Abstract
Human CD20 is a B-cell lineage–specific marker expressed by normal and leukemic B cells from the pre-B to the plasma-cell stages and is a target for rituximab (RTX) immunotherapy. A CD20 reverse transcriptase–polymerase chain reaction (PCR) on B-cell lines cDNA yielded a short PCR product (ΔCD20) corresponding to a spliced mRNA transcript linking the exon 3 and exon 7 ends. We established here that this novel, alternatively spliced CD20 transcript is expressed and detectable at various levels in leukemic B cells, lymphoma B cells, in vivo tonsil- or in vitro CD40L-activated B cells, and Epstein-Barr virus (EBV)–transformed B cells, but not in resting CD19+- or CD20+-sorted B cells from peripheral blood or bone marrow of healthy donors. The truncated CD20 sequence is within the reading frame, codes a protein of 130 amino acids (∼ 15-17 kDa) lacking large parts of the 4 transmembrane segments, suggesting that ΔCD20 is a nonanchored membrane protein. We demonstrated the translation into a ΔCD20 protein which is associated with the membrane CD20 protein and showed its involvement in RTX resistance. Study of patient samples before and after RTX resistance or escape confirms our in vitro findings.
Introduction
CD20 (MS4A1) is a 33- to 37-kDa nonglycosylated transmembrane (TM) phosphoprotein that is widely expressed throughout B-lymphocyte ontogeny, in normal or malignant B cells.1 The 16-kb gene encoding human CD20, consisting of 8 exons, has been mapped to chromosome 11 (11q12-q13) and belongs to the MS4A (membrane spanning 4A) gene family localized within a cluster of related genes (MS4A1 to MS4A11).2 Its transcription leads to 3 mRNA isoforms: a dominant 2.8-kb transcript, using exons 1 to 8; a second exon 1–spliced transcript shorter by 263 bp; and a third, minor 3.4-kb transcript,3 all encoding a full-length CD20 protein.
The CD20 protein consists of cytoplasmic N- and C-termini and 4 hydrophobic regions for anchoring the molecule in the membrane.4 A total of 3 isoforms have been identified, including a predominant 33-kDa molecule and 2 isoforms of 34.5 and 36 kDa, resulting from differential phosphorylation states (on serine and threonine residues) in relation to B-cell stimulation and proliferation.5 CD20 appears to play a role in Ca++ conductance6 and is also involved in cell-cycle progression by interaction with src family kinases.7 Finally, CD20 circulating form has been identified in chronic lymphocytic leukemia (CLL), Hodgkin disease, or non-Hodgkin lymphoma (NHL), and in healthy persons.8
CD20 expression at the cell surface of malignant B cells makes it a target for monoclonal antibody (mAb) therapy. Rituximab (RTX), the first US Food and Drug Administration (FDA)–approved mAb for clinical therapy, targets the CD20 antigen9 and leads to CD20-expressing B-cell depletion through different mechanisms9,10 (for review, see Cartron et al11 ). Thus, RTX is widely used against B-cell malignancies and also for autoimmune diseases such as rheumatoid arthritis,12 steroid refractory chronic graft-versus-host disease (GVHD),13 or posttransplantation lymphoproliferative disease,14 and for treatment of refractory kidney transplant humoral rejection.15
Although its clinical effectiveness is uncontested, some factors, directly linked to CD20 gene expression or related to apoptotic signaling,16 may influence its clinical benefit and sometimes lead to RTX resistance. These factors include FcγRIII polymorphism,17 CD20 cell-surface expression level,18,19 CD20 distribution within the membrane lipid rafts,20 the presence of a mutation/deletion in the CD20 coding region,21 epigenetic regulation of the CD20 gene,22 or CD20 protein phosphorylation rate.23
Retroviral CD20 gene transfer was proposed as an alternative suicide gene therapy to improve the system of genetically modified T-lymphocyte adoptive transfers.24–26 Because transcriptional regulation through aberrant alternative splicing is an emerging mechanism involved in cancer progression27 and was previously associated with resistance to transgenic T-cell depletion,28 we analyzed CD20 transcriptional regulation in CD20-transduced cells.
We indentified a novel CD20 alternative mRNA (ΔCD20), demonstrating that this splice variant mRNA encodes a truncated protein and providing evidence that ΔCD20 is directly correlated with RTX resistance. Moreover, ΔCD20 splice mRNA was absent from normal B cells isolated from healthy donors and present in malignant B cells, making it a molecular marker of choice for diagnosis or molecular minimal residual disease follow-up.
Methods
Patients, cell lines, B-cell isolation, and purification
Human cell lines were obtained from the DSMZ or ATCC cell banks. Cells were maintained in RPMI 1640 with 10% fetal calf serum added. We established Epstein-Barr virus (EBV)–transformed autologous lymphoblastic cell lines by coculturing peripheral blood mononuclear cells (PBMCs) with EBV-containing supernatant from the B95.8 EBV-producing cell line, as described.29 CD19+-sorted B cells were activated with irradiated CD40L-transfected cells for 3 days or Pokweed mitogen (Sigma-Aldrich). Cells were then split or fed with fresh medium until harvest at day 7. Human tonsil, peripheral blood, bone marrow, and lymph nodes and spleen were obtained, respectively, from clinical tonsillectomies, hematologic B-cell disease samples (B-CLL, B–acute lymphoblastic leukemia [ALL], follicular lymphoma [FL], mantle-cell lymphoma [MCL], diffuse large B-cell lymphoma [DLBCL], and marginal zone lymphoma [MZL]) for diagnostic assessment, clinical trial, or from a blood bank for the healthy PBMCs or bone marrow (BM) cells.
Informed consent for functional tests and genetic analysis was obtained from patients and healthy donors.
Human CD19+ or CD20+ cells were immunomagnetically purified using whole-blood CD19 or CD20 microbeads kits with an autoMACS (Miltenyi Biotec).
Molecular study: genomic DNA and RNA isolation, reverse-transcription and real-time PCR for mRNA quantification, and cycle sequencing
Genomic DNA was extracted using a standard salting-out method. Total cellular RNAs were extracted using the RNeasy Total RNA Isolation kit (QIAGEN), following manufacturer protocols. For qualitative PCR, 1 μg of total RNA was reverse-transcribed into cDNA as previously described.30 Conventional full-length cDNA PCR amplification (flCD20-PCR) was performed from 2 μL of cDNA using primers specific from the start (exon 3) and the stop codon (exon 8), respectively: Fw-hCD20-start (5-ATGACAACACCCAGAAATTC-3′) and R-hCD20-stop (5′-TTAAGGAGAGCTGTCATTTTCT-3′). (Bold characters indicate start and stop codons, respectively).
For specific and more sensitive amplification (ΔCD20-PCR) of the identified splice variant, a primer Fw-ΔhCD20 (5′-GATGTCTTCACTGG/AACT-3′) spanning the junction was used in combination with the R-hCD20 stop primer. PCR was performed using BIOTAQ polymerase (Bioline) in standard conditions. Annealing temperature was 58°C for both fl- and ΔCD20-PCR.
For CD20 mRNA transcript expression analysis, real-time quantitative PCR (RT-qCPR) was performed using pairs of primers to specifically amplify the full-length form of CD20, as follows: TQM-Fw-hCD20-wt (5′-GAGCCAATGAAAGGCCCTATT-3′) and TQM-R-hCD20-wt (5′-AAGAAGCTTTGCGTGGGGCC-3′; complementary of the spliced region). For specific analysis of the spliced forms of mRNA CD20, a reverse primer TQM-R-ΔhCD20 (5′-AGCTATTACAAGTT/CCAGTG-3′) spanning the junction was used in combination with the TQM-Fw-hCD20-wt. PCR products were revealed using a dual-labeled Fam/Tamra TaqMan probe: TQM-probe-hCD20 (5′-ATGCAATCTGGTCCAAAACCACTCTTCAGG-3′). To determine the sequence of the CD20 variant and of the site-directed mutated form (mutCD20), we performed cycle sequencing on a 3130 DNA analyzer, directly from PCR- or gel-purified products, in both directions using the Cycle Sequencing Kit Version 3.1 (Applied Biosystems).
