Abstract
PAX5 is the main target of somatic mutations in acute B lymphoblastic leukemia (B-ALL). We analyzed 153 adult and child B-ALL harboring karyotypic abnormalities at chromosome 9p, to determine the frequency and the nature of PAX5 alterations. We found PAX5 internal rearrangements in 21% of the cases. To isolate fusion partners, we used classic and innovative techniques (rolling circle amplification-rapid amplification of cDNA ends) and single nucleotide polymorphism-comparative genomic hybridization arrays. Recurrent and novel fusion partners were identified, including NCoR1, DACH2, GOLGA6, and TAOK1 genes showing the high variability of the partners. We noted that half the fusion genes can give rise to truncated PAX5 proteins. Furthermore, malignant cells carrying PAX5 fusion genes displayed a simple karyotype. These data strongly suggest that PAX5 fusion genes are early players in leukemogenesis. In addition, PAX5 deletion was observed in 60% of B-ALL with 9p alterations. Contrary to cases with PAX5 fusions, deletions were associated with complex karyotypes and common recurrent translocations. This supports the hypothesis of the secondary nature of the deletion. Our data shed more light on the high variability of PAX5 alterations in B-ALL. Therefore, it is probable that gene fusions occur early, whereas deletions should be regarded as a late/secondary event.
Introduction
Precursor B-cell acute lymphoblastic leukemia (B-ALL) is characterized both by a blockage of B-cell differentiation and uncontrolled proliferation of blastic cells. Adult and childhood B-ALL differs in terms of prognosis.1 Although 80% of children with ALL can be cured, only 30% of adults achieve long-term disease-free survival.1 These discrepancies in prognosis correlate with different occurrences of chromosomal abnormalities. Besides frequent recurrent translocations, such as t(9;22)(q34;q11), t(12;21)(p13;q22), t(1;19)(q23;p13), or MLL translocation, new oncogenes have been identified recently. Among them, the paired box domain gene 5 (PAX5), located on 9p13, has been reported as being frequently mutated in both childhood2 and adult B-ALL.3
PAX5 encodes a paired box domain (PBD) transcription factor considered as the guardian of B-cell identity. Its homozygous deletion in the mouse leads to total blockage of B-cell differentiation at the pro-B stage.4 Furthermore, Pax5 inactivation at later stages of differentiation entails transdifferentiation or dedifferentiation of B cells.5 Pax5, which activates crucial genes for B-cell lineage differentiation and represses genes important for commitment in other hematopoietic lineages, is expressed from early pro-B stage until final plasmacytic differentiation, where it is turned off.6,7
Fusion genes involving PAX5 have been associated with blockage of B-cell differentiation, the first reported example being PAX5-ETV6, the product of the dic(9;12)(p13;p13) rearrangement.8 A recent study has shown that PAX5 is rearranged in 2.6% of pediatric B-ALL cases,9 being fused, to date, with 17 different partners, thus demonstrating the variability of the fusion partners. In contrast, very few studies have been performed on adult B-ALL, only reporting fusion between PAX5 and ETV610 and the elastin gene (ELN).11 Cytogenetic abnormalities affecting the chromosome 9p arm and potentially involving PAX5 occur in 10% of child B-ALL.12 Using classic cytogenetic techniques and a newly developed molecular strategy (rolling circle amplification-rapid amplification of cDNA ends [RCA-RACE]), we investigated 9p abnormalities, focusing especially on the PAX5 locus. For this purpose, we collected 153 childhood and adult B-ALL with 9p rearrangements diagnosed and reviewed by members of the Groupe Francophone de Cytogénétique Hématologique (GFCH). Our data revealed that PAX5 deletions accounted for most of the 9p alterations and were often associated with complex karyotypes. On the contrary, PAX5 internal rearrangements were less frequent and were most of the time the sole chromosomal abnormality detected. This collaborative study also allowed the description of novel PAX5-fusion partners.
Methods
Patients
The criterion for inclusion, in this retrospective study of the GFCH, was the detection by karyotype analysis of acquired structural 9p rearrangement in patients with B-ALL. We selected the structural abnormalities with the largest variety of 9p chromosome partners because the aim of the study was to find new fusion partners of PAX5.
This retrospective study included 153 B-ALL (92 males and 61 females) recruited from 1989 to 2008 from 20 French and 2 Belgian cytogenetic centers (“Morphologic and immunologic validation”). Among them, we examined 140 patients at diagnosis and 13 at relapse. This cohort was composed of 89 children (median age, 6 years; range, 1 month to 15 years, 52 males and 37 females) and 64 adults (median age, 43 years; range, 16-82 years, 40 males and 24 females). Patients distributed according to the EGIL classification13 of B-ALL subtypes were: B-I ALL (n = 6), B-II ALL (n = 92), B-III ALL (n = 48), and B-ALL unclassified (n = 7).
Morphologic and immunologic validation
Morphologic and immunophenotypic studies of patients were carried out in each center and validated by groups of morphologists and immunologists responsible for data collection in various working groups, particularly involved in therapeutic protocols for children (EORTC, FRALLE, and IGLALL) and adults (GOELAL, GRAALL, GRAAPH, LALA, and OMAN). Patient material was provided by participative centers (Centre Hospitalier Universitaire Toulouse, n = 42; Strasbourg, n = 16; Dijon, n = 11; Nantes, n = 11; Lyon, n = 9; Grenoble, n = 8; Rouen, n = 8; Bordeaux, n = 8; St Etienne, n = 7; Montpellier, n = 6; Ghent, Belgium, n = 4; Reims, n = 4; Angers, n = 3; Bobigny n = 3; Paris, Necker, n = 3; Tours, n = 3; Marseille IPC, n = 2; Marseille, Timone, n = 1; Laboratoire Biomnis, Lyon, n = 1; Louvain, Belgium, n = 1; Paris, St Antoine, n = 1; Versailles, n = 1).
