Abstract
Haploinsufficiency of RUNX1 (also known as CBFA2/AML1) is associated with familial thrombocytopenia, platelet dysfunction, and predisposition to acute leukemia. We have reported on a patient with thrombocytopenia and impaired agonist-induced aggregation, secretion, and protein phosphorylation associated with a RUNX1 mutation. Expression profiling of platelets revealed approximately 5-fold decreased expression of 12-lipoxygenase (12-LO, gene ALOX12), which catalyzes 12-hydroxyeicosatetraenoic acid production from arachidonic acid. We hypothesized that ALOX12 is a direct transcriptional target gene of RUNX1. In present studies, agonist-induced platelet 12-HETE production was decreased in the patient. Four RUNX1 consensus sites were identified in the 2-kb promoter region of ALOX12 (at −1498, −1491, −708, −526 from ATG). In luciferase reporter studies in human erythroleukemia cells, mutation of each site decreased activity; overexpression of RUNX1 up-regulated promoter activity, which was abolished by mutation of RUNX1 sites. Gel shift studies, including with recombinant protein, revealed RUNX1 binding to each site. Chromatin immunoprecipitation revealed in vivo RUNX1 binding in the region of interest. siRNA knockdown of RUNX1 decreased RUNX1 and 12-LO proteins. ALOX12 is a direct transcriptional target of RUNX1. Our studies provide further proof of principle that platelet expression profiling can elucidate novel alterations in platelets with inherited dysfunction.
Introduction
RUNX1, also known as CBFA2 (core binding factor A2), is a member of a family of transcription factors that regulate the expression of several hematopoietic-specific genes through a highly conserved DNA-binding region called the RUNT homology domain (RHD).1 The RHD dimerizes with CBFβ to form a stable complex. The complex acts as an anchor to recruit other cofactors that bind in cis to adjacent sites or interact directly with RUNX1. RUNX1 plays a critical role in normal fetal hematopoiesis.2,3 Homozygous deletion of RUNX1 results in embryonic lethality related to absence of definitive hematopoiesis.2-4 In humans, haploinsufficiency of RUNX1 is associated with familial thrombocytopenia, platelet dysfunction, and predisposition to acute leukemia.5 Most of the point mutations identified in RUNX1 occur in the RHD leading to loss of DNA binding.6,7
We have previously reported8,9 studies in a patient with mild thrombocytopenia, impaired agonist-induced platelet aggregation, secretion and protein phosphorylation (myosin light chain and pleckstrin), and decreased platelet protein kinase C-θ (PKC-θ), associated with a mutation (haplodeficiency) in the conserved region of RUNX1. Expression profiling of patient platelets revealed an approximately 5-fold decreased mRNA expression of platelet-type 12-lipoxygenase (12-LO, gene ALOX12).10 Lipoxygenases are a family of non–heme iron-containing enzymes that catalyze the incorporation of molecular oxygen into polyunsaturated fatty acids, such as arachidonic acid (AA). The platelet 12-LO is expressed primarily in platelets, megakaryocytes, and epidermis and is present in human erythroleukemia (HEL) cells.11,12 Activation of platelets results in the release of free AA, which is metabolized by 2 major pathways (cyclooxygenase to thromboxane A2 and by 12-LO to 12-hydroperoxyeicosatetraenoic acid [12-HPETE]), which is further reduced to hydroxyeicosatetraenoic acid (12-S HETE).13 Thus, 12-LO mediates a major pathway in the metabolism of AA on platelet activation.
Although the role of 12-LO in platelets is not as well understood as that of cycloxygenase-1, LO products have been implicated in several aspects of platelet function. 12-LO has been reported to play a role in thrombin- and thromboxane-induced platelet aggregation and calcium signaling.14 12-HETE potentiates thrombin-induced aggregation of bovine15 and human platelets.16 Addition of nanomolar concentrations of 12-HPETE to platelets primed with nonaggregating concentration of AA or collagen potentiates platelet aggregation17,18 ; this is associated with increased mobilization of cellular AA and thromboxane formation. Inhibition of 12-LO has been reported to reduce glycoprotein (GP) IIb-IIIa activation and platelet aggregation in adenosine diphosphate (ADP), thrombin, or U46619-stimulated platelets.14,19 Interestingly, mouse platelets deficient in 12-LO had normal aggregation and secretion responses on activation with most agonists but enhanced aggregation on exposure to ADP, and increased mortality in a thrombosis model involving the injection of ADP.20 12-HETE amplifies p-selectin–induced tissue factor expression by monocytes.21 Lastly, 12-LO mediates the generation of peroxide and other reactive oxygen species in platelets by the nicotinamide adenine dinucleotide phosphate oxidase pathway and is a major player in antibody-induced peroxide lysis of platelets.22 Currently, little is known regarding the transcriptional regulation of 12-LO in platelets. Based on the findings in our patient of decreased platelet ALOX12 mRNA expression and RUNX1 haplodeficiency, we postulated that ALOX12 is a direct target of this transcription factor, which is implicated in platelet production and function. We provide the first evidence for this in the present studies and also show that platelet 12-HETE production is indeed decreased in our patient with RUNX1 haplodeficiency. Equally important, our studies validate the concept that platelet expression profiling in patients with inherited platelet dysfunction has the potential to provide new insights into specific gene/protein abnormalities.
Methods
Patient information
We have previously described8,9 the clinical presentation and detailed studies in this 24-year-old white man, documenting decreased platelet aggregation, secretion, activation of GPIIb-IIIa, pleckstrin and myosin light chain phosphorylation, and PKC-θ level. This patient has a single point mutation in intron 3 at the splice acceptor site for exon 4, leading to a frameshift with premature termination in the conserved Runt homology domain of RUNX1.9 Platelet expression profiling studies have been described10 and show decreased expression of ALOX12 (by 5-fold compared with normal platelets) and other genes. Control subjects used in the studies described here were healthy subjects not on any medications. These studies were performed after approval by Temple University's Institutional Review Board.