Computational analysis: splice-site prediction and site-directed mutagenesis
We performed computer-assisted analysis for splice-site and branched-point prediction using NetGene2 (release 2.4)31 and NNsplice Version 0.9.32 We designed site-directed mutagenesis primers for acceptor site (AS) mutation using the online program QuikChange Primer Design Program (http://www.stratagene.com/sdmdesigner/default.aspx) from Stratagene.
Generation of plasmids and retroviral constructs, site-directed mutagenesis, and production of packaging cell lines
flCD20 and ΔCD20 forms were cloned into a pcDNA 3.1/CT–green fluorescent protein (GFP) vector (Invitrogen) for expressing a CD20/GFP fusion protein to study subcellular protein localization.
The degeneracy of the genetic code allowed a change of the third nucleotide of the AS codon at position 612, from CAG (Gln) to CAA (Gln). Primers used were G612A-Fw 5′-GATCTTTGCCTTCTTCCAAGAACTTGTAATAGCTGGC-3′ and G612A-R 5′-GCCAGCTATTACAAGTTCTTGGAAGAAGGCAAAGATC-3′.
Finally, both these and the site-directed mutated form of the CD20 (mutCD20) were cloned into the retroviral pLXSN vector (Clontech). Supernatant was produced from the PG13 amphotropic packaging cell line.
Establishment of RTX resistance, immunophenotyping, and in vitro CDC assay
Establishment of RTX resistance was performed as previously described.33 Briefly, Ramos and Raji B-cell lines (group N), adjusted to 106 cells/mL, were serially (4 times) exposed for 24 hours to a low dose of RTX (0.5 μg/mL) to generate the R1 group, or 3 times in the presence of escalating high doses of RTX (2 and 64 μg/mL) for generating the R2 group. RTX exposure was done in the presence of 25% of newborn rabbit serum (NRS) or 50% human serum (HS) as a source of complement. Ficoll gradient centrifugation was applied after each RTX exposure to remove death cells. RTX was obtained from the Besançon Hospital Pharmacy Department.
We assessed resistance acquisition first by flow cytometric analysis, using a conjugated anti-CD20 antibody (mouse anti–human CD20 mAbs [IgG2b κ, clone 2H7]; BD Pharmingen) to visualize CD20 membrane expression change as a signature of the resistance, as described.20 Further, we performed an in vitro complement-dependent cytolysis (CDC) assay. Briefly, cells were incubated with an increased dose of RTX for 1 hour at 37°C in the presence or absence of NRS. The cell lysis percentage was calculated by blue trypan cell counting and reported as follows: % cell lysis = [1 − (viable cells after RTX + 25% NRS exposure) / (viable cells before RTX + 25% NRS)] × 100.
Experiments were performed 15 days after the last RTX exposure.
Confocal microscopy, slide preparation, and immunofluorescence staining and Western blotting
Cells were spread onto poly-L-lysine–coated slides (Sigma-Aldrich), fixed with paraformaldehyde 4%, and washed. After blocking with 20% fetal bovine serum and washing, cells were stained with the appropriate mAbs or directly visualized using GFP LASER excitation. Stacks of confocal images were collected with an FV1000 laser-scanning confocal microscope (Olympus). Cell nuclei were counterstained with DAPI.
For the Western blotting study, cells were lysed with sample buffer (2% SDS in 125mM Tris HCl, pH 6.8). Cytoplasm and membrane subcellular fractions were harvested after differential ultracentrifugation in adapted buffers; the presence or absence of subcellular-specific proteins by Western blot attested to subcellular separation. Proteins were extracted from 0.5 × 107 to 1 × 107 cells and by electrophoresis on 12.5% SDS-polyacrylamide gels and transferred to PDVF membranes (GE Healthcare).
The blots were then blocked for 1 hour in 6% milk before incubation with specific antibodies against human CD20 as follows: rabbit anti–human CD20 specific to the COOH-terminal region (Thermo Scientific), MS4A1 MaxPab mouse polyclonal antibody (B01; Abnova), RTX (Roche), or mouse polyclonal anti–human CD20 (7D1; AbD Serotec). Blotted proteins were detected and quantified on a bioluminescence imager and BIO-1D advanced software (Wilber-Lourmat) after incubating blots with a horseradish peroxidase–conjugated appropriate secondary antibody (Beckman Coulter). A synthetic CD20/GST 88-mer recombinant polypeptide and B-cell line known to express CD20 served as controls in every experiment.
Results
From alternative splicing, a novel isoform of human CD20 (ΔCD20) mRNA arises, devoid of the sequence coding for the 4 major TM domains
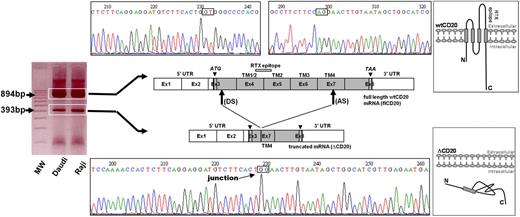
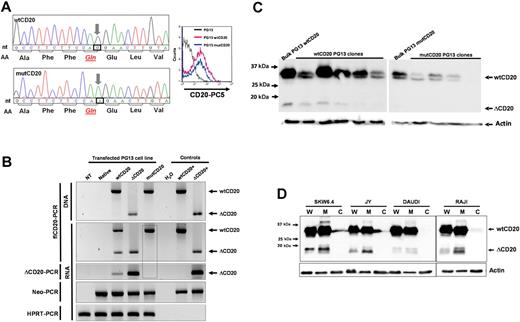
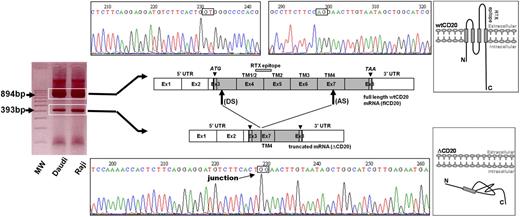
From the reverse transcription and PCR of full-length CD20 (flCD20) cDNA, synthesized from Raji or Daudi B-cell lines, using 2 primers respectively complementary of the start and stop codon regions, we observed a PCR product smaller (393 bp) than the expected size (894 bp; Figure 1). Sequencing analysis showed that the smaller transcript (ΔCD20) was identical to the wild-type (wt) MS4A1 sequence published in the National Center for Biotectnology Information (NCBI) GenBank34 (NM_021950.3), but lacked an internal 501-bp fragment, corresponding to part of exon 3 to part of exon 7. Therefore, the open reading frame remained conserved. This ΔCD20 splice form transcript differs from previously described 2.8- and 3.4-kb alternative dominant MS4A1 forms3 and from other known transcripts of the MS4A gene family. Furthermore, smaller PCR products we obtained from all of the screened B-cell lines were similar in length. In silico sequence analysis of the full-length CD20 sequence, using the NetGene2 and NNsplice Version 0.9 programs, showed putative donor (“GT,” localized in exon 3) and acceptor (“AG,” localized exon 7) sites, as well as a branched site (“A,” nt 594 of the coding sequence), corresponding exactly to the open reading frame of the smaller transcript ΔCD20. This deletion covered nucleotides 111 to 612 (from the +1 ATG nucleotide) and removed codons 37 to 204. Thus, this new in-frame cDNA encodes a putative novel isoform of CD20 (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article) lacking much of the 4 TM domains and the extracellular area. Consequently, the region of mRNA coding for the RTX epitope35 is deleted, suggesting that RTX cannot target this truncated spliced ΔCD20 protein.
