Abstract
Many clinically validated kinases, such as BCR-ABL, c-Kit, PDGFR, and EGFR, become resistant to adenosine triphosphate-competitive inhibitors through mutation of the so-called gatekeeper amino acid from a threonine to a large hydrophobic amino acid, such as an isoleucine or methionine. We have developed a new class of adenosine triphosphate competitive inhibitors, exemplified by HG-7-85-01, which is capable of inhibiting T315I- BCR-ABL (clinically observed in chronic myeloid leukemia), T670I-c-Kit (clinically observed in gastrointestinal stromal tumors), and T674I/M-PDGFRα (clinically observed in hypereosinophilic syndrome). HG-7-85-01 is unique among all currently reported kinase inhibitors in having the ability to accommodate either a gatekeeper threonine, present in the wild-type forms of these kinases, or a large hydrophobic amino acid without becoming a promiscuous kinase inhibitor. The distinctive ability of HG-7-85-01 to simultaneously inhibit both wild-type and mutant forms of several kinases of clinical relevance is an important step in the development of the next generation of tyrosine kinase inhibitors.
Introduction
The Abl inhibitor, imatinib mesylate (Gleevec, Novartis Pharma AG), is an effective, frontline therapy for chronic-phase chronic myeloid leukemia (CML).1,2 Imatinib resistance is rare in chronic-phase patients; however, for patients with blast crisis-phase CML or Philadelphia chromosome–positive CML, resistance is common after an initial response in the first year.3,4 Development of imatinib resistance is often the result of point mutations in the BCR-ABL kinase domain that reduce the binding affinity of imatinib by direct steric interference or by destabilizing the “DFG-out” confirmation of the activation loop that is required for high-affinity imatinib binding.5 Because of the clinical importance of these mutations, there has been intense interest in the synthesis of novel inhibitors that are able to circumvent these mutations.
Pharmacologic inhibitors of kinase activity are characterized as (1) type I, or “DFG-in” adenosine triphosphate (ATP) competitive inhibitors, which directly compete with ATP in the ATP-binding site, (2) type II, or “DFG-out” ATP competitive inhibitors, which, in addition to binding the ATP binding site, also engage an adjacent hydrophobic binding site that is only accessible when the kinase is in an inactivated configuration, and (3) non-ATP competitive inhibitors that bind at sites outside the ATP-binding site that affect the activity of the kinase.6 Clinically used second-generation Abl inhibitors, such as the type I inhibitor dasatinib and the type II inhibitor nilotinib are significantly more potent against BCR-ABL than imatinib and are active against most imatinib-resistant BCR-ABL mutants,7–9 with the exception of the T315I “gatekeeper” mutant, which is located at the boundary between the nucleotide and adjacent hydrophobic binding pocket that is exploited by type II kinase inhibitors.5,9 Both dasatinib and nilotinib make a hydrogen-bonding interaction to the side-chain hydroxyl group of T315, and their binding modes are incompatible with introduction of a large isobutyl side chain of isoleucine.10,11
Importantly, replacement of the “gatekeeper” threonine amino acid with a large hydrophobic amino acid, such as isoleucine or methionine, is a reoccurring mechanism of resistance in several clinically important tyrosine kinases. For example, the c-Kit-T670I mutation, which substantially modifies the Kit-binding pocket, occurs under the selective pressure of imatinib therapy and causes imatinib resistance in gastrointestinal stromal tumors (GISTs).12,13 PDGFRα-T674M/I in the context of the kinase oncogene FIP1LI-PDGFRα gives rise to imatinib resistance in HES,14–16 and the T790M mutation of EGFR is responsible for approximately 50% of cases of resistance to erlotinib and gefitinib.17,18
HG-7-85-01 was designed as a novel type II class kinase inhibitor that would traverse the gatekeeper position in a manner that would accommodate amino acid side chains of a variety of sizes. A cocrystal structure of HG-7-85-01 with Src demonstrates that HG-7-85-01 does indeed bind as a type II inhibitor. HG-7-85-01 inhibits the proliferation of cells expressing the major imatinib-resistant gatekeeper mutants, BCR-ABL-T315I, Kit-T670I, and PDGFRα-T674M/I, as well as Src-T341M/I. In each case, inhibition of proliferation is mediated by selective inhibition of kinase activity, induction of apoptosis, and inhibition of cell-cycle progression.
Methods
Cell lines and cell culture
A detailed description of the cell lines used in this study is provided in the supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Chemical compounds and biologic reagents
HG-7-85-01 was synthesized in the laboratory of N.G. Nilotinib and imatinib were synthesized by Novartis Pharma AG. Compounds were initially dissolved in dimethyl sulfoxide to make 10mM stock solutions and then were serially diluted to obtain final concentrations for in vitro experiments.
Effects of HG-7-85-01 on phosphorylation status of target kinases
Z-lyte format.
The Z-lyte Enzymatic Kinase Assay format (Invitrogen) was carried out using the SelectScreen Kinase Profiling Service (Invitrogen Discovery Sciences). HG-7-85-01 was assayed following the detailed procedures outlined in the SelectScreen. Assay protocol and conditions can be located at www.invitrogen.com/kinase profiling.
Radioenzymatic format.
In vitro kinase assays were carried out as previously described.19
Colony assays
Human bone marrow cells were obtained from normal donors after obtaining informed consent in accordance with the Declaration of Helsinki on a Dana-Farber Institutional Review Board-approved protocol. Mononuclear cells were isolated from normal bone marrow by density gradient centrifugation through Ficoll-Plaque Plus (GE Healthcare) at 824g for 30 minutes, followed by 2 washes in 1 times phosphate-buffered saline. Normal human bone marrow was analyzed in a colony assay: plates of 5 × 104 cells in “complete” methylcellulose medium containing recombinant cytokines (contents: fetal bovine serum, recombinant human [rh] stem cell factor, rh granulocyte-macrophage colony-stimulating factor, rh interleukin-3 [IL-3], bovine serum albumin, methylcellulose in Iscove modified Dulbecco medium, 2-mercaptoethanol, rh erythropoietin, L-glutamine; MethoCult GFH4434, StemCell Technologies) were prepared. The plates also contained HG-7-85-01 at the indicated concentrations. The plates were incubated at 37°C in 5% CO2 for more than 1 week, and then myeloid and erythroid colonies (early progenitors with erythroid and myeloid components: colony-forming units-granulocyte-macrophage, colony-forming units-erythroid, burst-forming units-erythroid, and colony-forming units-granulocyte erythrocyte macrophage megakaryocyte) were counted on an inverted microscope.
