expression was detected at day 2 in mock + CEBPA-ER (top panels) and MN1 + CEBPA-ER cells (bottom panels), treated with vehicle or 4-HT. Figure represents one of the duplicates. (F) In an independent experiment, without pre-5-fluorouracil treatment of the mice, lineage-depleted primary mouse BM cells were cotransduced with GFP-expressing mock or MN1- and YFP-expressing CEBPA retroviruses. Four days after transduction, 300 or 500 cells were FACS-sorted (mock or MN1, GFP+/YFP−; CEBPA, GFP−/YFP+; MN1 + CEBPA, YFP+/GFP+) and plated in methylcellulose media in duplicate. Colonies in each dish were counted 8 days later. Graph represents mean of duplicates ± SEM.  represents 100 cells/dish;
represents 100 cells/dish;  , 166 cells/dish.
, 166 cells/dish.
CEBPA overrides the MN1-induced inhibition of differentiation of primary mouse lin−BM cells. (A) The protocol used for generation of mock (YFP+/GFP−), MN1 (YFP+/GFP−), mock + CEBA-ER (YFP+/GFP+), and MN1 + CEBPA-ER (YFP+/GFP+) transduced primary mouse BM cell populations. (B) Aliquots of cells were analyzed using FACS after a second lineage depletion at the day 0 time point (before 4-HT treatment). Expression of Sca1 and c-Kit surface markers was determined of untreated YFP+ (top panels: mock; bottom panels: MN1) or YFP+/GFP+ (top panels: mock + CEBPA-ER; bottom panels: MN1 + CEBPA-ER) populations. (C) Similarly, Mac1/Gr1 expression was analyzed using the indicated gates at day 0 (before 4-HT treatment). The percentage of Mac1−/Gr1+(1) or Mac1+/Gr1+(2) granulocytes and Mac1+/Gr1−(3) monocytes is shown in mock and MN1 cells (without CEBPA-ER), and in mock + CEBPA-ER, MN1 + CEBPA-ER, or CEBPA-ER populations. (D) Day 0 cells were seeded (2 × 105/mL) in duplicate with vehicle or 4-HT (100 nM) in the presence of SCF, IL-3, and IL-6. Two days later, Mac1/Gr1 expression was determined using FACS of YFP+/GFP− gated cells in mock and MN1 cells treated with vehicle [4-HT (−)] or 4-HT (mean of duplicates ± SEM). *P < .04; **P = .007; ***P < .001. (E) Similarly, Mac1/Gr1 expression was detected at day 2 in mock + CEBPA-ER (top panels) and MN1 + CEBPA-ER cells (bottom panels), treated with vehicle or 4-HT. Figure represents one of the duplicates. (F) In an independent experiment, without pre-5-fluorouracil treatment of the mice, lineage-depleted primary mouse BM cells were cotransduced with GFP-expressing mock or MN1- and YFP-expressing CEBPA retroviruses. Four days after transduction, 300 or 500 cells were FACS-sorted (mock or MN1, GFP+/YFP−; CEBPA, GFP−/YFP+; MN1 + CEBPA, YFP+/GFP+) and plated in methylcellulose media in duplicate. Colonies in each dish were counted 8 days later. Graph represents mean of duplicates ± SEM.  represents 100 cells/dish;
represents 100 cells/dish;  , 166 cells/dish.
, 166 cells/dish.
In a separate experiment, we analyzed whether the increased proliferation of ectopic MN1-expressing primary hematopoietic cells could be prevented by reintroduction of CEBPA. We cotransduced lin− mouse BM cells with GFP and YFP-retroviruses, sorted GFP+, YFP+, and GFP+/YFP+ cells by FACS (mock, MN1, constitutive human CEBPA, MN1 + CEBPA), and plated the cells into methylcellulose media supporting myeloid colony formation. Similar to our previous data, MN1 cells showed up to 3-fold increase in colony formation, reflecting the increased numbers of HSPCs.9 The increased colony formation of MN1 cells was lost when CEBPA was reintroduced into MN1 cells (Figure 5F), which is consistent with the observed growth arrest/differentiation of U937-MN1 + CEBPA cells (Figure 4A). Therefore, in primary HSPC and U937 cells, forced CEBPA expression was sufficient to override the MN1-induced partial block in myeloid differentiation and to suppress MN1-mediated proliferation.