Platelet preparation
Blood was collected in one-tenth volume of acid-citrate-dextrose from the patient and control subjects, and platelets isolated as described.23 Platelet-rich plasma was prepared by centrifugation at 180g for 20 minutes at room temperature. Platelets were washed twice by pelleting at 500g for 20 minutes and resuspended in N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid modified Tyrode buffer (pH 7.4, containing 138mM NaCl, 2.9mM KCl, 12mM NaHCO3 5.5mM glucose, and 10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) with 14mM ethylenediaminetetraacetic acid. Washed platelets were resuspended in N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid-Tyrode buffer containing 1mM CaCl2, and the platelet count was adjusted to a concentration of 3 × 108/mL.
Determination of platelet 12-HETE production
A total of 1-mL aliquots of platelet suspensions were stimulated with AA (1mM) or thrombin (1 or 5 U/mL), both from Sigma-Aldrich, at 37°C for 30 minutes. Methanol (2 mL) was added to the samples on ice and then acidified with 2N HCl to a pH 3.5 for 15 minutes. Lyophilized samples were resuspended in 250 μL of assay buffer and used for 12-HETE measurements using an enzyme-linked immunoassay kit (Assay Designs).
Cell-line and culture conditions
HEL cells (ATCC) were grown at 37°C in 5% CO2. The culture media used was RPMI 1640 supplemented with 10% fetal bovine serum and 1% penicillin. Cells were stimulated with 50nM phorbol 12-myristate 13-acetate (PMA; Fisher Scientific) to induce megakaryocytic transformation.24
Construction of luciferase reporter vectors
To construct wild-type and deletion mutants, human ALOX12 promoter regions of various lengths with common 3′-prime end were prepared by polymerase chain reaction (PCR) amplification of human genomic DNA. The primers used for amplification are shown in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). To facilitate directional cloning, 2 restriction sites, Xho1 and Nhe1, were incorporated into the primer at the 5′ and 3′ ends, respectively. The PCR products were verified by sequencing (Genewiz) and cloned into promoterless luciferase reporter gene vector PGL3-Basic (Promega).
Mutagenesis in the RUNX1 consensus site (TGGGGT) was done by altering 3 nucleotides in the binding site by a PCR-based method using mutant primers (supplemental Table 1). The PCR products were verified by sequencing and then cloned into PGL3-Basic (Promega) luciferase expression vector.
Transfection of plasmids in HEL cells
HEL cells (5 × 105/well) were cotransfected with firefly luciferase reporter plasmid (2 μg) and Renilla luciferase reporter plasmid (Promega) in a ratio of 50:1 using Lipofectamine (Invitrogen) or FuGene 6 Transfection Reagent (Roche Diagnostics) as per the manufacturer's instructions. Transfected cells were incubated for 3 hours at 37°C in 5% CO2. At the end of incubation, 250 μL of growth media containing 2 times the concentration of serum and 50nM of PMA was added to each well. Cells were harvested after 24 hours and analyzed for luciferase activity using Dual-Luciferase assay system (Promega). The promoter activity was calculated by dividing the luciferase activity of the constructs by the internal Renilla luciferase activity and expressed relative to that of the promoterless vector. All transfection studies were performed in triplicate. To study the effect of overexpression of RUNX1, HEL cells (106) were cotransfected using Turbofectin 8.0 (OriGene) with 2 μg each of luciferase reporter constructs with ALOX12 promoter region and RUNX1 expression plasmid, RUNX1-pCMV6-XL4 (SC106348; OriGene). In parallel, luciferase reporter vectors were cotransfected with empty vector, pCMV6-XL4, as a control. Renilla luciferase reporter plasmid was cotransfected as an internal standard, as described in this section. After 3 hours of transfection, medium containing 50nM PMA was added to the cells. Cells were lysed at 48 hours and assayed for luciferase activities.
EMSA
Nuclear extracts were prepared from PMA-stimulated HEL cells using Pierce Kit (NE-PER Nuclear and Cytoplasmic Extraction kit) as per the manufacturer's instructions. Cell extracts were stored at −80°C until use. A 33-bp wild-type and 5 mutant IRDye 700 Infrared Dye Labeled oligonucleotides corresponding to each RUNX1-binding site were synthesized (Integrated DNA Technologies; supplemental Table 1). Electrophoretic mobility shift assay (EMSA) was performed using a commercially available kit (Light Shift EMSA Kit; Pierce Chemical) according to the manufacturer's instructions. Nuclear extracts, with or without 50-fold excess of unlabeled competitor oligo, were incubated for 15 minutes at room temperature in 1 times binding buffer. IRDye 700-labeled, double-stranded oligos were then added, and the binding reactions were performed for 30 minutes at room temperature. For supershift assays, antibody against RUNX1 (Santa Cruz Biotechnology; sc-8563) or nonspecific immunoglobulin G (IgG; Santa Cruz Biotechnology) was preincubated with nuclear extracts at room temperature for 30 minutes before addition of labeled probe. The DNA-protein complexes were resolved on native 4% polyacrylamide gels, and the protein-DNA complexes were detected using Odyssey Infrared Imaging System (LiCor Biosciences).
Binding of recombinant RUNX1 (Abnova Taiwan Corporation) to ALOX12 was conducted in 1 times LiCor binding buffer (LiCor Biosciences). A total of 50 ng of recombinant protein, diluted in buffer containing 0.1mM ethylenediaminetetraacetic acid, 100mM KCl, 20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.8, 5 μg/mL bovine serum albumin, and 20% glycerol25 was incubated with IRDye 700-labeled or unlabeled, double-stranded oligos and 20 ng dI:dC along with 2mM MgCl2 and 25mM dithiothreitol/2.5% Tween-20. For supershift assay antibody against RUNX1 (Santa Cruz Biotechnology; sc-8564X) or nonspecific IgG (Santa Cruz Biotechnology) was preincubated for 30 minutes in cold before addition of labeled probe. The rest of the procedure was performed as described in the previous paragraph.