Human bone marrow samples obtained from normal healthy donors were also investigated for responsiveness to HG-7-85-01 in liquid culture (Iscove modified Dulbecco medium, supplemented with 20% fetal calf serum, l-glutamine, and a cytokine cocktail containing rh Fms-like tyrosine kinase 3 ligand [100 ng/mL], rh stem cell factor [100 ng/mL], rh IL-3 [20 ng/mL], rh IL-6 [20 ng/mL], and rh granulocyte-macrophage colony-stimulating factor [10 ng/mL]) in the presence of different concentrations of drug.
Human CML peripheral blood samples were investigated for responsiveness to HG-7-85-01 in liquid culture (Iscove modified Dulbecco medium, supplemented with 20% fetal calf serum and l-glutamine) in the presence of different concentrations of drug. A fresh aliquot of the T315I-positive peripheral blood patient sample (“Cml-1”) was treated with HG-7-85-01, and results were later confirmed using a freeze-thawed aliquot in a subsequent liquid proliferation assay. The second of the 2 CML patient samples analyzed for responsiveness to HG-7-85-01 (“Cml-2”) was a sample freeze-thawed after prolonged storage in liquid nitrogen. Both CML samples had 80% to 90% blasts at the time of the assays.
Antibodies and immunoblotting
A detailed description of the antibodies used in this study is provided in the supplemental Methods. Protein lysate preparation, immunoblotting, and immunoprecipitation were carried out as previously described.20
Proliferation studies, cell-cycle analysis, and apoptosis assay
Growth and viability assays were carried out as previously described.20 A detailed description is provided in the supplemental Methods.
Drug combination studies
Drug combination studies were carried out as previously described.8 A detailed description is provided in the supplemental Methods.
Bioluminescent imaging
Bioluminescence imaging was carried out as previously described.21 Detailed descriptions are provided in figure legends and in the supplemental Methods.
Protein expression, crystallization, and structure determination
Chicken c-Src kinase domain was expressed and purified from bacteria for crystallographic purposes as previously described.22 Detailed descriptions of the crystallization and the structure determination are provided in the supplemental Methods and supplemental Table 1.
Results
Discovery of HG-7-85-01
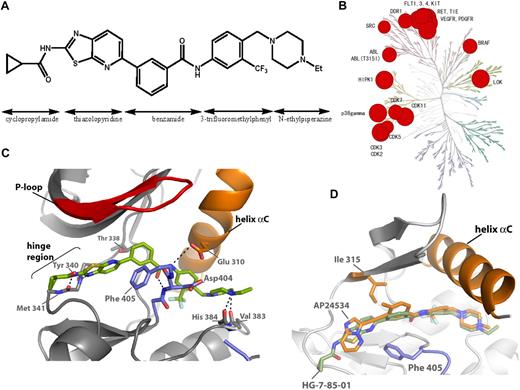
HG-7-85-01 was designed as a hybrid between the type I inhibitor dasatinib and the type II inhibitor nilotinib (Figure 1A; supplemental Figure 1A).23 Specifically, a superposition of the Abl-bound conformation of dasatinib (PDB code, 2GQG) and nilotinib (PDB code, 2CS9) guided the choice of how to connect the aminothiazole hinge-interacting motif of dasatinib with the N-(3-(trifluoromethyl)phenyl)benzamide substructure of nilotinib, which is known to be responsible for inducing the “DFG-out” flip that is characteristic of type II kinase inhibitors; details of synthesis are presented in the supplemental Methods (synthesis section) and supplemental Figure 2. Iterative synthesis and cellular assays of 120 analogs resulted in the selection of HG-7-85-01 as the most promising candidate for further analysis. Kinase selectivity was assessed using KINOMEscan (Ambit Biosciences), a high-throughput method for screening kinase inhibitors against a panel of 353 human kinases.24 The technology enables compounds of interest to be assayed for their ability to compete for the binding to the ATP site of a panel of 353 kinases, each fused to a proprietary tag. The quantity of each kinase bound to an immobilized, ATP site-directed ligand was measured in the presence and absence of a 10μM concentration of HG-7-85-01 (Figure 1B). Kinases that displayed greater than 90% displacement relative to the dimethyl sulfoxide control were subject to determination of a dissociation constant (Kd). These studies revealed that a number of wild-type and mutant forms of kinases, such as BCR-ABL, PDGFRα/β, VEGFR, FLT3, Ret, Tie-2, Kit, Src, DDR1, and b-raf, exhibited potent binding to HG-7-85-01 (Figure 1B; supplemental Figure 2). As there is no simple relationship between a Kd determined by these methods and a biochemical or cellular 50% inhibitory concentration (IC50), each putative interaction must be validated by an independent experimental method. To compare the selectivity to other Abl inhibitors, we calculated selectivity scores (S scores), which demonstrate that HG-7-85-01 (S = 0.056) is intermediate in selectivity relative to imatinib (S = 0.031) and dasatinib (S = 0.16).24
Discovery of HG-7-85-01. (A) Chemical structure of HG-7-85-01 with substructure names indicated. (B) Kinase selectivity of HG-7-85-01 based on screening 400 kinases. Kinases where significant binding affinity was detected at 10μM were retested in dose-response format to determine a Kd. The size of the red circle is proportional to Kd. Numerical dissociation constants are listed in the supplemental Table. (C) X-ray crystal structure of the c-Src wt/HG-7-85-01 complex. HG-7-85-01 is shown in green carbon atoms, P-loop of the protein in red, helix α C in orange, and activation loop in blue. Dotted lines indicate H-bonds. (D) Accommodation of gatekeeper Thr315Ile mutation by HG-7-85-01 and AP24534. The kinase domains of the Src wt/HG-7-85-01 complex and the Abl Thr315Ile/AP24534 complex were aligned in PyMOL. The protein structures are highly conserved around the gatekeeper residues, and for clarity only the structure of Abl Thr 315, AP24534 (orange carbon atoms), and HG-7-85-01 (green carbon atoms) is displayed.