CEBPA overexpression induces differentiation of MN1-overexpressing primary AML cells
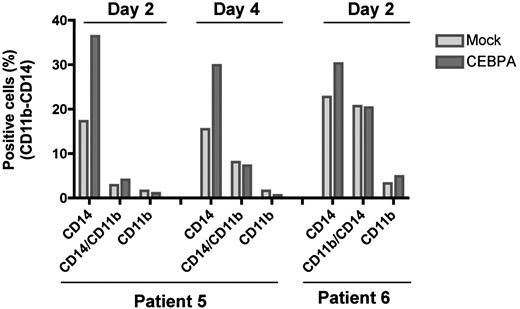
To determine whether ectopic CEBPA expression could also induce differentiation of MN1-overexpressing primary AML cells, we transduced 2 AML patients' BM samples (patients 5 and 6, Figure 1F) exhibiting 94% and 89% blast, respectively, with MIG or MIG-CEBPA retroviruses. FACS analysis showed that CEBPA overexpression increased the level of CD14 by 2-fold in patient 5 and 1.3-fold in patient 6, whereas the expression of CD11b showed only very moderate change (Figure 6). This indicates that CEBPA forces myeloid differentiation in AML patients who overexpress MN1.
Ectopic CEBPA induces differentiation of primary AML cells overexpressing MN1. BM samples of 2 inv(16) AML patients who overexpress MN1 (Figure 1F top panel, patients 5 and 6) were expanded for one day. Cells were transduced 3 times with MIG or MIG-CEBPA retroviruses during 2 consecutive days (retronectin-coated 24-well plates, 2 × 105 cells/well; same conditions described in CD34+ cells with addition of 0.1 mg/mL DNAse I to prevent cell clumping). Half of the media was replaced every other day. Expression of the CD11b and CD14 was determined 2 or 4 days after the last transduction using FACS.
Ectopic CEBPA induces differentiation of primary AML cells overexpressing MN1. BM samples of 2 inv(16) AML patients who overexpress MN1 (Figure 1F top panel, patients 5 and 6) were expanded for one day. Cells were transduced 3 times with MIG or MIG-CEBPA retroviruses during 2 consecutive days (retronectin-coated 24-well plates, 2 × 105 cells/well; same conditions described in CD34+ cells with addition of 0.1 mg/mL DNAse I to prevent cell clumping). Half of the media was replaced every other day. Expression of the CD11b and CD14 was determined 2 or 4 days after the last transduction using FACS.
Discussion
Our main conclusions of this study are as follows: (1) Ectopic MN1 expression not only inhibited granulocytic differentiation11 but also vitamin D3, M-CSF, or growth factor-induced monocytic differentiation of human CD34+ cells, primary mouse lin− hematopoietic cells, and AML cell lines in vitro. (2) Compared with CD34+ cells, endogenous MN1 expression is down-regulated on monocytic/granulocytic differentiation and forced expression of MN1 in human CD34+ cells down-regulated CEBPA expression, increased their self-renewal, proliferation, survival, and cell cycle traverse in response to hematopoietic growth factors in vitro. (3) Reintroduction of tamoxifen-inducible or -constitutive CEBPA bypassed these effects of MN1, suggesting that CEBPA repression contributes to the MN1-induced hematopoietic phenotype.
Forced MN1 expression in lin− mouse BM cells enhanced their self-renewal capacity, resulting in the generation of cell lines and the induction of myeloid disease in a mouse model.9,11 Here we showed that enforced expression of MN1 in human CD34+ hematopoietic cells also caused enhanced self-renewal (Figure 2F) and proliferative advantage of MN1 cells over that of control mock cells in liquid cultures promoting the growth of HSPCs (Figure 2D-E). Furthermore, subsequent analysis of CD34-mock and CD34-MN1 cells demonstrated that this was the result of increased survival and cell cycle traverse of MN1 cells (Figure 2G-H, respectively). We think that this activity is cell type and context dependent, given that, in human osteoblast30 and epithelial cell lines,31 increased MN1 expression is correlated with growth arrest.