Immunoblotting analysis
Total cell lysates were prepared from cotransfected HEL cells using M-Per Mammalian Protein Extraction Reagent (Pierce Chemical) with proteinase inhibitors (1mM ethyleneglycoltetraacetic acid, 1mM dithiothreitol, 1mM phenylmethylsulfonyl fluoride, 10 mg/mL leupeptin), and protein concentrations were determined by the bicinchoninic acid method using a commercial assay (Pierce Chemical); the aliquots were stored at −80°C. Samples were boiled, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (10% resolving gel) and electrophoretically transferred onto polyvinylidene fluoride nitrocellulose (Millipore). The membranes were washed in Tris-buffered saline with 0.1% Tween 20. The primary antibodies were used at a dilution 1:1000 for rabbit polyclonal anti-RUNX1, 1:400 for rabbit polyclonal anti–12-LO, and 1:7000 for anti–β-actin (Santa Cruz Biotechnology). Antibodies bound to nitrocellulose were detected using a peroxidase-conjugated goat anti–rabbit IgG as the secondary antibody (1:10 000; Promega). Blots were developed using Immobilon Western Chemiluminscent HRP Substrate (Millipore) and detected using a Luminescent Image Analyzer (Fuji Film Medical Systems). For studies on the effect of overexpression of RUNX1, HEL cell lysates were subjected to immunoblotting using anti-RUNX1 (sc-8563) and antiactin (I-19)–R (sc-1616-R) antibodies from Santa Cruz Biotechnology. Infrared labeled secondary antibodies were used to detect the expression on Odyssey Infrared Imaging System (LiCor Biosciences).
siRNA-mediated knockdown of RUNX1
siRNA against RUNX1 and unrelated mock siRNA were purchased from Santa Cruz Biotechnology. The RUNX1 siRNA used was a pool of 3 target-specific 19- to 25-nt siRNAs designed to knock down gene expression. For transfection 400nM of siRNA and 2 μg of wild-type construct were dissolved in 100 μL of Optimem media (Invitrogen). In another tube, transfection media was dissolved in 100 μL of Optimem media. The 2 mixtures were then combined and incubated for 45 minutes at room temperature. The mixture was added to 50 000 HEL cells resuspended in 400 μL of serum-free media, plated in a 24-well plate, and incubated for 5 hours at 37°C. After transfection, 2 times fetal bovine serum medium containing 50nM of PMA was added to the wells. On the following day, the 2 times medium was replaced with regular medium containing 50nM PMA. Cells were harvested at 48 hours after PMA stimulation for whole cell extract and luciferase assays as described.
ChIP analysis
This was performed using ChIP-It kit (Active Motif). PMA-stimulated (24 hours) HEL cells were cross-linked with 1% formaldehyde at room temperature for 10 minutes followed by addition of glycine to a final concentration of 0.125 M. Cell lysates were Dounce homogenized, and nuclear pellet was sheared enzymatically. The sheared chromatin was incubated with anti-RUNX1 antibody (sc-8564x; Santa Cruz Biotechnology) or control IgG (Santa Cruz Biotechnology) overnight at 4°C. This anti-RUNX1 antibody has been used previously in chromatin immunoprecipitation (ChIP) studies.26 The chromatin/antibody complex was eluted with 1% sodium dodecyl sulfate/1M NaHCO3 and reverse cross-linked by heating to 65°C. The DNA was treated with proteinase K and purified by spin columns as per kit instructions. The purified DNA samples were used as templates for PCR, with primer pairs spanning approximately 200 bp of ALOX12 gene promoter. As a negative control, glyceraldehyde-3-phosphate dehydrogenase was amplified with specific primers provided in the kit.
Bioinformatics
Potential binding sites for transcription factors were analyzed by computer program TFSEARCH (http://mbs.cbrc.jp/research/db/TFSEARCH.html).
Results
Activation-induced platelet 12-HETE production is decreased
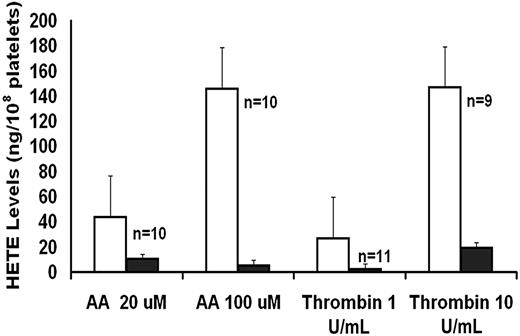
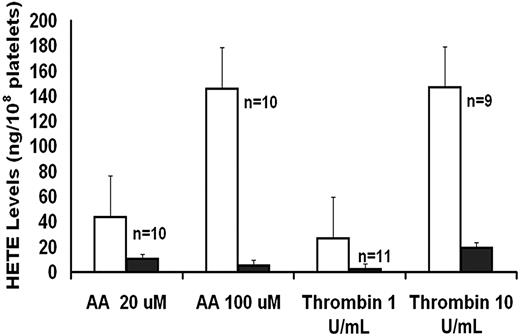
As an index of 12-LO activity, 12-HETE production was measured in platelets activated with AA and thrombin (Figure 1). These studies showed that platelet 12-HETE production was markedly diminished in the patient with both agonists.
AA and thrombin-induced platelet HETE production in patient (■, mean of 2 visits) and control subjects (□, mean ± SE).
AA and thrombin-induced platelet HETE production in patient (■, mean of 2 visits) and control subjects (□, mean ± SE).
Luciferase reporter studies in HEL cells
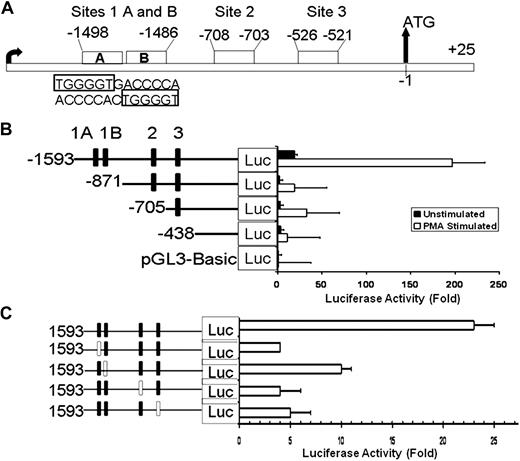
Analysis of ALOX12 5′ upstream region (2 kb) using the “TFSEARCH” program indicated the presence of 4 potential RUNX1-binding sites (Figure 2A): a palindromic sequence at −1498/−1486 with 2 RUNX1 consensus sites (labeled sites 1A and 1B) separated by a single nucleotide; at −708 (site 2), and −526 (site 3) bp upstream from ATG. None of the identified RUNX1 sites was the predominant consensus (5′-PuACCPuCA-3′) RUNX1 sequence but was a RUNX1 variant (TGGGGT) sequence that has previously been reported27,28 and shown to play a major role in regulation of αIIb promoter and megakaryocytic differentiation.27 A similar palindromic sequence with 2 RUNX1 sites has been reported in human urokinase plasminogen activator promoter.29
Luciferase reporter studies on ALOX12 promoter in PMA-treated HEL cells. (A) Schema showing 5′ upstream region of ALOX12 gene with 4 RUNX1 consensus sites. (B) Effect of 5′ sequential truncation of ALOX12 promoter region on expression of luciferase reporter in untreated (■) and PMA-treated (□) HEL cells. Fold increase in luciferase activity was measured as ratio of control vector to luciferase vectors carrying various lengths of promoter and normalized to Renilla luciferase constructs. Data are mean ± SEM of 3 experiments. (C) Effect of disruption of individual RUNX1 sites on ALOX12 expression. Each RUNX1 site was disrupted sequentially by introducing point mutations (□ boxes) in RUNX1-binding sites. Data are mean of 3 experiments.