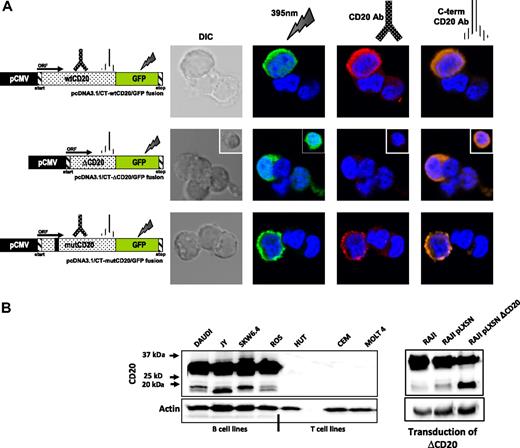
Identification of the ΔCD20 splice isoform mRNA. Agarose gel electrophoresis of full-length (fl) PCR of the CD20 coding sequence from the Daudi and Raji cell line cDNA. Sequencing electrophoregrams of both full-length wt and truncated (ΔCD20) CD20 forms, showing juxtaposed part of the end of exon 3 (Ex) and exon 7. Localization of splicing sites (DS indicates donor site; and AS, acceptor site) on the RNA sequence (Ex1 to Ex8; UTR indicates untranslated regions). Correspondence of CD20 protein segments is shown (TM indicates transmembrane domains), and RTX epitope localization is indicated on wtCD20 as a hatched box. A schematic figure of the intracellular putative ΔCD20 is given for comparison to the wt protein. MW indicates the 100-bp molecular marker. ATG and TAA indicate start and stop codons, respectively.
Identification of the ΔCD20 splice isoform mRNA. Agarose gel electrophoresis of full-length (fl) PCR of the CD20 coding sequence from the Daudi and Raji cell line cDNA. Sequencing electrophoregrams of both full-length wt and truncated (ΔCD20) CD20 forms, showing juxtaposed part of the end of exon 3 (Ex) and exon 7. Localization of splicing sites (DS indicates donor site; and AS, acceptor site) on the RNA sequence (Ex1 to Ex8; UTR indicates untranslated regions). Correspondence of CD20 protein segments is shown (TM indicates transmembrane domains), and RTX epitope localization is indicated on wtCD20 as a hatched box. A schematic figure of the intracellular putative ΔCD20 is given for comparison to the wt protein. MW indicates the 100-bp molecular marker. ATG and TAA indicate start and stop codons, respectively.
ΔCD20 mRNA is selectively expressed in different malignant or EBV-transformed B-cell lines, but not in resting B lymphocytes isolated from healthy donors
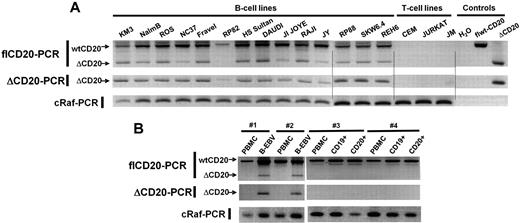
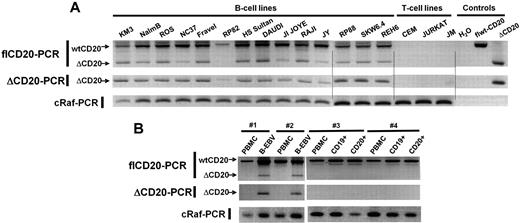
Having detected the new isoform of CD20 mRNA in Raji and Daudi, we decided to analyze ΔCD20 expression in other malignant B-cell lines. For this purpose, flCD20-PCR was performed in pre-B (n = 3), Burkitt (n = 3), B-ALL (n = 1), lymphoblastic (n = 1), plasmocytic (n = 3), EBV-transformed (n = 3), and T-CLL (n = 2) and T-ALL (n = 1) cell lines. In addition to the flCD20 transcript PCR products (894 bp), we detected ΔCD20 transcripts of PCR products (393 bp) in all screened malignant and EBV-transformed B-cell lines but not T-cell lines (Figure 2A). To confirm these results, we designed a PCR assay (ΔCD20-PCR) allowing specific and more sensitive detection of the spliced transcripts, using a forward primer spanning the splice junction. The expected 295-bp PCR product was detected in all previously screened B-cell lines. We confirmed specificity of the ΔCD20 RT-PCR assay by absence of amplification from T-cell line–derived cDNA and from a plasmid vector carrying the flCD20. As a control, each short PCR product was sequenced, enabling confirmation that each 393-bp amplicon corresponded to ΔCD20 (data not shown).
RT-PCR detection of the ΔCD20-spliced mRNA. Qualitative full-length RT-PCR (flCD20-PCR) and specific RT-PCR (ΔCD20-PCR) allowing detection, respectively, of both wt/Δ or specific Δ forms of CD20. cRaf PCR amplified a control gene. flCD20 and ΔCD20 controls consisted of plasmids carrying the respective cloned sequences (A) on different B- and T-cell lines and (B) on in vitro B-EBV–produced cell lines and their respective PBMCs and on CD19+- or CD20+-purified cells from healthy donors and their corresponding PBMCs.
RT-PCR detection of the ΔCD20-spliced mRNA. Qualitative full-length RT-PCR (flCD20-PCR) and specific RT-PCR (ΔCD20-PCR) allowing detection, respectively, of both wt/Δ or specific Δ forms of CD20. cRaf PCR amplified a control gene. flCD20 and ΔCD20 controls consisted of plasmids carrying the respective cloned sequences (A) on different B- and T-cell lines and (B) on in vitro B-EBV–produced cell lines and their respective PBMCs and on CD19+- or CD20+-purified cells from healthy donors and their corresponding PBMCs.
To determine whether this specific CD20 alternatively spliced form is expressed only in transformed B cells, we searched for the ΔCD20 isoform in cDNA synthesized from PBMCs (n = 7) and bone marrow mononuclear cells (BMMCs; n = 5) derived from healthy donors using both flCD20- and ΔCD20–RT-PCR. Moreover, we screened immunomagnetically CD20+-purified B cells, and because ΔCD20 may modulate CD20 cell-surface membrane expression, we also studied CD19+-purified B cells. We detected no alternative transcripts, even with the ΔCD20 PCR assay, either in BMMCs or in CD20+ and CD19+ PBMC–purified B cells (Figure 2B). In addition, we investigated ΔCD20 expression in B-EBV–transformed cell lines (n = 4; Figure 2B), in in vitro–activated B lymphocytes, and in CD19+ from tonsillectomy samples. All expressed the ΔCD20-spliced form. A kinetic study on B-EBV– or CD40L-activated B blasts revealed the truncated form of ΔCD20 all over the culture when stimuli were maintained. However, for B blasts, the signal decreased 72 hours after initial CD40L activation (data not shown).
Translation of in-frame ΔCD20 alternative transcript codes for a protein
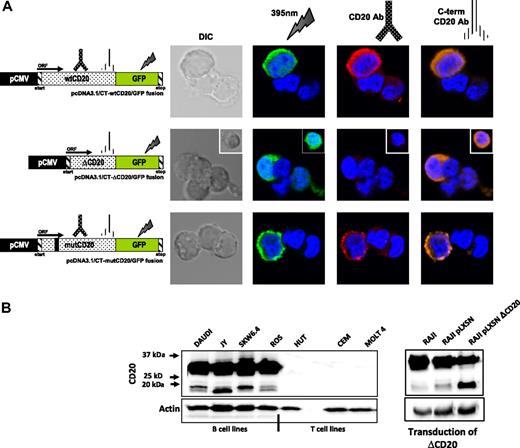
In silico translation analysis show that splicing of the CD20 gene results in in-frame transcript coding for a ΔCD20 protein. Splicing affected a cysteine amino acid, implying an effect in a disulfide bond in a major part of the TM domain (5 amino acids of the TM4 domain remain) but not the phosphorylation sites. To explore this issue and to confirm the existence of the ΔCD20 protein, we used transfected 293T cells with a vector carrying wt and Δ-CD20 cDNA fused with the GFP sequence, allowing expression of wt and Δ-CD20/GFP fusion proteins, respectively, examined under confocal microscopy. We thus detected protein expression with either anti-CD20 Ab staining or after GFP excitation. As expected, while GFP signal was detected for both constructs, the anti-CD20 antibody, recognizing the deleted region, emitted fluorescence only for the full-length protein. The anti-CD20 antibody, specific to the C-terminus (C-term) region, recognized both complete and truncated ΔCD20 proteins (Figure 3A).