Cytogenetics
Cytogenetic analysis was conducted on bone marrow in 139 cases, on peripheral blood in 12 cases, on the central nervous system in 1 case, and on lymph nodes in another. In this multicenter study, different cellular culture techniques were used. Short synchronized cultures (≤ 24 hours) were most frequently carried out. RHG or GTG banding techniques (or both) were applied and karyotypes described according to the International System for Human Cytogenetic Nomenclature 2005.14 All chromosomal data were reviewed by the members of GFCH during 2 successive workshops to sharpen karyotype interpretation.
FISH approach for the detection of PAX5 rearrangements
A first screening of all 9p rearrangements was performed with a commercial dual-color PAX5 probe (Dako Denmark). Direct involvement of PAX5 was verified using 2 overlapping bacterial artificial chromosome (BAC) clones: RP11-243F8 and RP11-344B23 and exon-specific fosmides G248P85962F9 (exon1) and G248P82268A1 (exon10; CHORI). Positioning of probes relative to PAX5 is depicted in Figure 1.
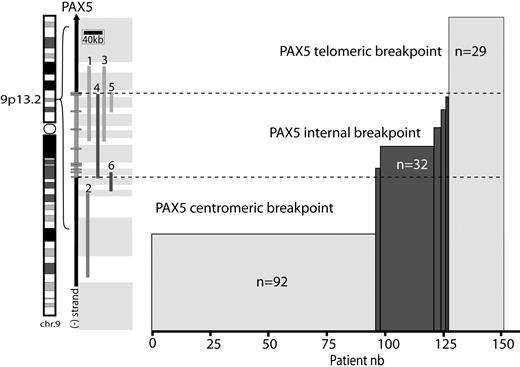
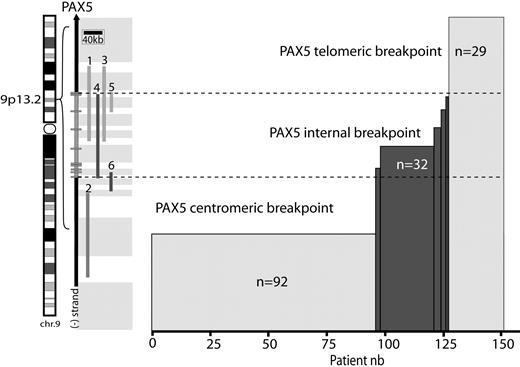
Repartition of breakpoints on chromosome 9p arm. Location of PAX5 on chromosome 9 (left). A schematic of PAX5 genomic structure (right); rectangles represent exons. (1-6) Probes used for FISH analyses: telomeric and centromeric Dako commercial probes (1,2), RP11-344B23 (3), and RP11-244F8 (4), BACs and G248P82268A1 (5), and G248P85962F9 (6) fosmids. These probes enable localization of the breakpoint regions (gray rectangles). Histogram represents the number of patients harboring these breakpoints.
Repartition of breakpoints on chromosome 9p arm. Location of PAX5 on chromosome 9 (left). A schematic of PAX5 genomic structure (right); rectangles represent exons. (1-6) Probes used for FISH analyses: telomeric and centromeric Dako commercial probes (1,2), RP11-344B23 (3), and RP11-244F8 (4), BACs and G248P82268A1 (5), and G248P85962F9 (6) fosmids. These probes enable localization of the breakpoint regions (gray rectangles). Histogram represents the number of patients harboring these breakpoints.
PCR approach for the detection of PAX5 fusion partners
Total RNA was extracted according to the TRIZOL method (Invitrogen) and 1 μg reverse-transcribed into cDNA by SuperScript III (Invitrogen) according to the manufacturer's instructions, using an oligo-dTanchor primer (RACE kit, second generation; Roche Applied Science). The cDNA product was used as a template in a polymerase chain reaction (PCR) experiment performed with the Advantage 2 PCR kit (BD Biosciences), a PCR anchor primer (5′-GACCACGCGTATCGATGTCGAC-3′), and the PAX5-specific primer PAX5-771F (5′-CCATGTTTGCCTGGGAGATCAGG-3′). The temperature cycling program used with a GeneAmp PCR system 9600 (PerkinElmer) was as follows: (1) 1 minute at 94°C; (2) 15 seconds at 94°C, 6 minutes at 68°C 35 times; and (3) 10 minutes at 68°C. A seminested PCR using an internal PAX5-846F (5′-GTTCCATCAACAGGATCATCCGG-3′) primer and the anchor primer was subsequently performed on 1 μL of the PCR product under the same cycling conditions. The obtained products were sequenced using the PAX5-846F primer and BigDye dideoxynucleotides. Products were separated on a 3130 XL sequencing apparatus (Applied Biosystems); electrophoregrams were analyzed using the Sequencher software (Version 4.1.2; Gene Codes Corporation). The presence of chimeric transcripts has been further verified by PCR using specific primers (PAX5-415F: 5′-CCCTGTCCATTCCATCAAGTCCTG-3′, DACH2-928R: 5′-TGGTTAGCAGGTGGTTCTGCTGC-3′, NCoR1-7605R: 5′-GAGATCCCTCTCCTGCACCCTG-3′, GOLGA6-2134R: 5′-ACGCAGGGTTTGCTGGGCAAGC-3′, ELN-2297R: 5′-ATGAGGTCGTGAGTCAGGGGTC-3′, JAK2-4020R: 5′-CACATCTTTGCTGGCTAGCATCATG-3′, FOXP1-2595R: 5′-CAATCTTCATTCTCGGGGTTGG-3′).