Luciferase reporter studies on ALOX12 promoter in PMA-treated HEL cells. (A) Schema showing 5′ upstream region of ALOX12 gene with 4 RUNX1 consensus sites. (B) Effect of 5′ sequential truncation of ALOX12 promoter region on expression of luciferase reporter in untreated (■) and PMA-treated (□) HEL cells. Fold increase in luciferase activity was measured as ratio of control vector to luciferase vectors carrying various lengths of promoter and normalized to Renilla luciferase constructs. Data are mean ± SEM of 3 experiments. (C) Effect of disruption of individual RUNX1 sites on ALOX12 expression. Each RUNX1 site was disrupted sequentially by introducing point mutations (□ boxes) in RUNX1-binding sites. Data are mean of 3 experiments.
Luciferase reporter studies were performed using the 5′ upstream region −1593/−1 and its serial truncations in HEL cells treated or not with PMA (Figure 2B). The luciferase activity was markedly greater in HEL cells treated with PMA than in its absence (Figure 2B), indicating that PMA-induced megakaryocyte transformation is associated with up-regulation in ALOX12 promoter activity. In PMA-treated HEL cells, truncation to −871 bases showed an 85% decrease in promoter activity, indicating the presence of a positive regulatory region. Further deletion to −705 did not lead to additional decrease in activity. Deletion to −438 bp, which eliminates all 4 potential RUNX1-binding sites, showed an approximately 90% decrease in promoter activity compared with the −1593/−1 construct. The initial large decline in activity with truncation at −871 with no further loss with subsequent truncations suggested that −1593/−871 region may be particularly important for maximal promoter activity.
In subsequent reporter studies using construct −1593/−1, we mutated each of the putative RUNX1-binding sites (Figure 2C). Site 1A mutant construct contained a 3-bp mutation (from TGGGGT to TGGATC) in the region of −1498/−1493. The site 1B mutant construct had a 3-bp mutation in −1491/−1485 region (from ACCCCA to ACGTAT). Site 2 (−708) mutant construct was mutated from ACCCCA to ATCCAG, and site 3 mutant (−526) was mutated (from ACCCCA to GATCCA). Compared with the wild-type sequence, there was a decrease in luciferase activity on mutating each of the aforementioned 4 sites, indicating their individual functional relevance. Taken together, the findings in studies with serial deletions (Figure 2B) and mutations of the individual RUNX1 sites (Figure 2C) are consistent with the conclusion that multiple RUNX1-binding sites (including sites 2 and 3) are required for optimal promoter activity. This has been previously noted with respect to regulation of αIIb promoter by multiple RUNX1 sites.27 To determine that these sites are specific to RUNX1, we carried out EMSA.
Binding of RUNX1 to consensus sites by EMSA
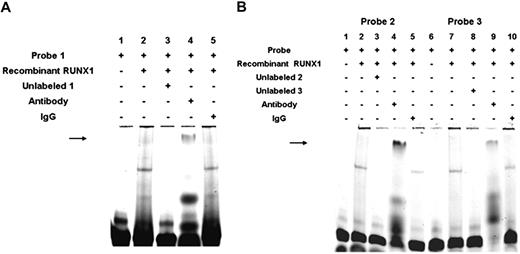
EMSA was performed with 30-mer probes (supplemental Table 1) with the RUNX1 consensus sites and nuclear extracts from PMA-stimulated HEL cells. Figure 3A shows the results with the DNA probe −1510/−1481, which has the palindromic sequence with 2 consensus (sites 1A and 1B). Protein binding was observed that was competed by excess unlabeled probe with loss of specific bands (Figure 3A lanes 2 and 3). Competition with excess unlabeled probes with either site 1A (lane 4) or site 1B (lane 5) mutated revealed inhibition of protein binding, consistent with the presence of the second RUNX1-binding site in each of these probes. In contrast, excess unlabeled probe with both sites mutated (lane 8) did not inhibit protein binding. The observed binding was inhibited by anti-RUNX1 antibody (lane 6), but not by a control IgG (lane 7). Binding studies were also performed with probes with site 1A or site 1B mutated (Figure 3B). With both probes, protein binding was observed (lanes 2 and 7) that was competed by excess unlabeled probe (lanes 3 and 8). Anti-RUNX1 antibody inhibited protein binding (lanes 5 and 10); control IgG had no effect (lanes 4 and 9). These findings indicate that RUNX1 can bind to both site 1A and site 1B.