ΔCD20 protein expression. (A) Confocal microscopy analysis of wt and ΔCD20 protein expression, on 293T cells transfected with different constructs carrying flCD20, ΔCD20, or mutCD20 cDNA fused with a GFP sequence leading to the expression of a CD20/GFP fusion protein. Cells were imaged using a Fluoview FV1000 (Olympus) and were stained with DAPI (blue) for nuclear staining and also with either monoclonal anti-CD20 antibody (recognizing wt and mutCD20 forms; red), C-term anti-CD20 antibody (recognizing all CD20 forms; orange), or by GFP 385-nm excitation (green). wtCD20 and mutCD20-transfected cells show membrane staining according to the presence of the 4 transmembrane domains allowing anchoring, whereas ΔCD20/GFP staining is localized mainly within the cytoplasm and absent within the membrane. Simultaneous staining (intracellular and membrane) was achieved with an anti–C-term CD20 antibody. DIC indicates differential interference contrast; and ORF, open reading frame. Untransduced cells were used as controls. (B) Western blot (WB) analysis, after denaturing acrylamide electrophoresis, with anti–C-term CD20 antibody of whole-cell lysates from B- and T-cell lines (left) and a retrovirally transduced Raji cell line with a vector carrying the ΔCD20 cDNA (right). As expected, we detected a signal at position 33 to 35 kDa, corresponding to the wtCD20 protein isoforms (differentially phosphorylated), but also 2 additional bands at 15 to 17 kDa, corresponding to the size of the translated spliced mRNA. The 2 bands at position 15 to 17 kDa could correspond to different phosphorylation states of the ΔCD20 protein. Moreover, detection of an increased signal at the same size length after CD20 transduction confirmed that the smaller band is the product of the ΔCD20 mRNA translation. Antiactin WB on the whole-cell lysates was performed as controls.
ΔCD20 protein expression. (A) Confocal microscopy analysis of wt and ΔCD20 protein expression, on 293T cells transfected with different constructs carrying flCD20, ΔCD20, or mutCD20 cDNA fused with a GFP sequence leading to the expression of a CD20/GFP fusion protein. Cells were imaged using a Fluoview FV1000 (Olympus) and were stained with DAPI (blue) for nuclear staining and also with either monoclonal anti-CD20 antibody (recognizing wt and mutCD20 forms; red), C-term anti-CD20 antibody (recognizing all CD20 forms; orange), or by GFP 385-nm excitation (green). wtCD20 and mutCD20-transfected cells show membrane staining according to the presence of the 4 transmembrane domains allowing anchoring, whereas ΔCD20/GFP staining is localized mainly within the cytoplasm and absent within the membrane. Simultaneous staining (intracellular and membrane) was achieved with an anti–C-term CD20 antibody. DIC indicates differential interference contrast; and ORF, open reading frame. Untransduced cells were used as controls. (B) Western blot (WB) analysis, after denaturing acrylamide electrophoresis, with anti–C-term CD20 antibody of whole-cell lysates from B- and T-cell lines (left) and a retrovirally transduced Raji cell line with a vector carrying the ΔCD20 cDNA (right). As expected, we detected a signal at position 33 to 35 kDa, corresponding to the wtCD20 protein isoforms (differentially phosphorylated), but also 2 additional bands at 15 to 17 kDa, corresponding to the size of the translated spliced mRNA. The 2 bands at position 15 to 17 kDa could correspond to different phosphorylation states of the ΔCD20 protein. Moreover, detection of an increased signal at the same size length after CD20 transduction confirmed that the smaller band is the product of the ΔCD20 mRNA translation. Antiactin WB on the whole-cell lysates was performed as controls.
Using Western blot analysis with the C-term region anti-CD20 Ab, we detected, in addition to the expected size length (35-37 kDa) of the wtCD20, an additional signal at 15 to 17 kDa in all screened B-cell lines (Figure 3B left). The 15- to 17-kDa size length correlated with the putative coded protein translated from the ΔCD20 alternative transcript described above. Signal was detected only in B-cell lines (n = 4) but not in T-cell lines (n = 3).We also detected the same signal in 5 native samples from patients with CLL (supplemental Figure 2) and MCL. More precisely, the 15- to 17-kDa signal split into 2 distinct bands, as generally detected for the wtCD20 protein; these bands correspond to different phosphorylation states, as previously described,23 in which the smaller band is the unphosphorylated form. Remarkably, band intensity corresponding to the ΔCD20 protein phosphorylation status differed between cell lines, as illustrated in Figure 3B; the lower band was more intense for JY, the upper band was more intense for SkW 6-4 and Daudi, and the intensity remained similar in the ROS cell line.
For confirmation that the 15- to 17-kDa protein correlated with ΔCD20 transcript translation, we transduced a Raji cell line with a retroviral vector carrying the ΔCD20 cDNA sequence and obtained a significant 2.96-fold increase in signal at position 15 to 17 kDa, indicating again that the ΔCD20 mRNA encodes a protein (Figure 3B right). Interestingly, the signal intensity of the wtCD20 band decreased (fold change [FC], 0.68). Altogether, these results demonstrated that ΔCD20 alternative mRNA is translated into protein.
Finally, to demonstrate that the smaller product is not a breakdown product, we analyzed propidium iodide (PI+; death) versus PI− (live) cells after RTX exposure. As reported in supplemental Figure 3, presence of ΔCD20 mRNA and protein was detected on both cell fractions.
ΔCD20 alternative transcripts are the source of the ΔCD20 protein with an abnormal intracellular compartmentalization
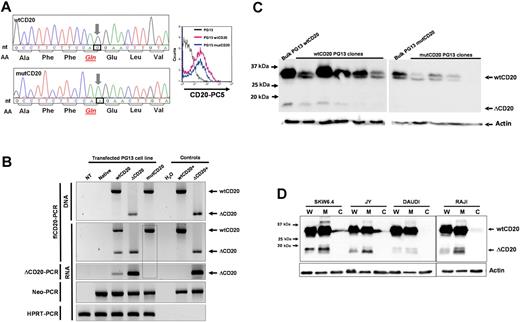
To confirm that the splice signals (donor site [DS] and AS) are the source of the ΔCD20 mRNA and protein, we modified the nucleotide sequence to delete one of these sites, the AS site, codon 612, from CAG (Gln) to CAA (Gln). Fluorescence-activated cell sorter (FACS) analysis indicated that this new mutated protein is expressed and addressed at the membrane (Figure 4A). RT-PCR analysis, with fl- or ΔCD20-PCR assays, confirmed that a packaging cell line transfected with retroviral vector carrying mutCD20 did not generate splice ΔCD20 mRNA (Figure 4B). Confocal analysis of mutCD20/GFP fusion–transfected 293T cells revealed that the mutCD20 protein was expressed and recognized by both anti-CD20 antibodies (Figure 3A). We did not detect a signal at the expected 15- to 17-kDa size length using different packaging of PG13 clones and Western blot analysis with the C-term anti-CD20 antibody that recognizes the ΔCD20 protein, confirming the absence of translation of splice ΔCD20 protein. We did, however, still detect the truncated protein within wtCD20-transfected PG13 clones (Figure 4C). Overall, the mutagenesis results with the splice AS sequence confirmed that the splice signal is the source of the ΔCD20 mRNA and protein.