Discovery of HG-7-85-01. (A) Chemical structure of HG-7-85-01 with substructure names indicated. (B) Kinase selectivity of HG-7-85-01 based on screening 400 kinases. Kinases where significant binding affinity was detected at 10μM were retested in dose-response format to determine a Kd. The size of the red circle is proportional to Kd. Numerical dissociation constants are listed in the supplemental Table. (C) X-ray crystal structure of the c-Src wt/HG-7-85-01 complex. HG-7-85-01 is shown in green carbon atoms, P-loop of the protein in red, helix α C in orange, and activation loop in blue. Dotted lines indicate H-bonds. (D) Accommodation of gatekeeper Thr315Ile mutation by HG-7-85-01 and AP24534. The kinase domains of the Src wt/HG-7-85-01 complex and the Abl Thr315Ile/AP24534 complex were aligned in PyMOL. The protein structures are highly conserved around the gatekeeper residues, and for clarity only the structure of Abl Thr 315, AP24534 (orange carbon atoms), and HG-7-85-01 (green carbon atoms) is displayed.
Biochemical kinase assays were performed on a panel of 17 kinases using the invitrogen Z-lyte format25 (supplemental Figure 2C) and a panel of 39 kinases using a radioenzymatic format (supplemental Figure 2D). HG-7-85-01 exhibited strong inhibition of gatekeeper mutant T315I of Abl (IC50 = 0.003μM) and inhibited KDR (IC50 = 0.02μM) and RET (IC50 = 0.03μM), whereas there is only weak or no inhibition of other kinases (IC50 > 2μM). Interestingly, the biochemical assays revealed no inhibition of CDK1, 5, and 7 despite significant binding being detected to these kinases using the Ambit approach. This discrepancy is potentially the result of the absence of the cyclin component in the ambit assay, which may be responsible for preventing a type II inhibitor from effectively inhibiting a CDK-cyclin complex. These results confirmed that HG-7-85-01 is a relatively selective inhibitor of wild-type and T315I Abl at a biochemical level that also inhibits KDR, RET, DDR1/2. Further cellular assays will be required to ascertain whether any of these additional kinase targets are efficiently inhibited in a cellular context.
Structural basis for recognition of Src by HG-7-85-01
To investigate the binding mode of HG-7-85-01, we cocrystallized HG-7-85-01 with Src. The general binding mode of HG-7-85-01 to Src is similar to that of imatinib and the DSA series of Src and Abl inhibitors.22,26 HG-7-85-01 binds to Src in the “DFG-out” inactive conformation and forms 5 hydrogen-bonding interactions: 2 hydrogen bonds to the hinge region (Figure 1C; from the cyclopropyl amide NH to the backbone carbonyl of M341 and between the thiazole N and backbone NH of Y340) and a pair of hydrogen bonds between the benzamide carbonyl and the backbone NH of D404 of the “DFG-motif” and the benzamide NH and side-chain carboxylate of E310 from the αC-helix and hydrogen bonds from the presumably protonated distal piperazine nitrogen and the backbone carbonyls of V383 and H384. The most obvious difference between the binding mode of HG-7-85-01 to Src relative to the Src/imatinib26 and Abl/dasatinib complexes27 is the lack of a hydrogen bond between HG-7-85-01 and the side-chain hydroxyl of the gatekeeper residue T338. This loss of a hydrogen bond to the side chain of T338 is compensated in HG-7-85-01 by the formation of an extra hydrogen bond to the backbone of M341 in the hinge region of the kinase (Figure 1C; supplemental Figure 1B). A similar exchange of a hydrogen bond to the mutation prone T338 side chain with a hydrogen bond to the nonmutatable backbone has been seen in the complexes of Src with the DSA series of compounds.22 Recently, another type II inhibitor, AP24534, which has potent activity against T315I was crystallized in complex T315I-Abl.28 A superposition of the AP24534-T315I Abl complex with the HG-7-85-01-Src complex demonstrates that both compounds use a very similar trajectory to traverse the larger gatekeeper residue (Figure 1D). However, the detailed interactions of the 2 inhibitors with the ATP site differ: whereas HG-7-85-01 forms 2 hydrogen bonds with the hinge segment, AP24534 only makes a single hydrogen bond. To further corroborate the ability of HG-7-85-01 to accommodate a large gatekeeper residue, a model of the T338I mutation in Src/HG-7-85-01 complex was prepared and shown to lack carbon-carbon distances of less than 2.8 Å between inhibitor and the side chain of I338 (supplemental Table 1; supplemental Figure 1C). These alignment and modeling studies indicate that it is possible to accommodate a larger side chain at the gatekeeper position without major alterations to the inhibitor-binding mode. Further crystallographic analysis of HG-7-85-01 with T338M Src or T315I Abl will be required to corroborate this prediction. The broader kinase specificity of HG-7-85-01 compared with imatinib or nilotinib is probably the result of the same factors that give HG-7-85-01 activity toward the gatekeeper mutant. First, the hydrogen bonding of HG-7-85-01 to the backbone of the kinase is less sequence specific than the hydrogen bonding of nilotinib and imatinib to the T338 side chain and can therefore be provided by more kinases. Second, the cyclopropylamide of HG-7-85-01 is required to provide the hydrogen bonding to the backbone of the hinge region but makes the compound larger and less hydrophobic than the equivalent regions of imatinib or nilotinib. In the complex of Abl with imatinib or nilotinib, the P-loop of the kinase kinks toward the C-lobe and shields hydrophobic regions of nilotinib and imatinib effectively from solvent exposure. The cyclopropylamide of HG-7-85-01 is probably preventing the P-loop of Abl from accessing this unusually kinked conformation, and it has less of a requirement for solvent protection because it is more hydrophilic than imatinib or nilotinib. This lack of a requirement for solvent protection was used to explain the activity of the DSA compounds against imatinib resistance mutations located in the P-loop of Abl kinase21 and could explain the activity of HG-7-85-01 against these mutations.