EMSA using oligonucleotide probes with RUNX1-binding sites 1A, 1B, 2, and 3 and nuclear extracts from PMA-treated HEL cells. (A) EMSA using wild-type oligonucleotide probe with sites 1A (−1498/−1493) and 1B (−1491/−1486), and nuclear extract from PMA-treated HEL cells. Lane 1 indicates probe alone; lane 2, probe with nuclear extract; lane 3, competition with excess unlabeled probe; lane 4, competition with unlabeled site 1A mutant probe; lane 5, competition with unlabeled site 1B mutant probe; lane 6, effect of RUNX1 antibody; lane 7, effect of nonspecific IgG; and lane 8, competition with unlabeled probe with sites 1A and 1B mutated. (B) EMSA using oligonucleotide probes with site 1A mutated (mutant probe 1A lanes 1-5) or site 1B mutated (mutant probe 1B lanes 6-10). Lanes 1 and 6 indicate probe alone; lanes 2 and 7, protein binding with nuclear extract; lanes 3 and 8, competition with excess respective unlabeled probe; lanes 4 and 9, effect of nonspecific IgG; and lanes 5 and 10, inhibition of binding with RUNX1 antibody. (C) EMSA using oligonucleotide probe with site 2 (−708/−703). Lane 1 indicates free probe; lane 2, protein binding with extract; lane 3, competition with excess unlabeled probe; lane 4, competition with unlabeled probe with site 2 mutated; lane 5, inhibition of binding with RUNX1 antibody; and lane 6, effect of nonspecific IgG. (D) EMSA using oligonucleotide probe with site 3 (−526/−521). Lane 1 indicates free probe; lane 2, protein binding with extract; lane 3, competition with excess unlabeled probe; lane 4, competition with unlabeled probe with site 3 mutated; lane 5, inhibition of binding with RUNX1 antibody: and lane 6, effect of nonspecific IgG.
EMSA using oligonucleotide probes with RUNX1-binding sites 1A, 1B, 2, and 3 and nuclear extracts from PMA-treated HEL cells. (A) EMSA using wild-type oligonucleotide probe with sites 1A (−1498/−1493) and 1B (−1491/−1486), and nuclear extract from PMA-treated HEL cells. Lane 1 indicates probe alone; lane 2, probe with nuclear extract; lane 3, competition with excess unlabeled probe; lane 4, competition with unlabeled site 1A mutant probe; lane 5, competition with unlabeled site 1B mutant probe; lane 6, effect of RUNX1 antibody; lane 7, effect of nonspecific IgG; and lane 8, competition with unlabeled probe with sites 1A and 1B mutated. (B) EMSA using oligonucleotide probes with site 1A mutated (mutant probe 1A lanes 1-5) or site 1B mutated (mutant probe 1B lanes 6-10). Lanes 1 and 6 indicate probe alone; lanes 2 and 7, protein binding with nuclear extract; lanes 3 and 8, competition with excess respective unlabeled probe; lanes 4 and 9, effect of nonspecific IgG; and lanes 5 and 10, inhibition of binding with RUNX1 antibody. (C) EMSA using oligonucleotide probe with site 2 (−708/−703). Lane 1 indicates free probe; lane 2, protein binding with extract; lane 3, competition with excess unlabeled probe; lane 4, competition with unlabeled probe with site 2 mutated; lane 5, inhibition of binding with RUNX1 antibody; and lane 6, effect of nonspecific IgG. (D) EMSA using oligonucleotide probe with site 3 (−526/−521). Lane 1 indicates free probe; lane 2, protein binding with extract; lane 3, competition with excess unlabeled probe; lane 4, competition with unlabeled probe with site 3 mutated; lane 5, inhibition of binding with RUNX1 antibody: and lane 6, effect of nonspecific IgG.
EMSA with an oligo encompassing the RUNX1 site 2 at −721 bp also showed protein binding (Figure 3C lane 2), which was competed by unlabeled probe (lane 3) but not by a mutant probe (lane 4). It was blocked by RUNX1 antibody (lane 5) but not control IgG (lane 6). Similarly, studies with the probe with the potential RUNX1 site at −537 bp (site 3) showed protein binding (Figure 3D lane 2) that was competed by excess unlabeled probe (lane 3) and by RUNX1 antibody (lane 5), but not by a mutant probe (lane 4) or control IgG (lane 6). These studies suggest that RUNX1 binds to the probes with sites 2 and 3. Overall, the gel-shift studies provide evidence of RUNX1 binding to all of the 4 sites in the promoter region studied.
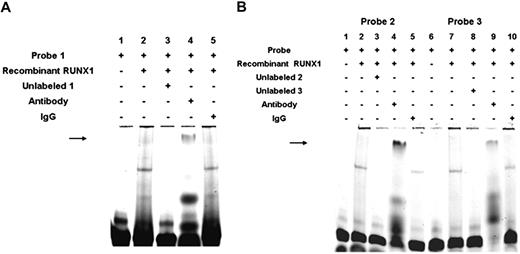
To obtain additional evidence that RUNX1 binds to the DNA elements, we performed studies using recombinant RUNX1 (Figure 4). Figure 4A shows RUNX1 bindings to the probe (−1510/−1481) containing 2 consensus sites (lane 2). This binding was competed by excess unlabeled probe (lane 3). The observed band was abolished by anti-RUNX1 antibody, and a supershifted band was noted (lane 4); this was not noted with control IgG (lane 5). Likewise, studies with the probes containing site 2 at −721 bp (Figure 4B lanes 1-5) and site 3 at −537 bp (Figure 4B lanes 6-10) also showed similar results with a loss of the observed band and a supershifted band observed with anti-RUNX1 antibody (lanes 4 and 9, respectively), but not IgG (lanes 5 and 10, respectively). These studies provide evidence that RUNX1 binds to the regions of ALOX12 promoter.
EMSA using oligonucleotide probes with RUNX1-binding sites 1A, 1B, 2, and 3 and recombinant RUNX1. (A) EMSA using oligonucleotide probe with sites 1A and 1B and recombinant RUNX1. Lane 1 indicates probe alone; lane 2, probe with recombinant RUNX1; lane 3, competition with excess unlabeled probe; lane 4, effect of RUNX1 antibody (arrow, supershift); and lane 5, effect of nonspecific IgG. (B) EMSA using recombinant RUNX1 and oligonucleotide probes with site 2 (−708/−703; lanes 1-5) and with site 3 (−526/−521; lanes 6-10). Lane 1 indicates free probe with site 2; lane 2, protein binding with recombinant RUNX1; lane 3, competition with excess unlabeled probe; lane 4, effect of RUNX1 antibody ( , supershift); lane 5, effect of nonspecific IgG; lane 6, free probe with site 3; lane 7, protein binding with recombinant RUNX1; lanes 8, competition with excess unlabeled probe; lane 9, effect of RUNX1 antibody (
, supershift); lane 5, effect of nonspecific IgG; lane 6, free probe with site 3; lane 7, protein binding with recombinant RUNX1; lanes 8, competition with excess unlabeled probe; lane 9, effect of RUNX1 antibody ( , supershift): and lane 10, effect of nonspecific IgG.