ΔCD20 alternative transcripts code for an intracellular ΔCD20 protein. (A) Site-directed mutagenesis was performed on the wtCD20 sequence, using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene), according to manufacturer recommendations to kill the AS, with respect to the amino acid sequence. Left panel shows electropherograms after site-directed mutagenesis, confirming that the third nucleotide of the CAG codon (Gln) is replaced by an A nucleotide. Right panel shows cytometry detection of the mutCD20 protein with a monoclonal anti-CD20 antibody at the cell surface of transfected PG13 cells. (B) flCD20- or ΔCD20-PCR on DNA or cDNA of transfected PG13 packaging cell line with wtCD20, ΔCD20, or mutCD20 retroviral plasmids. Neo-PCR was performed to control cell transfection and hypoxanthine-guanine-phosphoribosyl transferase (HPRT)–PCR to confirm the absence of inhibitors of the PCR reactions. flCD20 and ΔCD20 plasmids were used as positive controls. The dashed box highlights the absence of ΔCD20 PCR products, even with the ΔCD20-specific PCR, after transfection of PG13 with the mut-CD20 construct. (C) Western blot analysis with the C-term anti-CD20 on cell lysates from the bulk cell population or isolated cloned PG13 cells transfected with constructs carrying wtCD20 or mutCD20 cDNA sequences. Absence of detection at 15 to 17 kDa confirms that the splice sequence is the source of the ΔCD20 protein expression. (D) Western blot analysis on whole protein lysate (W) or subcellular fractions as cytoplasm (C) and membrane (M) for 4 different B-cell lines using the C-term CD20 antibody.
ΔCD20 alternative transcripts code for an intracellular ΔCD20 protein. (A) Site-directed mutagenesis was performed on the wtCD20 sequence, using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene), according to manufacturer recommendations to kill the AS, with respect to the amino acid sequence. Left panel shows electropherograms after site-directed mutagenesis, confirming that the third nucleotide of the CAG codon (Gln) is replaced by an A nucleotide. Right panel shows cytometry detection of the mutCD20 protein with a monoclonal anti-CD20 antibody at the cell surface of transfected PG13 cells. (B) flCD20- or ΔCD20-PCR on DNA or cDNA of transfected PG13 packaging cell line with wtCD20, ΔCD20, or mutCD20 retroviral plasmids. Neo-PCR was performed to control cell transfection and hypoxanthine-guanine-phosphoribosyl transferase (HPRT)–PCR to confirm the absence of inhibitors of the PCR reactions. flCD20 and ΔCD20 plasmids were used as positive controls. The dashed box highlights the absence of ΔCD20 PCR products, even with the ΔCD20-specific PCR, after transfection of PG13 with the mut-CD20 construct. (C) Western blot analysis with the C-term anti-CD20 on cell lysates from the bulk cell population or isolated cloned PG13 cells transfected with constructs carrying wtCD20 or mutCD20 cDNA sequences. Absence of detection at 15 to 17 kDa confirms that the splice sequence is the source of the ΔCD20 protein expression. (D) Western blot analysis on whole protein lysate (W) or subcellular fractions as cytoplasm (C) and membrane (M) for 4 different B-cell lines using the C-term CD20 antibody.
In silico translation of the in-frame ΔCD20 mRNA indicated that much of the 4 TM domains of the CD20 protein is deleted (5 amino acids remaining), signifying that the ΔCD20 may be a nonanchored protein. To address this issue, we performed Western blot analysis of different subcellular fractions of cytoplasm and membrane. As shown in Figure 4D for 4 B-cell lines, the ΔCD20 signal occurred in the membrane subfraction but not in the cytoplasm. Moreover, immunoprecipitation with an anti-wtCD20 antibody revealed by Western blot that the ΔCD20 protein coimmunoprecipitated, suggesting an association between ΔCD20 and wtCD20 protein (supplemental Figure 4).
The alternatively spliced ΔCD20 form is associated with resistance to RTX treatment
Although CD20 is the target of one of the most-used immunotherapy drugs in hematology, it is important to assess if the newly discovered ΔCD20 protein is involved in the response to treatment. To answer this question, we induced RTX resistance in Raji and Ramos B-cell lines by serial exposure to escalating doses of RTX (0.5-64 μg/mL). Resistance acquisition was confirmed by in vitro CDC lysis assay and assessment of cell-surface staining of CD20 by FACS analysis. Cell lysis in the presence of RTX plus complement was 96%, 60%, and 52%, respectively, for native Ramos, R2-2, and R2-64, whereas it was close to 10% in the absence of complement. CDC assay showed also that the RTX resistance is established at least for 21 days after the last RTX exposure (supplemental Table 1).
Western blot analysis using the anti–C-term CD20 region antibody revealed increased signal at 15 to 17 kDa, corresponding to the ΔCD20 protein (Figure 5B) in relation to RTX exposure and independently of the complement source, human or newborn serum. This finding was also confirmed, in addition to Burkitt (Ramos or Raji), in pre-B (ROS) cell lines (supplemental Figure 5), and also by quantitative analysis of the signal in which the protein ratio of ΔCD20/wtCD20 signals similarly increased (Figure 5C left; supplemental Figure 5). Quantitative experiments at the mRNA level, using RT-qPCR assay, showed that the level of ΔCD20 mRNA transcripts increased with RTX exposure, in line with the Western blot experiments (Figure 5C right). Moreover, the protein ratio of ΔCD20/wt CD20 as well as the percentage of ΔCD20 mRNA correlated with RTX (5 μg/mL) sensitivity (respectively, R2 = .9234 and R2 = .9688). Finally, ΔCD20 mRNA quantification was stable in different phases of the cell cycle (supplemental Figure 6). This important result might suggest that the portion of the B cells expressing ΔCD20 protein escaped RTX elimination.
RTX resistance and ΔCD20 protein expression. (A) CDC lysis assay reporting the percentage of lysis of in vitro RTX-resistant established B cells plotted against an increasing dose of RTX. Both experiments (decrease of CD20 mean fluorescence intensity [MFI] and decrease of percentage of cell lysis) favored RTX-resistance establishment after repeated RTX exposure. Error bars indicate SE of 3 experiments. (B) Representative (of 3 experiments) Western blot (WB) analysis, using the c-Term anti-CD20 (recognizing wt and truncated CD20 proteins) on whole lysates from RTX-resistant B-cell lines. Antiactin WBs on the whole-cell lysates were performed as controls. Dashed boxes highlight an increase of immunoreactive signal generated by ΔCD20 protein. (C) Quantification of the WB immunoreactive ΔCD20 signal with the BIO-1D advanced software and normalized with actin signal. Results are reported as the protein ratio of ΔCD20/wtCD20 (left). RT-qPCR quantification of ΔCD20 transcripts expressed as follows: relative percentage of ΔCD20 = (ΔCD20 / wtCD20 + ΔCD20) × 100. Mean of triplicate is reported (right panel).
RTX resistance and ΔCD20 protein expression. (A) CDC lysis assay reporting the percentage of lysis of in vitro RTX-resistant established B cells plotted against an increasing dose of RTX. Both experiments (decrease of CD20 mean fluorescence intensity [MFI] and decrease of percentage of cell lysis) favored RTX-resistance establishment after repeated RTX exposure. Error bars indicate SE of 3 experiments. (B) Representative (of 3 experiments) Western blot (WB) analysis, using the c-Term anti-CD20 (recognizing wt and truncated CD20 proteins) on whole lysates from RTX-resistant B-cell lines. Antiactin WBs on the whole-cell lysates were performed as controls. Dashed boxes highlight an increase of immunoreactive signal generated by ΔCD20 protein. (C) Quantification of the WB immunoreactive ΔCD20 signal with the BIO-1D advanced software and normalized with actin signal. Results are reported as the protein ratio of ΔCD20/wtCD20 (left). RT-qPCR quantification of ΔCD20 transcripts expressed as follows: relative percentage of ΔCD20 = (ΔCD20 / wtCD20 + ΔCD20) × 100. Mean of triplicate is reported (right panel).