Effects of HG-7-85-01 on growth of nonmutated and mutant BCR-ABL–expressing cells and BCR-ABL kinase activity
HG-7-85-01 potently and selectively inhibited the proliferation of 32D- and Ba/F3 cells expressing nonmutant BCR-ABL and the BCR-ABL-T315I gatekeeper mutant (Figure 2A; Table 1; supplemental Figure 3A-G). HG-7-85-01 showed higher potency against nonmutant BCR-ABL and BCR-ABL-T315I (IC50 = 0.06-0.14μM) compared with the majority of a panel of imatinib/nilotinib-resistant BCR-ABL point mutants tested (IC50 = 0.5-1μM; Figure 2; Table 1). The relative imatinib/nilotinib resistance of BCR-ABL-T315I was confirmed both in vitro and in vivo (supplemental Figure 4). Coculture of nonmutant and BCR-ABL point mutant-positive cells with HG-7-85-01 and WEHI (used as a source of IL-3) led to full cytoprotection, and no inhibitory effect on parental cells was observed at comparable concentrations of HG-7-85-01 (Figure 2; supplemental Figure 3A-E). In addition to potently inhibiting the growth of murine cells expressing BCR-ABL, HG-7-85-01 was also effective in inhibiting the growth of 2 human cell lines derived from blast crisis CML patients, K562 and KU812F (supplemental Figure 3H-I).
Effects of HG-7-85-01 on growth of nonmutated and mutant BCR-ABL–expressing cells and BCR-ABL kinase activity. (A) Determination of IC50 values for HG-7-85-01 against parental Ba/F3 cells, Ba/F3.p210 cells, and Ba/F3-T315I cells. (B) ABL immunoprecipitation and pTYR and ABL immunoblots, respectively, of whole cell lysates prepared from Ba/F3.p210 cells treated for 2 hours with vehicle or HG-7-85-01 (0.1, 1μM). (C-D) Effects of HG-7-85-01 on proliferation of imatinib- and nilotinib-resistant BCR-ABL point mutant-expressing cells; 2- to 2.5-day treatments of BCR-ABL point mutant-expressing Ba/F3 cells with HG-7-85-01 in the absence (C) or presence (D) of IL-3. HG-7-85-01 treatment of parental Ba/F3 cells in the presence of IL-3 is shown in panels A and B as a control. Error bars represent the SEM for proliferation studies performed in duplicate. Assays for Ba/F3 cells expressing M351T, F317L, and F486S were carried out for 2 days. Assays for Ba/F3 cells expressing all other BCR-ABL point mutants were carried out for 2.5 days.
Effects of HG-7-85-01 on growth of nonmutated and mutant BCR-ABL–expressing cells and BCR-ABL kinase activity. (A) Determination of IC50 values for HG-7-85-01 against parental Ba/F3 cells, Ba/F3.p210 cells, and Ba/F3-T315I cells. (B) ABL immunoprecipitation and pTYR and ABL immunoblots, respectively, of whole cell lysates prepared from Ba/F3.p210 cells treated for 2 hours with vehicle or HG-7-85-01 (0.1, 1μM). (C-D) Effects of HG-7-85-01 on proliferation of imatinib- and nilotinib-resistant BCR-ABL point mutant-expressing cells; 2- to 2.5-day treatments of BCR-ABL point mutant-expressing Ba/F3 cells with HG-7-85-01 in the absence (C) or presence (D) of IL-3. HG-7-85-01 treatment of parental Ba/F3 cells in the presence of IL-3 is shown in panels A and B as a control. Error bars represent the SEM for proliferation studies performed in duplicate. Assays for Ba/F3 cells expressing M351T, F317L, and F486S were carried out for 2 days. Assays for Ba/F3 cells expressing all other BCR-ABL point mutants were carried out for 2.5 days.
CML peripheral blood samples consisting of 80% to 90% blasts were highly responsive to HG-7-85-01 (supplemental Figure 3K-L). One of these samples was T315I-positive (supplemental Figure 3K). In contrast, normal bone marrow obtained from a human donor was considerably less responsive to HG-7-85-01, as demonstrated by both liquid culture assay and bone marrow colony assay (supplemental Figure 3J and supplemental 3M, respectively), with little drug effects observed at up to 0.5μM. These results suggest selectivity of HG-7-85-01 for BCR-ABL–positive cells versus normal cells and argue in favor of potential clinical usefulness.
HG-7-85-01 inhibited BCR-ABL kinase activity in a concentration-dependent manner (Figure 2B), suggesting selective targeting of the BCR-ABL kinase as the mechanism of action of HG-7-85-01. Inhibition of BCR-ABL autophosphorylation is not accompanied by a decrease in levels of BCR-ABL protein, suggesting that drug inhibition is selective for kinase activity and drug effects do not include decreasing levels of BCR-ABL protein expression. In contrast, levels of BCR-ABL protein appear to increase in a drug concentration-dependent manner, which is a finding consistent with previous studies carried out with ABL inhibitors, such as imatinib and nilotinib.8 These results suggest a possible drug-protein complex formation and stabilization, concurrent with inhibition of kinase activity.
In vivo effects of HG-7-85-01 combined with nilotinib against BCR-ABL–positive leukemia
The combination of more than one Abl inhibitor in the treatment of imatinib-resistant disease may have beneficial therapeutic value because clonal resistance could potentially be overcome by combining 2 agents with different resistance profiles. We investigated the ability of HG-7-85-01 to positively combine with nilotinib. HG-7-85-01 was observed in vitro to positively combine with imatinib and nilotinib, respectively, against BCR-ABL–expressing cells (Figure 3A-B). In contrast, HG-7-85-01 combined with imatinib or nilotinib against T315I-expressing cells in vitro did not result in significantly more cell killing than either agent alone (supplemental Figure 5).