, supershift): and lane 10, effect of nonspecific IgG.
EMSA using oligonucleotide probes with RUNX1-binding sites 1A, 1B, 2, and 3 and recombinant RUNX1. (A) EMSA using oligonucleotide probe with sites 1A and 1B and recombinant RUNX1. Lane 1 indicates probe alone; lane 2, probe with recombinant RUNX1; lane 3, competition with excess unlabeled probe; lane 4, effect of RUNX1 antibody (arrow, supershift); and lane 5, effect of nonspecific IgG. (B) EMSA using recombinant RUNX1 and oligonucleotide probes with site 2 (−708/−703; lanes 1-5) and with site 3 (−526/−521; lanes 6-10). Lane 1 indicates free probe with site 2; lane 2, protein binding with recombinant RUNX1; lane 3, competition with excess unlabeled probe; lane 4, effect of RUNX1 antibody ( , supershift); lane 5, effect of nonspecific IgG; lane 6, free probe with site 3; lane 7, protein binding with recombinant RUNX1; lanes 8, competition with excess unlabeled probe; lane 9, effect of RUNX1 antibody (
, supershift); lane 5, effect of nonspecific IgG; lane 6, free probe with site 3; lane 7, protein binding with recombinant RUNX1; lanes 8, competition with excess unlabeled probe; lane 9, effect of RUNX1 antibody ( , supershift): and lane 10, effect of nonspecific IgG.
, supershift): and lane 10, effect of nonspecific IgG.
Binding of RUNX1 in vivo using ChIP studies
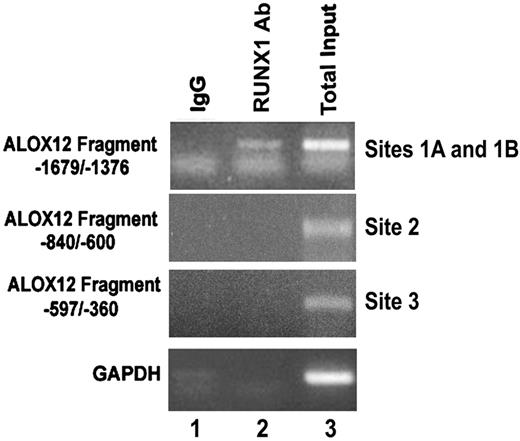
To demonstrate that RUNX1 binds to the region of interest in vivo as well, ChIP analysis was performed using PMA-stimulated HEL cells. The DNA fragment (−1679/−1376), which encompasses RUNX1-binding sites 1A and 1B, showed specific amplification by PCR of the RUNX1 precipitate (Figure 5 lane 2), whereas IgG control did not show any amplification. Fragments corresponding to region −840/−600 (containing site 2) and −597/−360 (containing site 3) showed no amplification in the DNA precipitated with the RUNX1 antibody or the control (IgG). These results indicate that RUNX1 binds in vivo to the region of −1498/−1485 bp with sites 1A and 1B, and suggest that the relevance in vivo of the other 2 sites may be less under the cellular conditions studied but does not exclude such a role under a different cellular state.
Binding of RUNX1 to ALOX12 promoter region in vivo. ChIP using control IgG (column 1) and RUNX1 antibody (column 2). Column 3 shows PCR of total input DNA. Data are PCR amplification of ALOX12 promoter regions encompassing sites 1A and 1B (−1679/−1376 nt), site 2 (−840/−600 nt), site 3 (−597/−360 nt), and glyceraldehyde-3-phosphate dehydrogenase.
Binding of RUNX1 to ALOX12 promoter region in vivo. ChIP using control IgG (column 1) and RUNX1 antibody (column 2). Column 3 shows PCR of total input DNA. Data are PCR amplification of ALOX12 promoter regions encompassing sites 1A and 1B (−1679/−1376 nt), site 2 (−840/−600 nt), site 3 (−597/−360 nt), and glyceraldehyde-3-phosphate dehydrogenase.
Effect of siRNA knockdown of RUNX1
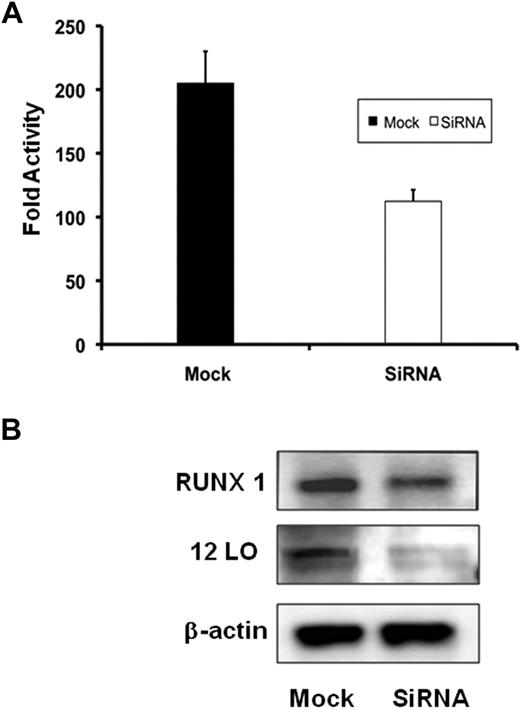
To determine whether knockdown of endogenous RUNX1 would have an effect on ALOX12 promoter activity, ALOX12 wild-type luciferase reporter construct (−1593/−1) was cotransfected with either RUNX1 siRNA or with mock siRNA. RUNX1 siRNA decreased promoter activity by approximately 50% compared with cells transfected with mock siRNA (Figure 6A). In parallel experiments, immunoblotting also showed a decrease in RUNX1 as well as 12-LO protein levels in cells transfected with siRNA compared with mock-transfected cells (Figure 6B).
Effect of siRNA RUNX1 knock down on ALOX12 promoter activity and protein. (A) Effect on ALOX12 promoter activity. HEL cells were cotransfected with RUNX1 or mock siRNA and ALOX12 luciferase-reporter construct (−1593/−1). Bar graphs represent activity as mean ± SD of 3 experiments. (B) Western blotting analysis of RUNX1, 12-LO, and actin in HEL cells transfected with mock or RUNX1 siRNA. Data are representative of 3 experiments.