RCA-RACE
Gene-specific RCA was performed as previously described.15 Briefly, starting from 100 to 250 ng of total RNA, SuperScriptTM III reverse transcription was performed according to the manufacturer's instructions (Invitrogen) replacing the oligo-dT primer by a 5′ phosphate oligo-dT anchor primer (5′P-GACCACGCGTATCGATGTCGAC(T)16V-3′) at a final concentration of 2.5μM. Circularization of purified cDNA was obtained using CircLigase kit (Epicentre Biotechnologies), according to the manufacturer's instructions. RCA was carried out by mixing the purified circularized product with 5 μL of dNTPs (10 mM each; Promega), 1 μL of 10 mg/mL bovine serum albumin, 5 μL of 10 times ϕ29 DNA polymerase buffer, 10 U of ϕ29 DNA polymerase (New England Biolabs), 2 μL of 100μM PAX5-Exo5 primer (5′-CTTGTGATGTTGGCGAGA*A*C - 3′ (*phosphorothiotates chemical bounds, Eurogentec) and water up to 50 μL. This mixture was incubated for 21 hours at 30°C and then for 10 minutes at 65°C.
Amplification of 50 ng of RCA product was performed with the Advantage 2 PCR kit (BD Biosciences), using PAX5-771F and PAX5-570R (5′-ATCCTCTGGCGGACTACATCCG-3′) primers, according to the manufacturer's instructions. This mixture was incubated under the same cycling condition than for the RACE PCR experiment. If necessary, nested PCR was performed using 1 μL of previous PCR product, replacing PAX5-771F by PAX5-846F and PAX5-570R by PAX5-510R (5′-TTCACTCCTCCATGTCCTGTCC-3′) primers. For negative control, the cDNA was replaced by water. For non-B control, cDNA of Karpas cell line, which does not express PAX5, was used.
Purification of BAC DNA
BACs (chosen on the website www.genome.ucsc.edu) were amplified in 3 mL LB medium (MP Biomedicals), containing 12.5 μg/mL of chloramphenicol (Sigma-Aldrich) under constant shaking at 180 rpm for 12 hours at 37°C. Each purified BAC DNA was amplified by hyperbranched (H)–RCA: from 20 to 50 ng of BAC DNA was added to 1 μL of exo-resistant random heptamers (Fermentas), 1 μL of annealing buffer (200mM Tris-HCl, pH 8, 50mM MgCl2), and water up to 5 μL. This mixture was incubated for 4 minutes at 94°C on a 2720 Thermal Cycler (Applied Biosystems) and cooled on ice for 2 minutes. Then was added 2 μL of ϕ29 10 times Buffer (New England Biolabs), 1 μL of dNTPs 10mM each (Promega), 7 U of ϕ29 DNA polymerase (New England Biolabs), 0.5 μL of 10 mg/mL bovine serum albumin (New England Biolabs), and water up to 20 μL. This mixture was incubated under the condition described above for gene specific RCA.
Labeling of BAC DNA
BAC DNA was labeled using the nick-translation kit (GE Healthcare) according to the manufacturer's instructions, with dUTP-fluoroscein or dUTP-tetramethylrhodamine (Roche Applied Science). Overnight ethanol precipitation of the labeled DNA was performed in the presence of 0.1 mg of Human Cot-1 DNA (Invitrogen) and 0.2 mg of salmon testis DNA (Sigma-Aldrich). Labeled BAC DNA was then pelleted, dried, and resuspended in 100 μL of a mixture of 50% formamide (Sigma-Aldrich), 2 times SSC (Eurobio), and 10% dextran sulfate (Sigma-Aldrich). BACs are detailed in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
CGH array
Genomic DNA was extracted with DNeasy (QIAGEN) from blood or bone marrow samples. Samples were analyzed for copy number changes using Affymetrix Genome-Wide Human SNP 6.0 arrays (Affymetrix). Sample preparation and hybridizations were performed using Genome-wide Human SNP Nsp/Sty assay kit (Affymetrix) according to manufacturer's protocol. Analysis of copy number state was done using BRLMM-P-Plus algorithm with regional GC correction, embedded in Genotyping Console 2.0 software (Affymetrix).
PAX5 vectors
Complete coding sequences of wtPAX5, trPAX5, and PAX5-NCoR1 were amplified from cDNA of patient #h125, #108, and # 101, respectively, using PfuUltraII Hotstart PCR Master Mix (Stratagene) with primers PAX5-forward: ATGGATTTAGAGAAAAATTATCCG and PAX5-reverse: ATGGGCTCTCTGGCTATCTTCAGG for wtPAX5; PAX5-forward and trPAX5-reverse: AGTCTCACCCTGCTGCCTGTCTG for trPAX5; and PAX5-forward and NCoR1-reverse: GAGATCCCTCTCCTGCACCCTG for PAX5-NCoR1. PCR products were cloned into pcDNA3.1 (+) expression vector (Invitrogen)
Luciferase assay
Experiments had been performed as previously described11 replacing pRenilla-CMV by pSV-β-galactosidase (Promega) as transfection efficiency reporter plasmid. HeLa cells (in 6-well plates) were transfected with 1.5 μg luc-CD19,16 1.5 μg pSV-β-galactosidase, 0.3 μg pcDNA3-PAX5, and increasing amounts of pcDNA3-truncated PAX5 or pcDNA3-PAX5-NCoR1 (0.1-0.9 μg). Luciferase and β-galactosidase activity were revealed on a Mithras LB940 (Berthold Technologies) using Luciferase Reporter assay system and β-galactosidase enzyme assay system, respectively (Promega). Luminescence and absorbance were read by a Mithras LB940 (Berthold Technologies).
Firefly luciferase activity was normalized to the measured β-galactosidase activities and are shown as average values relative to the basal activity observed with pcDNA3 alone (mean ± SD); statistical significance of normalized luciferase activity was assessed by Wilcoxon Mann-Whitney U test for each point compared with wtPAX5 luciferase activity (P < .01, n = 3).