Combination studies between nilotinib and HG-7-85-01 against BCR-ABL–positive leukemia. (A) In vitro combination study: 2-day treatment of 32D.p210-luc+ cells with imatinib, HG-7-85-01, or a combination. Calcusyn results: 25% effective dose (ED25) = 0.96795 (nearly additive); ED50 = 0.85587 (slight synergism); ED75 = 0.78257 (moderate synergism); and ED90 = 0.73747 (moderate synergism). (B) Two-day treatment of 32D.p210-luc+ cells with nilotinib, HG-7-85-01, or a combination. (C-D) In vivo analysis of the combination of nilotinib and HG-7-85-01. (C) Plotted are bioluminescence values shown as percentage of baseline control values. In vivo combination study between HG-7-85-01 and nilotinib against nonmutant BCR-ABL: Day 8 after intravenous injection of 800 000 32D.p210-luc+ cells/mouse. Vehicles (n = 4): 2 vehicles were treated 2 times daily, 2 vehicles were treated 1 time daily. Nilotinib treatment group: Treatment mice were administered 1 time daily 20 mg/kg nilotinib (n = 4), 1 time daily 100 mg/kg HG85 (n = 2), or 1 time daily a combination of nilotinib and HG85 at the aforementioned doses (n = 2). Baseline imaging was performed 1 day after intravenous injection of 32D.p210-luc+ cells. Mice were treated for a total of 7 days before imaging. (D) Values plotted are percentage spleen/total weights. Mice were killed and spleens dissected 21 days after the last imaging day (day 8 after intravenous injection of cells).
Combination studies between nilotinib and HG-7-85-01 against BCR-ABL–positive leukemia. (A) In vitro combination study: 2-day treatment of 32D.p210-luc+ cells with imatinib, HG-7-85-01, or a combination. Calcusyn results: 25% effective dose (ED25) = 0.96795 (nearly additive); ED50 = 0.85587 (slight synergism); ED75 = 0.78257 (moderate synergism); and ED90 = 0.73747 (moderate synergism). (B) Two-day treatment of 32D.p210-luc+ cells with nilotinib, HG-7-85-01, or a combination. (C-D) In vivo analysis of the combination of nilotinib and HG-7-85-01. (C) Plotted are bioluminescence values shown as percentage of baseline control values. In vivo combination study between HG-7-85-01 and nilotinib against nonmutant BCR-ABL: Day 8 after intravenous injection of 800 000 32D.p210-luc+ cells/mouse. Vehicles (n = 4): 2 vehicles were treated 2 times daily, 2 vehicles were treated 1 time daily. Nilotinib treatment group: Treatment mice were administered 1 time daily 20 mg/kg nilotinib (n = 4), 1 time daily 100 mg/kg HG85 (n = 2), or 1 time daily a combination of nilotinib and HG85 at the aforementioned doses (n = 2). Baseline imaging was performed 1 day after intravenous injection of 32D.p210-luc+ cells. Mice were treated for a total of 7 days before imaging. (D) Values plotted are percentage spleen/total weights. Mice were killed and spleens dissected 21 days after the last imaging day (day 8 after intravenous injection of cells).
We assessed the pharmacologic properties of HG-7-85-1 in mice and rats after oral and intravenous delivery before performing the murine tumor models. HG-7-85-01 was demonstrated to have limited oral bioavailability (BAV% F mouse = 5%, rat = 19%), a moderate half-life (t1/2 mouse = 1.1 hours, rat = 5.8 hours), a relatively low maximal serum concentration (Cmax mouse = 106 ng/mL at 10 mg/kg, rat = 292 ng/mL at 2 mg/kg), and a relatively high clearance (Cl mouse = 23 mL/minute per kilogram, rat = 13 mL/minute per kilogram; supplemental Table 2). However, these pharmacokinetic properties were deemed sufficient for performing murine efficacy studies.
Using a bioluminescent model of CML, the combination of HG-7-85-01 and nilotinib in vivo was investigated. Combined treatment of mice harboring BCR-ABL–positive leukemia with nilotinib and HG-7-85-01 led to a greater suppression of leukemic cell growth in mice than either drug alone as assessed by bioluminescence and percentage spleen/total weights (Figure 3C-D).
Effects of HG-7-85-01 on Kit, PDGFRα, and PDGFRβ gatekeeper mutants
HG-7-85-01 potently inhibited the proliferation of Ba/F3 cells expressing the Kit-T670I gatekeeper mutation (Ba/F3- Kit-T670I), with the majority of cells killed at concentrations between 0.5 and 1μM and no effect on parental Ba/F3 cells (Figure 4A; Table 1). In contrast, Ba/F3-Kit-T670I cells were significantly less responsive to nilotinib, with 50% cell proliferation inhibited at higher concentrations between 2 and 4μM (Figure 4A). The relative imatinib/nilotinib resistance of the Kit-T670I mutant was confirmed in vitro (Figure 4B) and in vivo (supplemental Figure 6). HG-7-85-01 inhibited Kit phosphorylation in a concentration-dependent manner (Figure 4C). HG-7-85-01 treatment led to a G0G1 arrest (Table 2) and induction of apoptosis (Table 3).