ΔCD20 mRNA is found in activated B cells and in different human B-cell malignancies
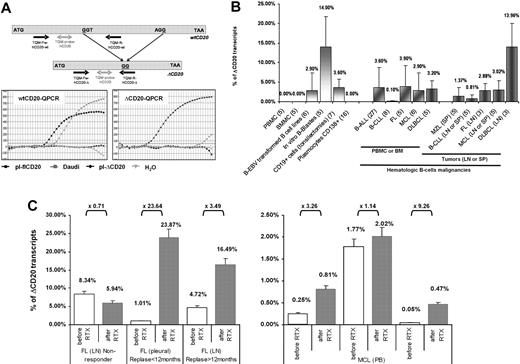
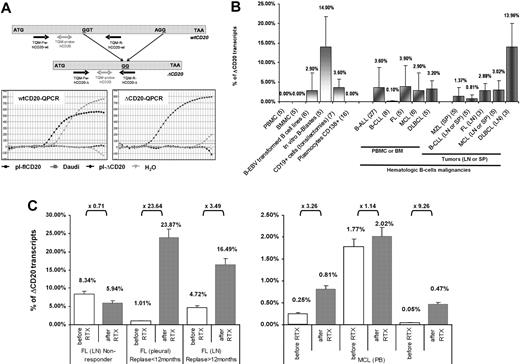
To quantify the spliced form of CD20 in different B-cell malignancies, we designed an RT-qPCR assay allowing specific quantification of both flCD20 and ΔCD20 transcripts, assessed using respective reverse primers complementary to the deleted area or spanning the splicing junction (Figure 6A). A cross-amplification experiment was performed from Daudi cDNA and plasmids carrying each form of transcript. flCD20-qPCR detected PCR product from Daudi (Ct = 18) and flCD20 plasmid (Ct = 12.7) but not from ΔCD20 plasmid, while ΔCD20 qPCR gave an amplification signal from Daudi (Ct = 34) and ΔCD20-plasmid (Ct = 11.8), but not from flCD20 plasmid. These results confirmed the specificity of both assays and showed that the ΔCD20 form is less abundant that the flCD20 (Ctwt = 18 vs CtΔ = 34 in Daudi). Both assays have the same sensitivity of detection of one copy of target CD20 among one equivalent genome.
Quantification of the spliced ΔCD20 mRNA in activated B cells and hematologic malignancies, and illustration of clinical relevance through 2 cases. (A) Design of the RT-qPCR with schematic localization of primers and bifluorescent FAM/TAMRA TaqMan probe. Both PCRs (ΔCD20- and wtCD20-specific) were performed using an iCycler thermocycler (Bio-Rad) under standard TaqMan PCR conditions with the TaqMan Universal PCR Master Mix (Applied Biosystems). Copy number of both forms of CD20 mRNA was assessed by comparison against serial plasmid dilutions carrying either the flCD20 or the ΔCD20 cloned cDNA. Representative qPCR curves confirming absence of cross-amplification between wtCD20- and ΔCD20-qPCR allowing, respectively, detection of both wtCD20 and ΔCD20mRNA. (B) ΔCD20 mRNA quantification in normal PBMCs or BMMCs from healthy donors as well as in in vitro EBV-transformed B-cell lines, in vitro–generated B blasts, or in CD19+ cell-sorted cells from tonsillectomy samples and CD138+ plasmocytes from multiple myeloma. Quantification of ΔCD20 mRNA, performed in duplicate, in different normal (PBMCs, BMMCs) or hematologic B malignancies or tumor samples from lymph node (LN), spleen (SP), or pleural effusion (PL) was also reported. Number of cases analyzed for each normal or neoplasia cases are given in brackets in the sample's names of the x-axis. (C) RT-qPCR of CD20 transcripts on pre-rituximab (sensitive) and post-rituximab (resistant) primary cells. Left panel shows human samples (LN or PL) of patients (n = 3) with FL and treated with 3 or 4 courses of RTX. Among these 3 representative cases, 1 is a nonresponse to RTX, whereas the other 2 are early (< 12 months) or late (> 12 months) relapses. Right panel shows PB quantification on 3 patients with MCL (n = 3) from a clinical trial of the French GOELAMS group and treated with 4 courses of RTX (375 mg/m2 of RTX). Fold changes (× FC) are indicated. ΔCD20 transcript quantification is reported as relative percentage of ΔCD20: R = (ΔCD20 / wtCD20 + ΔCD20) × 100. Error bars in panels B and C represent SE of RT-qPCR replicates.
Quantification of the spliced ΔCD20 mRNA in activated B cells and hematologic malignancies, and illustration of clinical relevance through 2 cases. (A) Design of the RT-qPCR with schematic localization of primers and bifluorescent FAM/TAMRA TaqMan probe. Both PCRs (ΔCD20- and wtCD20-specific) were performed using an iCycler thermocycler (Bio-Rad) under standard TaqMan PCR conditions with the TaqMan Universal PCR Master Mix (Applied Biosystems). Copy number of both forms of CD20 mRNA was assessed by comparison against serial plasmid dilutions carrying either the flCD20 or the ΔCD20 cloned cDNA. Representative qPCR curves confirming absence of cross-amplification between wtCD20- and ΔCD20-qPCR allowing, respectively, detection of both wtCD20 and ΔCD20mRNA. (B) ΔCD20 mRNA quantification in normal PBMCs or BMMCs from healthy donors as well as in in vitro EBV-transformed B-cell lines, in vitro–generated B blasts, or in CD19+ cell-sorted cells from tonsillectomy samples and CD138+ plasmocytes from multiple myeloma. Quantification of ΔCD20 mRNA, performed in duplicate, in different normal (PBMCs, BMMCs) or hematologic B malignancies or tumor samples from lymph node (LN), spleen (SP), or pleural effusion (PL) was also reported. Number of cases analyzed for each normal or neoplasia cases are given in brackets in the sample's names of the x-axis. (C) RT-qPCR of CD20 transcripts on pre-rituximab (sensitive) and post-rituximab (resistant) primary cells. Left panel shows human samples (LN or PL) of patients (n = 3) with FL and treated with 3 or 4 courses of RTX. Among these 3 representative cases, 1 is a nonresponse to RTX, whereas the other 2 are early (< 12 months) or late (> 12 months) relapses. Right panel shows PB quantification on 3 patients with MCL (n = 3) from a clinical trial of the French GOELAMS group and treated with 4 courses of RTX (375 mg/m2 of RTX). Fold changes (× FC) are indicated. ΔCD20 transcript quantification is reported as relative percentage of ΔCD20: R = (ΔCD20 / wtCD20 + ΔCD20) × 100. Error bars in panels B and C represent SE of RT-qPCR replicates.