Results
Cytogenetic characteristics of the cohort
The criterion for inclusion in this retrospective study was the detection on karyotype of acquired structural 9p rearrangement in B-ALL patients. Karyotypes were mostly pseudodiploid (53% children and 52% adults). A total of 32% had a modal number of 45 chromosomes (31% children and 33% adults). Hypodiploid karyotypes with 44 chromosomes were rare, only found in 2 cases. In the hyperdiploid group, modal numbers ranged from 47 to 55. Only 2 cases (1%) with a high hyperdiploid karyotype (> 50 chromosomes) were detected. Details of karyotypes are given in supplemental Table 2.
Karyotype and fluorescence in situ hybridization (FISH) analyses revealed that 14.5% of the patients harbored a t(9;22)(q34;q11)/BCR-ABL1 (4.5% in children, 28% in adults), 6.5% a t(1;19)(q23;p13)/TCF3-PBX1 (9% in children, 3% in adults), 9 pediatric cases a t(12;21)(p13;q22)/ETV6-RUNX1 (10%), 2 cases a 11q23/MLL translocation (2% children), and 3.5% a 14q32/IGH translocation (2% children and 5% adults).
To assess the distribution and the combination of the most frequent abnormalities associated with the 9p alteration, we reported all the chromosomal changes in Table 1, supplemental Table 3, and supplemental Figure 1. The most frequent abnormalities were in decreasing order: 20q deletion, 12p deletion, 9q translocation different from 9q34/ABL1, 13q abnormalities (deletion or translocation), 6q deletion, trisomy 8 or 8q gain, 7p deletion, 11q abnormalities different from 11q23/MLL, 7q translocation, 17p deletion, 5q translocation, and high hyperdiploidy.
Complex karyotypes (≥ 3 unrelated chromosomal abnormalities) were frequent, found in 48% (44% children and 54% adults, P = .3).
However, the chromosomal abnormalities that rendered these karyotypes complex differed in the 2 age groups. In children, complexity was mainly the result of the contribution of known recurrent changes in B-ALL (12p-, 20q-, 6q-, etc), whereas in adults, complexity mainly resulted from nonrecurrent changes indicating a higher heterogeneity of genome alterations (Table 1; supplemental Table 3; supplemental Figure 1).
The 9p abnormalities found in our cohort were translocations (resulting in 9p deletions when unbalanced), dicentrics, and isochromosome 9q leading to partial or total monosomy 9p, inversions, insertions, and simple deletions. Only a small number of cases of simple deletions were included.
Various PAX5 alterations and their association with karyotypic features
All patients were screened with a commercial dual-color PAX5 probe (Figure 1). When the 2 colors were separated, indicating a possible PAX5 internal rearrangement, we performed further analyses to pinpoint the location of the breakpoint (Figure 1).
Because our study was based on cytogenetic methods, the submicroscopic intragenic alterations of PAX5 (limited to a few exons) were not investigated.
PAX5 was not altered (telomeric breakpoint) in 29 cases (19%; 21.5% children and 16% adults, P = .4, male/female ratio = 1.2). Complex karyotypes were frequent (54.5%), and the 2 cases of high hyperdiploid karyotypes belonged to this group (Table 1). Only 2 patients had a partial loss of the PAX5 probe signal. Of the 27 remaining patients, 18 harbored unbalanced 9p rearrangement on the karyotype without losing the PAX5 probe signal.
Centromeric breakpoints were observed in 92 patients (60%, 54% children and 69% adults, P = .068, male/female ratio = 1.5). This 9p rearrangement generated a loss of PAX5 in 86 patients (93%). The entire PAX5 locus was translocated onto another chromosome in all other cases. The majority of recurrent fusions found in our cohort belonged to this group: 91% of BCR-ABL1 (20 of 22), 90% of TCF3-PBX1 (9 of 10), 80% of IGH translocation (4 of 5), 56% of ETV6-RUNX1 (5 of 9), and 50% of MLL translocation (1 of 2). In addition, this group composed 60% of the complex karyotypes (n = 30 of 50; Table 1; supplemental Table 3).
In addition, we detected a breakpoint localized inside the genomic region of PAX5 resulting in an internal rearrangement of PAX5 in 32 patients (25% children and 16% adults, P = .23, male/female ratio = 1.9). In contrast with the telomeric or centromeric breakpoint cases, a majority of these patients had a simple karyotype (73%, 22 of 30; P = .024). Only 2 patients had additional recurrent rearrangement: one case of t(10;14)(p12;q32) implicating the IGH locus and another case with MLL/MLLT3 fusion. Notably, no other recurrent fusion (BCR-ABL1, TCF3-PBX1, and ETV6-RUNX1) was detected in this group.
PAX5 internal rearrangements showed heterogeneous patterns of breakpoints (Figure 1; supplemental Table 2). The majority of the patients (n = 24) harbored PAX5 breakpoints located between exon 4, corresponding to the end of the DNA binding domain, and exon 6, coding the homeodomain. This breakpoint position was more frequent in children (19 of 89, 21%) than in adults (5 of 64 cases, 8%; P = .025). Furthermore, the PAX5 telomeric probe signal was deleted for 22 of 24 patients (92%). Among them, 21 of 24 (88%) appeared unbalanced on the karyotype (mainly dicentric and derivative chromosomes), the 3 others had a partial deletion. Three patients (2 adults and 1 male child) harbored breakpoints located in intron 7/intron 8. These patients showed a simple pseudodiploid karyotype and had a balanced rearrangement. Two patients (1 adult and 1 child) harbored a balanced translocation with PAX5 breakpoint located before the third exon, just before the DNA binding domain. The last 3 cases (2 adult females and 1 female child) showed an unbalanced translocation with breakpoints located in the last 2 exons of PAX5.