Effects of HG-7-85-01 on Kit, PDGFRα, and PDGFRβ gatekeeper mutants. (A) Approximately 3-day treatment of parental Ba/F3 cells and Ba/F3-Kit-T670I cells with nilotinib and HG-7-85-01, respectively. Error bars represent the SEM for proliferation studies performed in triplicate. (B) Comparison of the effects of imatinib (1000nM) and nilotinib (1000nM) on Ba/F3-T670I-luc+ cells. (C) Effect of HG-7-85-01 on gatekeeper mutant Kit-T670I phosphorylation: immunoblot of protein lysates prepared from Ba/F3-delWK+T670I cells treated for 2 hours with various concentrations of HG-7-85-01 (0-1μM). (D) Approximately 3-day treatment of T674M- and T674I-PDGFRα gatekeeper variants expressed in Ba/F3 with HG-7-85-01. (E) Comparison of the effects of HG-7-85-01, imatinib, and nilotinib on Ba/F3-T674M-PDGFRα cells. Error bars represent the SEM for proliferation studies performed in duplicate. (F) Effects of HG-7-85-01 on total cellular tyrosine phosphorylation in PDGFRα-T674M–expressing cells.
Effects of HG-7-85-01 on Kit, PDGFRα, and PDGFRβ gatekeeper mutants. (A) Approximately 3-day treatment of parental Ba/F3 cells and Ba/F3-Kit-T670I cells with nilotinib and HG-7-85-01, respectively. Error bars represent the SEM for proliferation studies performed in triplicate. (B) Comparison of the effects of imatinib (1000nM) and nilotinib (1000nM) on Ba/F3-T670I-luc+ cells. (C) Effect of HG-7-85-01 on gatekeeper mutant Kit-T670I phosphorylation: immunoblot of protein lysates prepared from Ba/F3-delWK+T670I cells treated for 2 hours with various concentrations of HG-7-85-01 (0-1μM). (D) Approximately 3-day treatment of T674M- and T674I-PDGFRα gatekeeper variants expressed in Ba/F3 with HG-7-85-01. (E) Comparison of the effects of HG-7-85-01, imatinib, and nilotinib on Ba/F3-T674M-PDGFRα cells. Error bars represent the SEM for proliferation studies performed in duplicate. (F) Effects of HG-7-85-01 on total cellular tyrosine phosphorylation in PDGFRα-T674M–expressing cells.
The PDGFRα-T674M and PDGFRα-T674I gatekeeper mutant variants were highly responsive to HG-7-85-01, with almost complete killing of cells at 25nM and significant IL-3 rescue (Figure 4D; Table 1; supplemental Figure 7). In contrast, the proliferation of Ba/F3 cells expressing the imatinib- and nilotinib-resistant PDGFRβ-T681I gatekeeper mutant was not inhibited at concentrations up to 1μM (supplemental Figure 7; Table 1). The PDGFRα-T674M mutant was highly resistant to imatinib yet responsive to HG-7-85-01 and nilotinib (Figure 4E). HG-7-85-01 inhibited PDGFR phosphorylation (Figure 4F), and drug treatment led to a G0G1 arrest (Table 2) and induction of apoptosis (Table 3).
The ability of HG-7-85-01 to inhibit the proliferation of other PDGFR mutations was investigated, including chronic myelomonocytic leukemia-associated TEL/PDGFRβ, and the imatinib-resistant GIST-associated PDGFRα mutants D842V (point mutation in the kinase activation loop domain) and V561D (point mutation in the juxtamembrane domain). HG-7-85-01 potently and selectively killed Ba/F3 cells expressing TEL/PDGFRβ and, to a lesser extent, D842V, with no effect against the V561D mutant (Table 1; and data not shown).
The ability of HG-7-85-01 to inhibit the proliferation of Ba/F3 cells transformed with nonmutated Src or either of the T341I and T341M Src gatekeeper mutants14 was investigated. HG-7-85-01 inhibited the proliferation of Ba/F3 cells transformed with human c-Src (half-maximal effective concentration [EC50] = 190nM), T338I Src (EC50 = 290nM), and T338M Src (EC50 = 150nM; chicken c-Src numbering). Wild-type Src was confirmed to be highly sensitive to the potent Src inhibitor dasatinib (EC50 < 10nM), whereas both gatekeeper mutants were insensitive (EC50 > 10μM). Likewise, neither nonmutated nor mutated Src showed sensitivity to imatinib (EC50 > 10μM).
Differential responsiveness of exon 9 and exon 11 kit mutations to HG-7-85-01
Exon 9 Kit primary mutations are reportedly less sensitive to imatinib than exon 11 Kit primary mutations.29 HG-7-85-01 was thus tested for its ability to inhibit a panel of exon 9 and exon 11 primary Kit mutant-expressing lines, with or without a variety of exon 13, exon 14, and exon 17 secondary Kit mutants. Similar to imatinib, whereas HG-7-85-01 potently inhibited the proliferation of exon 11 Kit mutant-expressing cells, exon 9 kit mutant-expressing cells were significantly less responsive (Figure 5A-C). The inhibitory effect of HG-7-85-01 was selective, as shown by partial or complete WEHI (IL-3) protection of Kit mutants in the presence of HG-7-85-01 (Figure 5D).
Differential responsiveness of exon 9 and exon 11 Kit mutations to HG-7-85-01. (A) Comparison of HG-7-85-01 responsiveness of primary Kit mutants, exon 9 kit (insAY) and exon 11 Kit (delWK, V559), with or without secondary Kit mutants, exons 13, 14, and 17 (2.5- to 3-day treatments). (B) HG-7-85-01 treatment of delWK (± IL-3), delWK + V654A (−IL-3), and delWK+T670I (−IL-3) (2.5-day treatment). (C) HG-85-01 treatment of V559D and delWK + Y823D (−IL-3) (2.5-day treatment). (D) HG-7-85-01 treatment of Kit mutants in the absence and presence of IL-3. Error bars represent the SEM for proliferation studies performed in duplicate.
Differential responsiveness of exon 9 and exon 11 Kit mutations to HG-7-85-01. (A) Comparison of HG-7-85-01 responsiveness of primary Kit mutants, exon 9 kit (insAY) and exon 11 Kit (delWK, V559), with or without secondary Kit mutants, exons 13, 14, and 17 (2.5- to 3-day treatments). (B) HG-7-85-01 treatment of delWK (± IL-3), delWK + V654A (−IL-3), and delWK+T670I (−IL-3) (2.5-day treatment). (C) HG-85-01 treatment of V559D and delWK + Y823D (−IL-3) (2.5-day treatment). (D) HG-7-85-01 treatment of Kit mutants in the absence and presence of IL-3. Error bars represent the SEM for proliferation studies performed in duplicate.