Although we do not know how MN1 induces growth of hematopoietic cells, we think that direct or indirect repression of CEBPA and its target genes (microarray analysis, Figure 3A; and data not shown), all critical regulators of self-renewal/proliferation and lineage commitment,12,16 is one of the important consequences of MN1 overexpression. Our findings raise the possibility that MN1 might be involved in self-renewal/myeloid commitment decisions of primitive hematopoietic cells, which is further supported by the following observations: (1) Endogenous MN1 expression is down-regulated in primary or in vitro differentiated human monocytes and granulocytes (Figure 2A-B). (2) Microarray analysis of cord blood-derived human CD34+ hematopoietic cells showed higher MN1 expression in the CD34+/CD38− fraction (highly enriched in long-term engrafting cells32 ), than in the committed CD34+/CD38+ cells,33 and knockdown of CEBPA inhibits myeloid differentiation of human CD34+ cells.34 (3) MN1 and CEBPA exhibit inversed expression patterns in primitive cells; CEBPA expression is lower in hematopoietic stem cells and higher in more committed cells.35
AML is characterized by unlimited self-renewal activity and impaired differentiation of hematopoietic cells, which results in accumulated myeloid blasts in the BM and periphery.3 Enforced expression of MN1 fulfills these criteria by deregulating both activities9,11 (Figures 1C-D, 2C,E,F, and 5E-F). Given that CEBPA is down-regulated in CD34-MN1 cells (Figure 3A) and is indispensable for granulocytic and monocytic maturation,15,26 we first analyzed whether CEBPA expression is also down-regulated in U937-MN1 cells. However, ectopic MN1 only moderately down-regulated endogenous CEBPA expression at the transcriptional level in U937 cells (Figure 3B), and whether MN1 directly represses CEBPA remains elusive. Although CEBPA expression only changed moderately, ATRA-induced activation of the CEBPA target genes, G-CSFR and miR223, was impaired in U937-MN1 cells, whereas activation of RAR/RXR, another well-established transcription factor regulating myeloid differentiation, was not altered as judged by similar expression of its direct target RARβ (Figure 3F-G). This is consistent with our previous findings showing that MN1 and ATRA synergistically activated RAR/RXR-induced transcriptional activity in Hep3B cells.36 This makes a stronger case for functional involvement of MN1-mediated repression of CEBPA in the inhibition of differentiation of hematopoietic cells. Supporting this notion, we found that reintroduction of constitutive human CEBPA or tamoxifen-inducible rat Cebpa26 bypassed the inhibitory effect of MN1 and promoted both monocytic and granulocytic differentiation in U937 cells, mouse lin− BM cells, and primary AML cells that overexpress MN1 (Figures 4,Figure 5–6).
Currently, we do not know how MN1 exerts its effect on CEBPA expression and activity, but our data clearly indicate that reintroduction of CEBPA is sufficient to limit the effect of MN1 on proliferation and myeloid differentiation in vitro. This is similar to CEBPA's effect on human wild-type CD34+ and AML-CD34+ cells.17 Based on our results, it is tempting to speculate that, in hematopoietic cells, MN1 represses CEBPA expression via factors required for its transcription, or perhaps via epigenetic modifications of the CEBPA promoter region. CEBPA exerts its function via direct binding to promoter/enhancer regions of its target genes as well as via posttranslational modifications and protein-protein interactions.29,37 Because MN1 can bind the transcriptional coactivator p300,36 we analyzed whether recruitment of MN1 to the CEBPA/p300 protein complex would inhibit CEBPA's transcriptional activity in U937-MN1 cells, but we were unable to detect any interaction between CEBPA and MN1 (data not shown). Furthermore, the amount of p300 bound to CEBPA remained unaltered (data not shown). We cannot exclude that binding of other proteins/coactivators to CEBPA might be impaired in MN1 cells. Therefore, future studies, including detailed identification of MN1 interacting proteins, might provide clues about the molecular consequences of MN1 expression.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Alan D. Friedman for kindly providing MIGR1-CEBPA-ER construct; Dr Mihaela Onciu for the analysis of cytospin preparations; Yvan Campos for subcloning of CEBPA; Dr Youngsoo Lee for providing help for real-time RT-PCR analysis; Dr Richard Ashmun, Dr Geoff Neale, Martha Holladay, and members of FACS and microarray facility for expert data analysis; Drs Jan Jacob Schuringa, Thasia Leimig, Hakan Cam, and John Easton for helpful suggestions; Sheila Shurtleff for providing patient cells; the St Jude blood donor center for providing apheresis rings; and Charlette Hill for secretarial assistance.