These results showed that PAX5 internal rearrangements were not associated with the most common B-ALL recurrent translocations, as these were, most of the time, the sole chromosomal abnormality. On the other hand, PAX5 deletion was associated with complex karyotypes and classic recurrent translocations.
Frequent alterations of PAX5 in dicentric chromosomes involving the 9p arm
Childhood and adult cohort presented the same frequency of dicentric rearrangements in 26 of 89 (29%) and 14 of 64 (22%), respectively, P = .18. The majority (23 of 40, 58%) resulted in monoallelic deletion of PAX5 [10 dic,(9;20) 4 dic,(9;12) 5 dic,(7;9) 1 dic, (9;16) 1 dic, (9;17) the dic(8;9), and the dic(9;15)]. Only 5 patients did not delete PAX5 [3 dic, (9;20) the idic(9;9), and the dic(9;13)], and 12 patients had a PAX5 intragenic breakpoint associated with partial deletion [4 dic,(9;20) 3 dic, (9;12) 2 dic(9;16), and 2 dic(9;17)] or without deletion of the gene [1 dic(7;9)]. Interestingly, dicentric cases with internal PAX5 breakpoint were much more frequent in children than in adults (11 of 26 vs 1 of 14 case, P = .024). Identical cytogenetic abnormalities proved to result in different genomic breakpoints. Thus, we showed that dicentric rearrangements involving 9p are associated with PAX5 deletion or internal rearrangement in approximately 90% of cases.
Molecular consequences of PAX5 internal breakpoints
According to the material available, we could investigate the presence of PAX5 fusion transcripts in 23 of the 32 patients with internal rearrangement. We identified an in-frame fusion of exon 4 of PAX5 with exon 3 of ETV6 by specific reverse-transcribed PCR and confirmed the rearrangement by FISH using ETV6 probes in 3 cases of dic(9;12)(p13;p13) (#99, #100, and #105; data not shown). The t(7;9)(q11;p13) translocation, which fused exon 7 of PAX5 to exon 2 of ELN, was previously described by our group11 in 2 of the 3 patients of this cohort (#119, #120). The presence of PAX5-ELN was confirmed in the third patient by FISH with ELN specific BAC probes (#121; supplemental Figure 2). We also confirmed the presence of known chimeric transcripts in 2 cases of t(3;9)(p14;p13) (#97, #98). This rearrangement yielded a chimeric transcript, which fused in-frame exon 6 of PAX5 to exon 7 of FOXP1 in one case (no RNA was available for the second patient). This fusion transcript was detected by RCA-RACE and confirmed by FISH using FOXP1 specific BAC probes for both (supplemental Figure 2). We detected PAX5-JAK2 fusion transcript in a del(9)(p13;p24) case (#118). This translocation led to in-frame chimeric transcript, which fused exon 5 of PAX5 to exon 19 of JAK2. The 2 genes being originally in opposite orientation on the 9p arm, we speculated that a deletion occurred before/or concomitant with JAK2 inversion. In agreement with these data, FISH experiments revealed that the JAK2 probe signal was slightly decreased whereas the PAX5 signal kept only the centromeric part of the probe (supplemental Figure 2). We identified the PAX5-POM121 chimeric transcript in 1 patient with a t(7;9)(q11;p13) (#93). This case expressed an in-frame fusion transcript, which composed the 5 first exons of PAX5, 77 noncontiguous nucleotides from PAX5 intron 5/intron 6, and POM121 sequence from base 14 of exon 4 (located in the 5′-untranslated region) to the end. The corresponding predicted protein juxtaposed PAX5 N-terminal sequence, included nuclear localization signal (NLS), 72 amino acids, which do not form any predictive structural domain, and the complete POM121 sequence (supplemental Table 4).
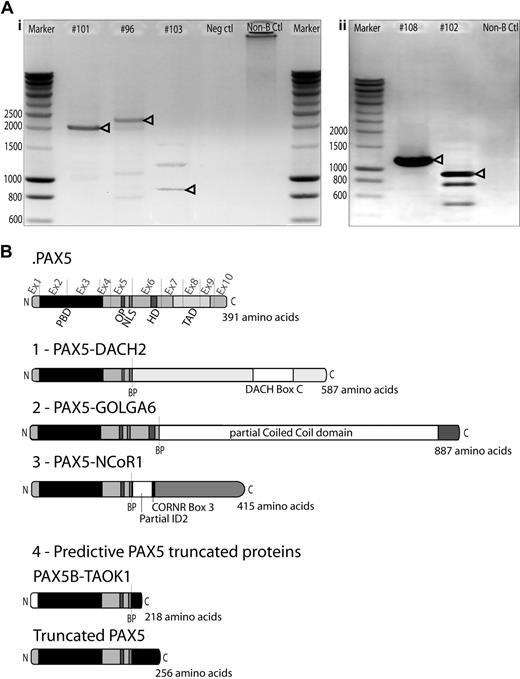
In addition, 3 new PAX5 rearrangements have been molecularly characterized in this study using RCA-RACE technique (Figure 2). In the chimeric transcript resulting from the t(X;9)(q21;p13) (#96), the exon 5 of PAX5 was fused in-frame to exon 3 of Dachshund 2 (DACH2). FISH analysis with DACH2 specific BAC probes was performed and confirmed its involvement in this case (supplemental Figure 2). The t(9;17)(p13;p11) led to generation of a chimeric transcript, which fused exon 5 of PAX5 in-frame with exon 43 of Nuclear receptor Co-Repressor 1 (NCoR1; #101). We also detected an in-frame fusion between exon 6 of PAX5 and exon 3 of golgi autoantigen, golgin subfamily a, 6 (GOLGA6) in a case of t(9;15)(p13;q24) translocation (#103).