The insAY Kit mutants were found to be generally resistant to imatinib, nilotinib, and HG-7-85-01, with the one exception being approximately 50% killing of the insAY + T670I Kit mutant at 1μM HG-7-85-01 (supplemental Figure 8A). The exon 11 (V559D) Kit mutant was approximately equally sensitive to all 3 inhibitors, whereas the exon 11 (delWK) Kit mutant was insensitive to imatinib and nilotinib, although potently inhibited by HG-7-85-01 (supplemental Figure 8B). The differential responsiveness of the exon 9 and exon 11 Kit mutants could not be attributed to significant differences in overall levels of Kit expression (supplemental Figure 9). HG-7-85-01 was also tested against wild-type Kit-expressing Ba/F3 cells prestimulated with Kit ligand and cultured under “starvation” (IL-3 deprivation) conditions; HG-7-85-01 displayed an IC50 between 0.1 and 1μM against these cells (supplemental Figure 8C).
Effects of HG-7-85-01 on EGFR-T790M gatekeeper mutant
The EGFR-T790M is a gatekeeper mutation associated with gefitinib and erlotinib (EGFR inhibitor)–resistant small cell lung cancer.18 Consistent with the Ambit kinase screening results that had suggested that EGFR is not a potential target of HG-7-85-01, no activity was observed with HG-7-85-01 against cells expressing this mutant (supplemental Figure 10).
Effects of HG-7-85-01 on imatinib-resistant cells overexpressing BCR-ABL and MDR1
HG-7-85-01 showed activity against imatinib-sensitive LAMA84S and imatinib-resistant LAMA84R cells, the latter made resistant to imatinib and found to overexpress BCR-ABL and MDR130,31 (supplemental Figure 11). HG-7-85-01 was also able to inhibit the growth of LAMA84S cells cocultured with conditioned media from LAMA84R cells, which renders LAMA84S cells resistant to imatinib (supplemental Figure 11).
Discussion
Relapse in CML patients treated with imatinib is often the result of the development of resistance caused by point mutations within the kinase domain of BCR-ABL that diminish the binding affinity of imatinib.5 In up to 50% of cases, imatinib resistance is attributed to clonal expansion of mutant BCR-ABL–positive cells, with more than 40 BCR-ABL kinase domain mutants identified at this point. This suggests a need for the development of new BCR-ABL inhibitors that have a different binding mode or a completely different mode of action from imatinib and that can overcome imatinib resistance caused by BCR-ABL point mutations.
Nilotinib and dasatinib have been approved for the treatment of imatinib-resistant CML patients and have shown considerable promise in the clinic. However, the inability of nilotinib and dasatinib to override the highly imatinib-resistant T315I mutation has spurred the development of third-generation inhibitors, which are able to inhibit cells expressing T315I.32
HG-7-85-01 is an early example of a type II ATP competitive inhibitor that opens up a new chemical space in the field of kinase inhibitor development because of its ability to potently inhibit kinases that possess either a threonine or a larger hydrophobic residue, such as isoleucine, methionine, or phenylalanine. Despite this tolerance at the gatekeeper position, HG-7-85-01 is not a promiscuous kinase inhibitor and, for example, is a considerably more selective inhibitor than dasatinib or AP24534, which displays a strong preference for kinases with a threonine gatekeeper residue.24 Also in contrast to HG-7-85-01, the majority of T315I inhibitors that are reported in the literature are either type I ATP-competitive inhibitors (ie, SGX-70393, VX-680, PPY-A, and PHA-739358) or substrate-site targeted inhibitors (ie, ON012380; supplemental Table 3). Several other type II kinase inhibitors have recently been reported to be capable of inhibiting T315I Bcr-abl: the DSA-compound series,22 AP24163,33 AP24534,28 and compound 14.14 The DSA and AP24163 compounds were primarily developed as dual Src/Abl inhibitors and exhibit only moderate potency toward T315I Bcr-Abl (EC50 = 300-500nM), and their pharmacologic potential against gatekeeper mutants of cKit and PDGFR has not been reported. Compound 1422 is a very promiscuous kinase inhibitor and does not possess the restricted selectivity profile of the HG-7-85-01 compound class. AP24534 exhibits excellent potency against wild-type (IC50 = 0.5nM) and T315I Bcr-Abl (IC50 = 11nM) and reasonable kinase selectivity and is currently in a phase 1 clinical study for imatinib-resistant CML. In addition, whereas HG-7-85-01 is a selective tyrosine kinase inhibitor, many of the other reported T315I inhibitors affect normal as well as malignant cells, through targeting of enzymatic pathways that regulate cell survival (ie, HSP90 and PP2A), interaction with substrate-binding sites of multiple kinases, and targeting of aurora kinases, which are involved in normal cell signaling. Such mechanisms may translate into toxicity problems, thus suggesting the need for improved third-generation inhibitors that can specifically override T315I-associated resistance.
A potentially valuable feature of third-generation Abl inhibitors would be an ability to synergize with other Abl kinase inhibitors so that they could be used simultaneously in highly resistant patients or to prevent initial emergence of resistance. Mice harboring nonmutated BCR-ABL treated with a combination of HG-7-85-01, and the type II Abl inhibitor nilotinib showed less tumor burden than mice treated with vehicle or either agent alone.
In addition to BCR-ABL–positive CML, HG-7-85-01 is a potent inhibitor of the Kit-T670I gatekeeper mutation, as well as several secondary Kit mutations that play a key role in imatinib-resistant GIST. Although the majority of GIST patients initially respond to imatinib, most patients progress at multiple sites within 2 to 5 years of starting therapy. Mutations in the kinase domain of Kit are associated with the development of imatinib resistance in 46% to 67% of patients, such as Kit-V654A and Kit-T670I.13 These mutations stabilize the active conformation of Kit, thus rendering the kinase imatinib resistant.