This work was supported by the National Cancer Institute (grant CA72999), the Cancer Center (support grant CA021765), and the American Lebanese Syrian Associated Charities.
Authorship
Contribution: A.K. designed and performed research, analyzed data, and wrote the paper; and G.C.G. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gerard C. Grosveld, Department of Genetics and Tumor Cell Biology, St Jude Children's Research Hospital, 262 Danny Thomas Place, Memphis, TN 38105; e-mail: gerard.grosveld@stjude.org.

![Figure 5. CEBPA overrides the MN1-induced inhibition of differentiation of primary mouse lin−BM cells. (A) The protocol used for generation of mock (YFP+/GFP−), MN1 (YFP+/GFP−), mock + CEBA-ER (YFP+/GFP+), and MN1 + CEBPA-ER (YFP+/GFP+) transduced primary mouse BM cell populations. (B) Aliquots of cells were analyzed using FACS after a second lineage depletion at the day 0 time point (before 4-HT treatment). Expression of Sca1 and c-Kit surface markers was determined of untreated YFP+ (top panels: mock; bottom panels: MN1) or YFP+/GFP+ (top panels: mock + CEBPA-ER; bottom panels: MN1 + CEBPA-ER) populations. (C) Similarly, Mac1/Gr1 expression was analyzed using the indicated gates at day 0 (before 4-HT treatment). The percentage of Mac1−/Gr1+(1) or Mac1+/Gr1+(2) granulocytes and Mac1+/Gr1−(3) monocytes is shown in mock and MN1 cells (without CEBPA-ER), and in mock + CEBPA-ER, MN1 + CEBPA-ER, or CEBPA-ER populations. (D) Day 0 cells were seeded (2 × 105/mL) in duplicate with vehicle or 4-HT (100 nM) in the presence of SCF, IL-3, and IL-6. Two days later, Mac1/Gr1 expression was determined using FACS of YFP+/GFP− gated cells in mock and MN1 cells treated with vehicle [4-HT (−)] or 4-HT (mean of duplicates ± SEM). *P < .04; **P = .007; ***P < .001. (E) Similarly, Mac1/Gr1 expression was detected at day 2 in mock + CEBPA-ER (top panels) and MN1 + CEBPA-ER cells (bottom panels), treated with vehicle or 4-HT. Figure represents one of the duplicates. (F) In an independent experiment, without pre-5-fluorouracil treatment of the mice, lineage-depleted primary mouse BM cells were cotransduced with GFP-expressing mock or MN1- and YFP-expressing CEBPA retroviruses. Four days after transduction, 300 or 500 cells were FACS-sorted (mock or MN1, GFP+/YFP−; CEBPA, GFP−/YFP+; MN1 + CEBPA, YFP+/GFP+) and plated in methylcellulose media in duplicate. Colonies in each dish were counted 8 days later. Graph represents mean of duplicates ± SEM. represents 100 cells/dish; , 166 cells/dish.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/8/10.1182_blood-2009-02-205443/4/m_zh89990940870005.jpeg?Expires=1767710903&Signature=SCCahjG9EL4pU--M7LC~2-fO~3bl1lKWxXuZE0lZ-f9cCF4X1KhOvAWQpReCjR4fLcqjZJy6Kpe3lgZNjHGKJzD0bNSl~YBqkabMekXy~twHh3rRGCJzyfSZtE8BhUnHLqFZPFFzVFOjSkViQrt7HFu4ZdsFBPAZV8hW8mAgJUuyWS3~GzgWQ6h4gmQ3IJ6C9OQCaIo3ZlbskxasP9GOKN7Wspas4dtiBoIq484VAb9DFI-BSXR~702ReGYAXkdTTOGcb7LGfrGXYn91RCmDJaPI2E4FjPzB14dVYCB-kNnLz2YpBBQIDKqBNOersZPsiJRXQszaGtefXQOmwGxAGQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)