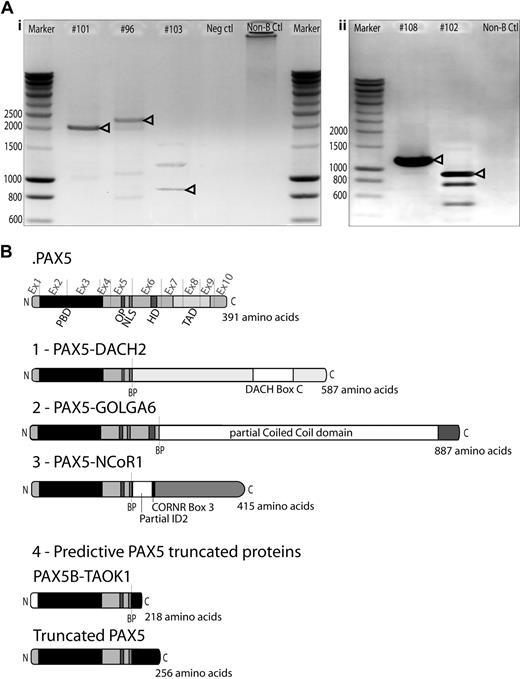
Cloning of novel PAX5 fusion partners by RCA-RACE. (A) RCA-RACE products were run on 1% agarose gel. Bands corresponding to amplified transcripts of fusion (i) or truncated genes (ii) are indicated by arrowheads. Additional bands correspond to amplification of fragments of wild-type PAX5 DNA. (i) Patient #101 amplified product corresponds to PAX5-NCoR1 fusion cDNA, #96 to PAX5-DACH2 fusion, #103 to PAX5-GOLGA6 fusion cDNA. (ii) Patient #108 product corresponds to truncated PAX5 cDNA, #102 to PAX5-TAOK1 fusion. (B) Schematic illustration of the fusion proteins predicted from cDNA sequencing, breakpoints (BP) are indicated by dashed lines. Ex1-10 indicates PAX5 exons. All fusion proteins retain the PAX5 DNA-binding domain (PBD, amino acids 16-142), the OP motif (amino acids 179-186), and NLS (amino acids 195-201). (1) PAX5-DACH2 contains the conserved coiled-coil domain of DACH2 (DACH-box C). (2) PAX5-GOLGA6 sequence contains a large part of a predicted coiled-coil domain of GOLGA6. (3) PAX5-NCoR1 retains part of the inhibitor of DNA-binding 2 (ID2) domain and the corepressor-nuclear receptor box 3 (CoRNR box 3). (4) PAX5-TAOK1 predictive protein contains an alternative amino acid sequence encoded by exon1B of PAX5 and an additional 17-amino acid tail, which does not correspond to any predictive functional domain. Truncated PAX5 contains N-terminal part of PAX5 (amino acids 1-201) and a 55-amino acid tail coded by the contiguous intron 5/intron 6 nucleotide sequence, which does not correspond to any predictive functional domain.
Cloning of novel PAX5 fusion partners by RCA-RACE. (A) RCA-RACE products were run on 1% agarose gel. Bands corresponding to amplified transcripts of fusion (i) or truncated genes (ii) are indicated by arrowheads. Additional bands correspond to amplification of fragments of wild-type PAX5 DNA. (i) Patient #101 amplified product corresponds to PAX5-NCoR1 fusion cDNA, #96 to PAX5-DACH2 fusion, #103 to PAX5-GOLGA6 fusion cDNA. (ii) Patient #108 product corresponds to truncated PAX5 cDNA, #102 to PAX5-TAOK1 fusion. (B) Schematic illustration of the fusion proteins predicted from cDNA sequencing, breakpoints (BP) are indicated by dashed lines. Ex1-10 indicates PAX5 exons. All fusion proteins retain the PAX5 DNA-binding domain (PBD, amino acids 16-142), the OP motif (amino acids 179-186), and NLS (amino acids 195-201). (1) PAX5-DACH2 contains the conserved coiled-coil domain of DACH2 (DACH-box C). (2) PAX5-GOLGA6 sequence contains a large part of a predicted coiled-coil domain of GOLGA6. (3) PAX5-NCoR1 retains part of the inhibitor of DNA-binding 2 (ID2) domain and the corepressor-nuclear receptor box 3 (CoRNR box 3). (4) PAX5-TAOK1 predictive protein contains an alternative amino acid sequence encoded by exon1B of PAX5 and an additional 17-amino acid tail, which does not correspond to any predictive functional domain. Truncated PAX5 contains N-terminal part of PAX5 (amino acids 1-201) and a 55-amino acid tail coded by the contiguous intron 5/intron 6 nucleotide sequence, which does not correspond to any predictive functional domain.
Five cases led to the production of truncated PAX5 transcripts. In a t(9;17)(p13;q11) case (#102), PAX5 was disrupted inside the intron 5/intron 6 and fused to intron 19/intron 20 of TAO kinase 1 (TAOK1) in reverse orientation to its open reading frame. Chimeric PAX5 transcript conserved exon 1B to exon 5 and an additional coding sequence of 54bp. The 3′-untranslated region of this transcript contained part of intron 19/intron 20 of TAOK1. This was further confirmed by FISH using TAOK1 specific BAC probes. On comparative genomic hybridization-single nucleotide polymorphism (CGH-SNP) chip, biallelic deletion of PAX5 exon 1A was detected, confirming the absence of PAX5A truncated isoform (supplemental Figure 3). This translocation was associated with an unbalanced t(17;20)(q11;q11), and SNP-CGH array revealed that the locus modified by this latter rearrangement involved an intergenic region. In case #108 with a dic(9;16)(p13;q11), we isolated a truncated PAX5, conserving exon 1A to exon 5 and 162 bp of additional coding sequence corresponding to the contiguous intron 5/intron 6 sequence (supplemental Table 4). CGH-SNP array analyses were performed and revealed that partner region on 16q11 did not contain any gene, suggesting that the dic(9;16)(p13;q11) results only in forced expression of the truncated transcript (supplemental Figure 3). This truncated form was also detected in 2 cases (#107, #122, data not shown) for which we could not detect any PAX5 fusion transcript. CGH-SNP analyses on patient #112 with a dic(9;20)(p13;q11) revealed breakpoints in intron 6/intron 7 of PAX5 and in the last exon of Pleiomorphic adenoma gene-like 2 (PLAGL2), leading to a putative truncated form of PAX5 (supplemental Figure 3).