Several agents have been reported to inhibit the Kit-T670I gatekeeper mutant, including sunitinib (SU11248; brand name Sutent), which has been Food and Drug Administration approved for GIST, and Sorafenib (BAY 43-9006; brand name Nexavar), which is in clinical trials for imatinib- and sunitinib-resistant GIST. Sunitinib inhibits the proliferation of delWK557-558/T670I–expressing cells with an EC50 between 50 and 100nM34 ; this is compared with an EC50 for HG-7-85-01 of approximately 25nM for delWK557-558/T670I. Sorafenib inhibits the proliferation of Kit-T670I–expressing cells with an EC50 of 1μM35,36 ; this is compared with an EC50 for HG-7-85-01 of approximately 250nM for T670I. Finally, PKC412 (midostaurin) has shown activity against delWK557-558 + T670I.37
HG-7-85-01 also shows activity against the imatinib-resistant PDGFRα-T674M/I gatekeeper mutant variants. Agents that are active against PDGFRα-T674I include sorafenib, novel inhibitors Ki1150238 and EXEL-0862,39 nilotinib,40,41 and PKC412.42 Sorafenib is probably of limited utility against the T674I mutant, however, as sorafenib treatment of FIP1L1-PDGFRα T674I CEL causes rapid emergence of FIP1L1-PDGFRα D842V.43 D842V is highly resistant to sorafenib, imatinib, dasatinib, and PKC41243 ; however, it is sensitive to HG-7-85-01.
HG-7-85-01 targets a distinctive broad spectrum of tyrosine kinases that are integral to the pathogenesis of both chronic and acute myeloid leukemias, as well as GIST and hypereosinophilic syndrome. The versatility of HG-7-85-01 is illustrated by its ability to inhibit multiple tyrosine kinase targets that play key roles in a number of malignancies, both imatinib-sensitive and imatinib-insensitive. The ability of HG-7-85-01 to potently and selectively inhibit both wild-type and gatekeeper mutations that are resilient to frontline therapies further highlights its widespread therapeutic potential for drug-resistant disease. The ability to develop a kinase selectivity profile that is appropriate for simultaneously targeting numerous “gatekeeper” mutant kinases provides a potential solution to the problem of having to develop separate drugs for each individual kinase.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank George Q. Daley for his development and provision of PDGFR mutant-expressing cells, Jonathan Fletcher and George Demetri for their kind development and provision of KIT mutant-expressing cell lines used for our studies, Jinyan Du for development and provision of the T670I-KIT plasmid and the T681I-PDGFRβ plasmid, Tina Davis for her help with in vivo analysis, and Jayasree Sundaram for technical assistance.
M.S. was supported by the National Institutes of Health (5K99GM080097). The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, US Department of Energy (contract DE-AC02-05CH11231). J.D.G is supported by the National Institutes of Health (grants CA66996, CA36167, and DK50654) and the Leukemia & Lymphoma Society (Specialized Center of Research Award). N.S.G. is supported by the National Institutes of Health (grant R01 CA130876-01A1).
National Institutes of Health
Authorship
Contribution: E.W. generated research findings (designed/performed in vitro and in vivo imaging experiments), was responsible for integrity and analysis of the data, and wrote the manuscript; H.G.C. designed and synthesized HG-7-85-01; A.R. assisted with AML patient proliferation assays, development of 32D-T315I-luc+– and Ba/F3-T315I-luc+–expressing lines used for in vivo bioluminescence imaging, and histopathologic analysis of mice used in in vivo imaging experiments, and developed the T670I-KIT– and the T681I-PDGFRβ–expressing Ba/F3 lines via electroporation of plasmid and culturing; R.B. performed many of the in vitro experiments, including cell proliferation assays, cell-cycle and apoptosis assays, as well as immunoblotting and immunoprecipitation, and assisted with bioluminescence imaging; J.Z. performed all of the biologic assays that resulted in the selection of HG-7-85-01 as a lead compound and assisted in the further characterization of the compound; T.S. designed HG-7-85-01 and designed synthetic routes to prepare HG-7-85-01; W.Z. assisted with synthesis of compounds tested and characterized, as well as Ambit screen analysis and pharmacokinetics data analysis; M.S. was responsible for solving the HG-7-85-01-Src cocrystal structure and for rationalizing the ability of the compound to bind to gatekeeper mutant kinases; M.C. assisted with pharmacokinetics data generation; M.A. developed and provided mutant PDGFR-expressing cells and contributed significant scientific input that helped in manuscript preparation; J.A.F., M.D.-R., and M.M. characterized KIT-expressing cells and contributed significant scientific input that helped in manuscript preparation; D.M. assisted with in vivo bioluminescence studies, specifically intravenous injection of cells into mice and ear-tagging, and oral gavage; A.L.K. provided scientific advisement for the in vivo bioluminescence studies; P.A.J. generated proliferation data for HG-7-85-01 tested against EGFR gatekeeper mutants; R.K.-F. and J.V.M. developed the imatinib-sensitive and imatinib-resistant LAMA84 lines; S.A. helped perform several of the in vivo bioluminescence imaging experiments; C.W. prepared and provided normal patient bone marrow cells for colony assays and liquid culture proliferation assays; P.W.M. performed enzymatic assays that characterized the kinase selectivity profile of HG-7-85-01; and N.G. and J.D.G. conceived the research and were responsible for the integrity and analysis of the data.
Conflict-of-interest disclosure: J.D.G. and A.L.K. have a financial interest with Novartis Pharma AG. P.W.M. is an employee of Novartis. N.G. has received funding from Novartis. The remaining authors declare no competing financial interests.
Correspondence: Ellen Weisberg, Department of Medical Oncology, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: Ellen_Weisberg@dfci.harvard.edu; or Nathanael Gray, Department of Cancer Biology, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: Nathanael_Gray@dfci.harvard.edu.
References
Author notes
E.W., H.G.C., and A.R. contributed equally to this study.