Effect of siRNA RUNX1 knock down on ALOX12 promoter activity and protein. (A) Effect on ALOX12 promoter activity. HEL cells were cotransfected with RUNX1 or mock siRNA and ALOX12 luciferase-reporter construct (−1593/−1). Bar graphs represent activity as mean ± SD of 3 experiments. (B) Western blotting analysis of RUNX1, 12-LO, and actin in HEL cells transfected with mock or RUNX1 siRNA. Data are representative of 3 experiments.
Effect of overexpression of RUNX1 on ALOX12 promoter activity in HEL cells
The promoter activity of wild-type ALOX12 construct was markedly enhanced by RUNX1 overexpression (Figure 7). Promoter activity was markedly decreased with mutations in site 1A or site 1B, and overexpression of RUNX1 did not increase the activity. Mutations of site 2 or site 3 also decreased the activity relative to wild-type construct; with overexpression of RUNX1, there was a minimal increase, but the activity was markedly less than the effect noted with the wild-type construct. We interpret the minimal increases observed with sites 2 and 3 as indicating an effect of RUNX1 overexpression on intact sites 1A and 1B. Together, these results are also consistent with the conclusion that the multiple RUNX1 sites participate in the overall regulation of ALOX12.
Effect of transient overexpression of RUNX1 on ALOX12 promoter activity. (A) Effect on ALOX12 promoter activity. HEL cells were cotransfected with RUNX1-pCMV6 expression vector (■), empty pCMV6 vector ( ), or neither (□), along with ALOX12 luciferase-reporter construct (−1593/−1), wild-type, or with mutations in sites 1A, 1B, 2, or 3. Reporter activity was measured at 48 hours. Bar graphs represent activity as mean ± SD of 3 experiments. (B) Western blotting analysis of RUNX1 and actin (control) in HEL cells transfected with pCMV6 vector alone or with RUNX1-pCMV6 vector.
), or neither (□), along with ALOX12 luciferase-reporter construct (−1593/−1), wild-type, or with mutations in sites 1A, 1B, 2, or 3. Reporter activity was measured at 48 hours. Bar graphs represent activity as mean ± SD of 3 experiments. (B) Western blotting analysis of RUNX1 and actin (control) in HEL cells transfected with pCMV6 vector alone or with RUNX1-pCMV6 vector.
Effect of transient overexpression of RUNX1 on ALOX12 promoter activity. (A) Effect on ALOX12 promoter activity. HEL cells were cotransfected with RUNX1-pCMV6 expression vector (■), empty pCMV6 vector ( ), or neither (□), along with ALOX12 luciferase-reporter construct (−1593/−1), wild-type, or with mutations in sites 1A, 1B, 2, or 3. Reporter activity was measured at 48 hours. Bar graphs represent activity as mean ± SD of 3 experiments. (B) Western blotting analysis of RUNX1 and actin (control) in HEL cells transfected with pCMV6 vector alone or with RUNX1-pCMV6 vector.
), or neither (□), along with ALOX12 luciferase-reporter construct (−1593/−1), wild-type, or with mutations in sites 1A, 1B, 2, or 3. Reporter activity was measured at 48 hours. Bar graphs represent activity as mean ± SD of 3 experiments. (B) Western blotting analysis of RUNX1 and actin (control) in HEL cells transfected with pCMV6 vector alone or with RUNX1-pCMV6 vector.
Discussion
Our studies provide evidence that ALOX12 is a direct target of transcription factor RUNX1 in megakaryocytes/platelets. RUNX1 is a transcription factor that regulates the expression of hematopoietic-specific genes and plays a major role in hematopoiesis.1-4 Our previous transcript profiling studies10 have shown that ALOX12 expression is decreased in platelets from the patient with RUNX1 haplodeficiency. Computer-based analysis identified 4 RUNX1-binding sites in the proximal 2-kb ALOX12 promoter region. In luciferase reporter studies, mutations in the individual sites resulted in substantial loss of transcriptional activity (Figure 2C), suggesting that each site is functionally important and that multiple sites may interact to produce optimal promoter activity.27,30 Gel-shift assays using nuclear extracts revealed that each of the 4 sites binds RUNX1 (Figure 3). In additional EMSA studies, recombinant RUNX1 also bound to the DNA regions of interest (Figure 4). The presence of multiple RUNX1 consensus sites is a frequent finding in RUNX1-regulated promoters,31 and they could be closed spaced or separated by hundreds of base pairs. RUNX1 can also form homodimers that modulate RUNX1 activity.30 ChIP analysis showed enrichment of the region −1676/−1376 that encompasses sites 1A and 1B (Figure 5) indicating in vivo binding of RUNX1 to this region. Consistent with this, the EMSA studies showed RUNX1 binding to these 2 sites (Figure 3) and in luciferase studies mutations inhibited promoter activity (Figure 2). Our studies using siRNA to down-regulate RUNX1 showed a decrease in promoter activity of ALOX12 (Figure 6) along with a concomitant decrease in RUNX1 and 12-LO protein. In addition, overexpression of RUNX1 markedly enhanced promoter activity with a loss of this effect with mutation of the 4 RUNX1 sites (Figure 7A). Lastly, but most importantly from the perspective of physiologic relevance, we show a decrease in agonist-induced 12-HETE production (Figure 1) in the platelets from the patient with the decreased ALOX12 expression, providing strong evidence from the primary cell (platelet) and a direct functional validation of the decreased transcriptional regulation. Together, these studies constitute the first evidence that platelet ALOX12 is regulated by RUNX1 at the transcriptional level.