Because PAX5 fusion proteins have been shown to act as competitive inhibitor of wtPAX5 transactivation activity,2,11,17 we assessed the functional consequence of one of the new fusion (PAX5-NCoR1) and of the truncated form (cloned from patient #108) using a luciferase reporter assay.16 We showed that these 2 types of PAX5 alterations (fusion and truncated form) behaved as competitive inhibitors of wild-type PAX5 transactivating activity (supplemental Figure 4).
In summary, we detected 5 recurrent and 3 new partner genes, thus confirming the wide diversity of PAX5 fusion partners. We also showed that truncated PAX5 can be a recurrent consequence of different chromosomal translocations.
Discussion
We investigated PAX5 alterations in a large cohort of 153 adult and childhood B-ALL patients harboring a 9p abnormality, using classic and innovative molecular and cytogenetic techniques. We detected PAX5 alteration in 81% of the patients with structural abnormality of the short arm of chromosome 9. Internal rearrangements were found in 21% of the cases, this alteration being significantly associated with simple karyotypes. Furthermore, PAX5 internal rearrangements and most recurrent translocations commonly found in B-ALL appeared to be mutually exclusive. This implies that PAX5 internal rearrangement might be sufficient to initiate leukemogenesis, through blockage of B-cell differentiation. Mechanistically, this hypothesis is emphasized by the fact that at least PAX5-ELN,11 PAX5-FOXP1,2 and PAX5-ETV617 have a trans-dominant negative effect on normal PAX5.
To identify PAX5 fusion partners, we implemented a new molecular biology method, until now only used in plant biology.15 The strategy is based on RCA to generate concatenated cDNA containing multiple copies of the target gene. This concatenated cDNA is subsequently used as a template for inverse PCR with 2 specific primers of the known part of the chimera (see principle in supplemental Figure 5). Thus, we isolated additional cases of rare translocations, as described previously (Figure 3). For example, PAX5-POM121 generated by the t(7;9)(q11;p13) translocation can now be classified as recurrent because we added a second case to a translocation.9 However, the fusion sequence we described is slightly different from that reported, even though the predictive protein structures are similar. It is noteworthy that the same fusion partner can give rise to either in-frame or out-of-frame chimeric transcripts (eg, PAX5-FOXP1).2,18 The PAX5 internal breakpoint can also lead to the production of truncated proteins resulting from an out-of-frame fusion (PAX5-FOXP1), fusion with a gene in opposite orientation (PAX5-TAOK1), or fusion with noncoding sequences. In most instances, a premature stop codon is generated immediately after PAX5 exon 5. The predicted chimeric proteins conserved the PBD, octapeptide (OP), and NLS of PAX5 (breakpoint after exon 5), suggesting that the chimera is located in the nucleus and able to bind PAX5 targets. It is probable that these truncated PAX5-X proteins block B-cell differentiation in a dominant negative manner, similarly to that reported for other PAX5 fusion products.2,11,17
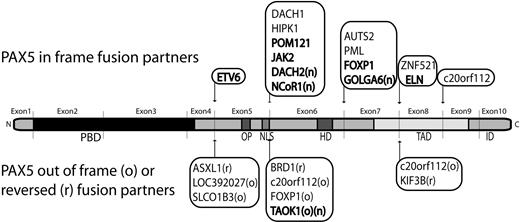
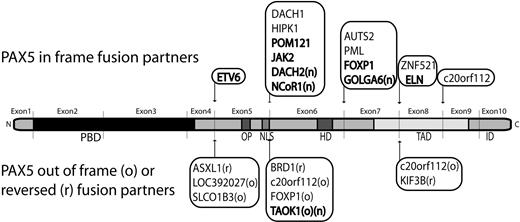
3′-PAX5 fusion partners in B-ALL. Partners previously described (references in supplemental Table 5) and detected in our series are indicated in bold, new PAX5 fusion partners are highlighted with an “n.” In-frame PAX5 fusions are indicated on the upper side. Fusion partners in reverse orientation (r) or out-of-frame fusion (o), leading to PAX5 predictive truncated protein are indicated on the lower side. PBD, OP, NLS, homeodomain (HD), transactivation domain (TAD), and inhibitory domain (ID) are shown.
3′-PAX5 fusion partners in B-ALL. Partners previously described (references in supplemental Table 5) and detected in our series are indicated in bold, new PAX5 fusion partners are highlighted with an “n.” In-frame PAX5 fusions are indicated on the upper side. Fusion partners in reverse orientation (r) or out-of-frame fusion (o), leading to PAX5 predictive truncated protein are indicated on the lower side. PBD, OP, NLS, homeodomain (HD), transactivation domain (TAD), and inhibitory domain (ID) are shown.