With this RT-qPCR assay, we quantified the ΔCD20 spliced form (expressed as relative percentage of total CD20 mRNA: R = (ΔCD20/wtCD20 + ΔCD20) × 100) in in vitro EBV-transformed B-cell lines (2.9% ± 4.51%; n = 6) as well as in CD19+-sorted cells from tonsillectomy samples (9% ± 2.2%; n = 7), in vitro B blast cells (14% ± 7.8%; n = 5; Figure 6B) or Pokweed-activated B cells (2.71% ± 1.48%; n = 2), without evidence of correlation with percentage of activated B cells (supplemental Figure 7). Interestingly, screening of a panel of B-cell hematologic malignancies in PB or BM showed that the spliced form is detectable at various levels. We found a mean of 3.6% (± 5.1%) in B-ALL (n = 27); 3.9% (± 5.3%) in FL (n = 5); 2.9% (± 4.5%) in MCL (n = 6); 3.2% (± 2.2%) in DLBCL (n = 5); and 0.1% (± 0.2%) in B-CLL (n = 8). In diagnosis tumor samples (lymph nodes, spleen, or pleural effusion), quantification of the splice form of CD20mRNA (mean ± SE) showed, compared with BM or PB, a similar level as 2.8% (± 1.7%) in FL (n = 3), 3% (± 2.3%) in MCL (n = 5), 1.3% (± 2.3%) in MZL (n = 5), and 0.81% (± 0.8%) in CLL (n = 5), whereas there was a higher level in DLBCL (13.9% ± 5.9%; n = 3; Figure 6B). Finally, RT-qPCR confirmed qualitative screening in which the spliced form was not detected in PBMCs (n = 5) or BMMCs (n = 5) from healthy donors as well as in CD138+ plasmocytes from multiple myeloma (n = 16; Figure 6B).
In addition, we quantified ΔCD20 mRNA on pre-RTX (sensitive) and post-RTX (resistant) primary cells from patients with FL (n = 3) or MCL (n = 3). We observed an increase of ΔCD20 mRNA in all cases of RTX escape (FC of ×1.14 to ×23.64; median, ×3.38) except for the case of FL that did not initially respond to RTX treatment and where the ΔCD20 mRNA levels on post-RTX sample biopsies remain similar or slightly lower to the pre-RTX sample (FC [FC] = ×0.71; Figure 6C).
Discussion
We describe here a new splice CD20 variant (ΔCD20) that differs from 2 previously reported variants and also from those identified in 2 genome-wide analyses describing different MS4A variants and their putative translated proteins.2,36 This novel ΔCD20 mRNA is expressed in-frame from the ATG start codon within exon 3 and fuses part of exon 3 to exon 7, leading to a coded protein lacking the extracellular domain (including the RTX epitope sequence35 ) and much of the 4 TM-spanning domains.
We also provide here, for the first time, substantial evidence that this spliced ΔCD20 mRNA variant is naturally translated into a protein. Confocal microscopy and Western blot results on B-cell lines, using a C-term CD20 antibody, showed that in-frame mRNA ΔCD20 transcript codes for a putative protein of 15 to 17 kDa. Second, the 15- to 17-kDa signal band was consequently increased after transduction of the Raji cell line with a retroviral vector carrying the ΔCD20 cDNA sequence. We also demonstrated, with site-directed mutagenesis of the AS that results in an absence of splicing, that the CD20-spliced mRNA is the source of this new ΔCD20 protein. Interestingly, this mutated/corrected CD20 cDNA sequence may also be an interesting tool for gene suicide therapy using gene-modified T cells to modulate alloreactivity after BM graft.25 To our knowledge, these results are the first demonstration that the ΔCD20 exists.
As shown after Raji cell transduction by a retroviral vector carrying the ΔCD20 cDNA, while ΔCD20 protein expression increased, wtCD20 protein expression decreased, suggesting a relation between the two. Moreover, Western blot on subcellular fractions showed that the ΔCD20 protein is harvested within the membrane compartment. In addition, anti-CD20 immunoprecipitation with an anti-CD20 antibody recognizing only the wtCD20 protein indicated an association of both proteins, ΔCD20/wtCD20, rather than an anchor by the remnant of the TM domains (5 amino acids).
A careful review of previous studies identifies reports of lower additional bands through Western blot using an anti-CD20 antibody, but none of these studies addressed these additional smaller molecular species as the product of the translation of a potential alternative ΔCD20 transcript. Czuczman et al20 assigned this additional lower band to the light chains of IgM, while Kennedy et al,37 in the case of a patient with CLL treated with RTX, concluded that this lower band may have corresponded to a partial digestion product of the CD20 protein.20
Although CD20 is the cell-surface target of the best-known immunotherapy (RTX) for treating B-cell malignancies or various autoimmune diseases, its function remains unclear. RTX is administered, with or without chemotherapy, for various CD20+ B-cell lymphoproliferative disorders. Despite the known efficiency of RTX, some patients with NHL exhibit poor or no clinical response to RTX monotherapy,38 and repeated exposure leads to relapse/resistance to RTX therapy.
Many factors influence RTX response at the protein level.16,19,20,23,33 One interesting and important finding in the current work is that RTX resistance leads to increased ΔCD20 signal, suggesting that some CD20-expressing B cells persist despite RTX treatment. Many examples of gene-expression modulation by an associated mRNA variant have been reported in B cells.39,40 As shown here, after Western blot on the ΔCD20-transduced Raji cell line, ΔCD20 protein seemed to modulate wtCD20 expression, thus leading to reduced wtCD20 expression as evaluated on patient samples in DLBCL and correlated with inferior survival.41 The ΔCD20 protein may also interact with the wtCD20 protein and modulate CD20 reorganization within the lipid raft and contribute to the development of RTX resistance. Moreover, detection by Western blotting of 2 forms of the ΔCD20 protein may correspond to different states of phosphorylation, as previously described for the wtCD20 protein;5,23 this finding suggests that kinases and phosphatases may regulate the ΔCD20 protein. The fact that splicing does not affect remaining putative serine/threonine phosphorylation domains42 in the ΔCD20 protein supports this hypothesis. Furthermore, persistence of phosphorylation sites after splicing and thus potential activation of ΔCD20 protein through protein kinases make this molecule an indirect potential target for kinase inhibitors to improve treatment.
Molecular events have also been associated with response modulation or RTX resistance.21,41,43,44 More recently, deletions/mutations were described within the C-term region of the CD20,21 but no previous study identified the presence of this alternative ΔCD20 transcript. Thus, we report here another possible phenomenon of RTX resistance associated with CD20 gene splicing and ΔCD20 expression.
Finally, it is now well known that splice variants are differentially expressed in tumors,45 and some are used as cancer biomarkers.46 The initial phase of this study thus targeted investigation of hematologic B-cell malignancies for the presence or absence of the splice ΔCD20 alternative transcripts to assess its potential use in a molecular assay for monitoring disease. ΔCD20 transcripts were found and quantified at different levels in all screened malignancies, as well as in EBV-transformed B cells, in vitro B blasts, or B lymphocytes purified from tonsillectomies, but was not detected in PBMCs, BMMCs, plasmocytes, or purified CD19+- or CD20+-sorted cells from healthy donors. All of these observations and a low-versus-high level found respectively in chronic or acute diseases (CLL vs ALL) favor a relationship between the activation state of B cells and ΔCD20 presence rather than between a potential malignant transformation with ΔCD20 presence. Our designed RT-qPCR molecular tool, allowing ΔCD20 discrimination from the wtCD20 form, may help in investigations of B-cell lymphoproliferative disorders or in monitoring RTX treatment in cases of GVHD47 or refractory kidney transplant rejection.15,48 Our data of ΔCD20 mRNA quantification before (sensitive) and after (resistant) RTX treatment in FL and MCL clinical cases provide some interesting support for future investigation of both potential clinical approach and future evaluation in a larger cohort.
In addition to describing this new spliced CD20 transcript, there are many arguments that presence of the ΔCD20 protein related to RTX resistance constitutes a potential target for therapeutic issues49 to improve efficiency of standard RTX treatment.11 Splicing factors are differentially expressed in tumors,45 thus constituting an interesting focus for therapeutic studies. The splicing signal may be modulated upstream by aiming for proteins involved in the splicing event or downstream by targeting the splicing products themselves. In our work, preliminary data from western blots with an antibody targeting the key and central splice factor ASF/SF2 revealed an increase in this factor in RTX resistance, making it a candidate for future therapies in B-cell lymphoproliferative disorders.