Computer-based analysis also revealed the presence of multiple consensus sites for GATA-1, a prototypic transcription factor that serves a major role in megakaryocytic differentiation32 and cooperates with RUNX1.27 Interestingly, the 4 RUNX1 sites present in the 2-kb upstream region are flanked by GATA1/GATA2 consensus sites located at −1926, −1538, −1408, −1328, −1152, −1068, −857, −729, −690, −531, and −421. The role of these flanking sites in transcription regulation of ALOX12 remains to be determined. Other significant elements, such as AP2 (−558 and −707), nuclear factor-κB (−539), SP1 (−158 and −594), CCAAT enhancer-binding protein (−433), CACC (−289), Ets (−1479), p300 (−488), and cMyb (−891 and −278) binding sites have also been predicted in the 2-kb upstream region. Nuclear factor-κB has been reported as negative regulator of 12-LO.33 Full transcriptional activity of RUNX1 is dependent on cofactors, such as Ets, cMyb, and p300, which can bind in cis to adjacent DNA sequences to form a complex. Clustering of these sites around RUNX1-binding sites suggests that these transcription factors may work in conjunction with RUNX1 to regulate the ALOX12. These interactions remain to be elucidated.
Thrombin-stimulated platelet 12-HETE production is decreased in our patient (Figure 1). Although the anucleate platelets possess a functional spliceosome and the ability to synthesize some proteins on activation,34,35 the bulk of the proteins present in platelets come from megakaryoytes. Therefore, down-regulation of ALOX12 at the megakaryocyte level as a result of the RUNX1 haplodeficiency and associated decreased enzyme levels provide a cogent explanation for the impaired 12-HETE production in platelets.
Deficiency of platelet 12-LO activity has been reported in patients with myeloproliferative diseases associated with thrombohemorrhagic symptoms,36,37 and in 1 report mRNA levels were also decreased.37 To our knowledge, an inherited deficiency of platelet 12-LO has hitherto not been described. A number of platelet abnormalities have been described in patients with RUNX1 haplodeficiency, including in platelet aggregation and secretion,6,8,38 dense and α granule contents,6,39 Mpl receptors,28 activation of GPIIb-IIIa,8 phosphorylation of myosin light chain and pleckstrin,8 and platelet PKC-θ.9 To this we now add decreased 12-LO activity and 12-HETE production (Figure 1). Platelet function defect in RUNX1 haplodeficiency is therefore complex and arises by multiple mechanisms mediated by distinct gene products presumably regulated by the transcription factor RUNX1. Additional studies on the targets of RUNX1 should provide insights into those specific genes.
Another major hallmark of RUNX1 haplodeficiency is familial thrombocytopenia and impaired megakaryopoiesis.5,6 Lipoxygenase products have been implicated in aspects of hematopoiesis. Lipoxygenase inhibitors block myelopoiesis induced by colony-stimulating factor and PMA, and erythropoietin-stimulated erythropoiesis.40 15-LO has been implicated in maturation of reticulocytes to mature red cells.41 12-LO is actively expressed in immature megakaryocytes.42 In human bone marrow cells, there is strong expression in megakaryocytes but not erythroblasts.27 PMA induces megakaryocytic transformation of HEL cells,24 and our promoter studies show a striking up-regulation of ALOX12 expression during such a transformation (Figure 2B). These observations suggest a potential role of 12-LO in megakaryocyte biology. Moreover, RUNX1 interacts with GATA-1 and enforced RUNX1 expression in K562 cells enhances induction of megakaryocytic-specific integrin αIIb, suggesting an important RUNX1 role in megakaryocytic lineage commitment.27 Because of altered megakaryopoiesis associated with RUNX1 mutations,5 the contribution of 12-LO to megakaryocyte biology needs to be explored.
Several studies have established the feasibility of platelet expression profiling43-45 despite the limited amount of mRNA present in these anucleate cells. Our present studies provide further proof of concept that this technology can indeed elucidate specific and novel molecular aberrations in patients with inherited platelet dysfunction, in the majority of whom we currently have no understanding of the molecular mechanisms.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Denise Tierney for assistance in manuscript preparation.
This work was supported by the National Institutes of Health (grants R01 HL85422 and R01 HL56724, A.K.R.). G.K. was supported by the National Institutes of Health (T32 training grant HL007777).
National Institutes of Health
Authorship
Contribution: G.K. performed the research, analyzed the data, and wrote the paper; G.J. and G.M. performed the research; and A.K.R. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: A. Koneti Rao, Sol Sherry Thrombosis Research Center, Temple University School of Medicine, 3400 N Broad St, OMS-300, Philadelphia, PA 19140; e-mail: koneti.rao@temple.edu.







 , supershift); lane 5, effect of nonspecific IgG; lane 6, free probe with site 3; lane 7, protein binding with recombinant RUNX1; lanes 8, competition with excess unlabeled probe; lane 9, effect of RUNX1 antibody (
, supershift); lane 5, effect of nonspecific IgG; lane 6, free probe with site 3; lane 7, protein binding with recombinant RUNX1; lanes 8, competition with excess unlabeled probe; lane 9, effect of RUNX1 antibody (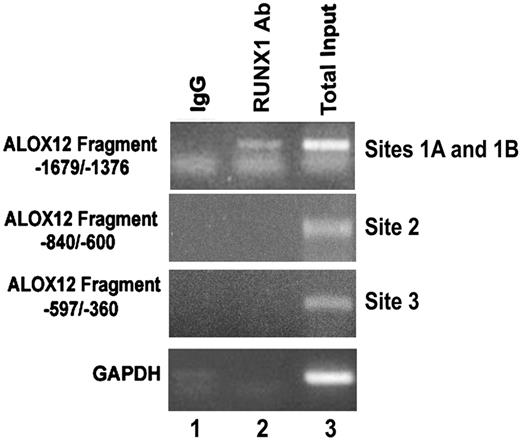

 ), or neither (□), along with ALOX12 luciferase-reporter construct (−1593/−1), wild-type, or with mutations in sites 1A, 1B, 2, or 3. Reporter activity was measured at 48 hours. Bar graphs represent activity as mean ± SD of 3 experiments. (B) Western blotting analysis of RUNX1 and actin (control) in HEL cells transfected with pCMV6 vector alone or with RUNX1-pCMV6 vector.
), or neither (□), along with ALOX12 luciferase-reporter construct (−1593/−1), wild-type, or with mutations in sites 1A, 1B, 2, or 3. Reporter activity was measured at 48 hours. Bar graphs represent activity as mean ± SD of 3 experiments. (B) Western blotting analysis of RUNX1 and actin (control) in HEL cells transfected with pCMV6 vector alone or with RUNX1-pCMV6 vector.