In addition, the RCA-RACE approach enabled us to clone new fusion genes: PAX5-NCoR1, PAX5-DACH2, and PAX5-GOLGA6 increasing the total number of partners of PAX5 in B-ALL to 20 (Figure 3). NCoR1 is a component of histone deacetylase complexes and acts as transcriptional repressor by modulating chromatin, which impedes the accessibility of basal transcription factors to genomic regulation sites.19 Interestingly, NCoR1 partial inhibitor of DNA-binding 2 domain replaces the trans-activating domain of PAX5 and the putative protein may act as a constitutive repressor of PAX5 targets. DACH2 is a transcription factor implicated in organogenesis and is supposed to be involved in proliferation, notably by repressing cyclin-dependent kinase inhibitor expression.20 As the patient harboring the t(X;9) translocation was a male, the DACH2-PAX5 fusion is associated with a total loss of wild-type DACH2. Moreover, DACH1, structurally close to DACH2, has been found fused to PAX5 in a t(9;13)(p13;q22).9 These results strongly suggest that DACH family genes could be involved in oncogenesis. The third in-frame fusion led to the expression of PAX5-GOLGA6 transcript. GOLGA6 is highly expressed in human testis, but its function is still unknown.21
Remarkably, even if the nature of the partners is very variable, 19 of 23 of the predicted chimeric proteins conserved the PBD, OP motif, and the nuclear localization signal of PAX5 (breakpoint after exon 5), strongly suggesting functional selection of the chimera.
Our data showed that 60% of the patients (adults and children) harbored a monoallelic deletion of PAX5. These patients mainly displayed a complex karyotype and classic recurrent fusion genes (BCR-ABL1, TCF3-PBX1, ETV6-RUNX1, and MLL rearrangements). We assumed that the complexity of karyotypes in patients with PAX5 deletion reflects the secondary nature of this event, contrary to internal PAX5 translocations. Miller et al22 have recently shown the interplay between BCR-ABL1 and the deletion of both PAX5 and INK4a/ARF in the leukemia process. Thus, the deletion of PAX5 might occur concomitantly with the loss of INK4a/ARF locus located in 9p21.
Dicentric cases represented 26% of the 9p abnormality, most often involving chromosomes 20, 12, and 7 and frequently leading to the deletion of PAX5. We observed that PAX5 internal rearrangements in dicentric chromosomes were much more frequent in children than in adults. Concerning the dic(9;12) cases, Heerema et al have reported one group of patients harboring PAX5-ETV6 fusion and another group of patients with both ETV6-RUNX1 rearrangement and PAX5 deletion, the 2 groups being mutually exclusive.23 We characterized a new situation in which patients harbored a PAX5 deletion without ETV6-RUNX1 fusion (cases #19, #20, and #106). We assumed that simple karyotypes with dicentric chromosomes are cytogenetic entities with heterogeneous breakpoints. Contrary to PAX5-ETV6 in the dic(9;12) rearrangement, Strefford et al have suggested that the dic(9;20) are not associated with an identical recurrent gene rearrangement.24 Our additional cases are in line with this assumption. Interestingly, we noted that dicentric cases can be masked by a complex rearrangement. Two dicentric rearrangements in this series (#101 and #107) involved a third chromosome (17 and 16, respectively). This third partner chromosome was split between chromosome 9 and chromosome 20, without loss of material. The final result was a loss of 9p and 20q in these 2 patients who could be structurally comparable with a dic(9;20).
In conclusion, we showed that PAX5 was altered in 81% of the cases with a 9p abnormality. These alterations were diverse in terms of nature and associated karyotypic features. The different types of alterations seemed to act at different levels of the leukemia process: PAX5 internal breakpoints, leading to the expression of fusion proteins or PAX5 truncated proteins, might occur early in the process, by way of contrast, deletions of PAX5, which were much more frequent, might appear later in the oncogenic process and could be a secondary event occurring in a cell already undergoing transformation. It is worth noting that PAX5 chimeric transcripts shared common features regardless of the fusion partners. The majority retained the DNA-binding domain and the NLS of PAX5, strongly suggesting that they may modulate wild-type protein activity in a dominant negative manner. Nonetheless, partner gene impact on leukemogenesis requires further investigation. It would be interesting to determine whether fusion partners are randomly or functionally selected because, to date, it has been impossible to identify features common to all fusion partners.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Eric Delabesse, Estelle Espinos, and José Meija for fruitful discussions; Roland Heilig and Gabor Gyapay (CNS, Evry, France) for kindly providing the BACs; Prof Meinrad Busslinger for kindly providing the luc-CD19 reporter plasmid; and the tumor bank of CHU Bordeaux, Bernardine Favre, Patrick Callier, Nathalie Marle (CRB Centre Hospitalier Universitaire Dijon), and Barbara De Moerloose (Department of Pediatric Hemato-Oncology, Ghent University Hospital, Ghent, Belgium) for technical support.
This work was supported by the Institut National du Cancer, Laurette Fugain foundation, the Cooperación de investigation transpirenaica en la terapia innovadora de la leukemia, the Recherche et Innovation en Thérapeutique Cancéreuse, Fondation de France, and l'association pour la recherche sur le Cancer (no. 4841).
Authorship
Contribution: C. Broccardo, N.D., and P.B. designed the study, C. Broccardo, E.C., S.S., N.D., and P.B. wrote the paper; E.C., S.S., N.P., J.F., R.E., C.Q., and M.B. performed the research and analyzed the data; and N.D., F.M., P.T., M.-P.P., C.L., D.P., E.L., N.N., S.T., B.P., I.L., L.B., V.E., I.R., C. Barin, M.-J.M., M.L.-P., H.A.-P., C.C., C.P., and C.T. provided patient samples, performed the karyotype analyses, and gave critical suggestions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cyril Broccardo, Inserm U563, CHU Purpan, CPTP BatB, BP3028, 31024 Toulouse Cedex 3, France; e-mail: cyril.broccardo@inserm.fr.
References
Author notes
E.C. and S.S. contributed equally to this study.