In other ways, more specific targeting of splice variants products at the mRNA or protein levels may be assessed respectively by RNA interference50 or immunotherapy approaches. As reported, a clinical trial evaluation (EPIC Study)51 of peptide vaccination against a BCR-ABL e14a2 junctional peptide revealed the potential efficiency of this approach. Affinity-autoreactive cytotoxic T lymphocytes (CTLs) have already been described, especially against peptide CD19 or CD20 antigens.52 These different strategies could apply to the ΔCD20 protein. Based on our preliminary data for vaccination, in a transgenic murine model expressing human HLA, we have demonstrated that the splice junction area of the protein could be targeted to direct a specific CTL response, and that autoreactive CTLs against ΔCD20 peptide junctions exist in healthy people. These results support continued investigation of a vaccination immunotherapy approach or of a redirection of CD8+ T primary lymphocytes against ΔCD20 by T-cell receptor (TCR) transfer52 of our isolated anti-ΔCD20 T-cell clones. This potential should be evaluated in a murine model53 to assess method efficiency.
In conclusion, we have demonstrated that a new, spliced CD20 mRNA variant codes for a novel, modified intracellular/submembrane CD20 protein correlated with RTX resistance, which may be targeted by in vivo autoreactive CTLs after immunization for improving standard RTX treatments. This alternative transcript may also be an interesting diagnostic, prognostic, or predictive molecular marker for monitoring B-cell malignant diseases, a possibility that requires evaluation in murine models as well as in larger cohorts of patients treated by RTX.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Delphine Sauce-Larsen and Dr Martin Larsen for providing the B blasts. We also thank Prof Thierry Fest (Rennes) and Dr Laurence Lode (Nantes) for supplying purified CD19+ cells from tonsillectomies and CD138+ plasmocytes from multiple myeloma samples, respectively. We thank Prof Françoise Berger (Lyon), Prof Bernadette Kantelip (Besançon), and Martine Chauvet (Grenoble) for preparing and providing samples and helpful discussion, and we thank Katy Billot for participating in this study while earning her Master 1 and Anne Duperrier and Claire Latruffe for their technical support.
C.H. received a fellowship from the Ministère de la Recherche et de la Technologie. This work was also supported by Inserm, the Ligue Contre le Cancer, Comité du Doubs, The Fondation de Transplantation, the Etablissement Français du Sang (EFS) national grant, and the associations Centpoursanglavie and Capucine (grant no. 2-2008).
Authorship
Contribution: C.H., M.D., and C.G. executed the molecular study, cloned the cDNA, expressed the protein, performed the vector constructs and the functional studies, and wrote the original draft of the manuscript; C.H., M.D., and J.-R.P. performed the immunofluorescence confocal microscopy analysis; C.H. and J.-P.R.-M. performed the Western blot analysis; P.-S.R., M.C., A.T.-G., R.G., E.D., and G.S. provided the clinical samples of hematologic B-cell malignancies and participated to the discussion; P.S.-R., F.G.-O., E.R., P.T., and C.B. contributed to improving the manuscript and gave final approval on the manuscript; and C.F. initiated and designed the study, participated in every step of the study, managed the whole project, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christophe Ferrand, Laboratoire de Thérapeutiques Immuno-Moléculaires et Cellulaires des Cancers, Inserm UMR645/IFR133, Etablissement Français du Sang–Bourgogne/Franche-Comté, 1 Blvd Alexandre Fleming, 25020 Besançon cedex, France; e-mail: christophe.ferrand@efs.sante.fr.
References
Author notes
C.H. and M.D. contributed equally to this work.

![Figure 5. RTX resistance and ΔCD20 protein expression. (A) CDC lysis assay reporting the percentage of lysis of in vitro RTX-resistant established B cells plotted against an increasing dose of RTX. Both experiments (decrease of CD20 mean fluorescence intensity [MFI] and decrease of percentage of cell lysis) favored RTX-resistance establishment after repeated RTX exposure. Error bars indicate SE of 3 experiments. (B) Representative (of 3 experiments) Western blot (WB) analysis, using the c-Term anti-CD20 (recognizing wt and truncated CD20 proteins) on whole lysates from RTX-resistant B-cell lines. Antiactin WBs on the whole-cell lysates were performed as controls. Dashed boxes highlight an increase of immunoreactive signal generated by ΔCD20 protein. (C) Quantification of the WB immunoreactive ΔCD20 signal with the BIO-1D advanced software and normalized with actin signal. Results are reported as the protein ratio of ΔCD20/wtCD20 (left). RT-qPCR quantification of ΔCD20 transcripts expressed as follows: relative percentage of ΔCD20 = (ΔCD20 / wtCD20 + ΔCD20) × 100. Mean of triplicate is reported (right panel).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/12/10.1182_blood-2009-06-229112/5/m_zh89991050010005.jpeg?Expires=1769130750&Signature=o86PSQrnNJ3NfbE8cIASkXycwS~q16i6~82hQkW8wPKriFnLF~Y5hQGBUwB5j0Tv92ruc4Q-VJqnPEBP~R6eykVlF1byp24BFggHtj0Nk1da30ApptF2soeGXhueFu8Mzwp-iUFkW~bbRi3Q-T7TtazbNCKrBPOzSEpdXyFx2aHf-ZwHqbEinBGPn7sKiI6X2Iw-ZO90i~3d~hczHV7YTbG~WD6nXTyLJKcBAFmRkKCKzci561ZQM-jkZEKTua~lUeueayVLZ6WWIqKV2Dc02mKTGGV9YzGoXlhMOZdV3lzfDq-kGOkvteZdMjCkjBiiW847KHQ-7xGU4RT45lIIrg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 5. RTX resistance and ΔCD20 protein expression. (A) CDC lysis assay reporting the percentage of lysis of in vitro RTX-resistant established B cells plotted against an increasing dose of RTX. Both experiments (decrease of CD20 mean fluorescence intensity [MFI] and decrease of percentage of cell lysis) favored RTX-resistance establishment after repeated RTX exposure. Error bars indicate SE of 3 experiments. (B) Representative (of 3 experiments) Western blot (WB) analysis, using the c-Term anti-CD20 (recognizing wt and truncated CD20 proteins) on whole lysates from RTX-resistant B-cell lines. Antiactin WBs on the whole-cell lysates were performed as controls. Dashed boxes highlight an increase of immunoreactive signal generated by ΔCD20 protein. (C) Quantification of the WB immunoreactive ΔCD20 signal with the BIO-1D advanced software and normalized with actin signal. Results are reported as the protein ratio of ΔCD20/wtCD20 (left). RT-qPCR quantification of ΔCD20 transcripts expressed as follows: relative percentage of ΔCD20 = (ΔCD20 / wtCD20 + ΔCD20) × 100. Mean of triplicate is reported (right panel).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/12/10.1182_blood-2009-06-229112/5/m_zh89991050010005.jpeg?Expires=1769524208&Signature=K~g0C~HA07GTUpCXo1tdnPE2kY32yemSQ2iZaP3K6n44Fn99G81aPk7Vu18fsKpwig~gS4Ttb~DPWN4gp3JN4v3juMlyYG~jzZETRi-~93H5MmpT7AObFmS3fCi97R8YBd62NcTxfTfzlYSIdZTylLK3JriWk-ruu63rcPVQqmsRbgS9tgC~TkYqZltWH3t5Dyhdja0A~Y9yqrAO6oRF2s~TfwB6KtjnfNq5HsMAUfT~l5eMQt7UYTkznCCMGhQnTtpCiAtMDTRvCVrzr0wBV7NZhzrmVR7ZiV4N4E1YWz-YAUAGqhkJ8Eyu354mLCN-hdmKXiSIP0Dx3i4IEZeQyg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)