In this issue of Blood, Schafer and colleagues report their findings on MLL-r infant ALL.1 They examined global promoter methylation in infants and found hypermethylation in cases with MLL-r ALL compared with both the normal and non-MLL-r ALL cases. When treating MLL-r cell lines with decitabine, they observed a reexpression of silenced genes and a cytotoxic effect. This may open up the prospect of new treatment options for a disease entity with a dismal prognosis.
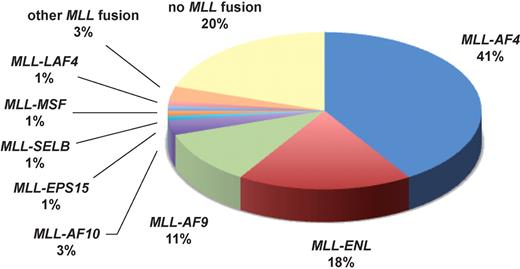
Hardly any other gene in cancer medicine is involved in as many different chromosomal translocations as the mixed lineage leukemia (MLL) gene. It has thus far been possible to identify more than 60 different genes dispersed throughout the genome and to cytogenetically characterize another 35 loci that are involved in MLL translocations.2 Around 10% of acute leukemia (AML and ALL) patients carry MLL aberrations. These patients have been shown to exhibit a distinct gene expression signature.3 In acute lymphoblastic leukemia (ALL), MLL aberrations are usually associated with an adverse prognosis in all age groups. More intensified treatment schedules, especially allogeneic transplantation, have brought some improvements in adults. However, these toxic regimens are often considered unacceptable for affected children. While the overall prognosis of pediatric ALL has steadily improved and is considered particularly good in children around the age of 4, the prognosis of infant ALL (ie, younger than 1 year) is still unsatisfactory with event-free survival rates below 50%.5 The main reason for this unfavorable outcome is the extremely high prevalence (around 80%) of MLL translocations in this age group (see figure).
Infant ALL has a unique genetic background with approximately 80% of patients carrying MLL aberrations (data according to Jansen et al4 ).
Infant ALL has a unique genetic background with approximately 80% of patients carrying MLL aberrations (data according to Jansen et al4 ).
The MLL protein has a highly complex function in normal and malignant hematopoiesis. It is part of a large macromolecular nuclear complex that has histone methyltransferase and histone acetyltransferase function and thus epigenetically regulates the expression of several genes and remodels nucleosomes.6 In particular, several homeobox (HOX) genes are regulated via MLL. Chromosomal translocations involving MLL lead to deregulated HOX gene expression and a different transcription pattern.
Schafer et al from Patrick Brown's research group focused on a specific aspect of infant MLL-r ALL. They investigated the “epigenome” in these patients, particularly the methylation of promoter islands. They used a specific assay (HELP = HpaII tiny fragment enrichment by ligation-mediated PCR).7 Basically, this assay utilizes the fact that the 2 restriction enzymes MspI and HpaII both recognize the same restriction site (5′–C|CGG–3′), but the first enzyme is methylation-insensitive, while the second one is not and will only cut when both cytosines are unmethylated. Thus, these enzymes cut genomic DNA into different fragments depending on its methylation status. The resulting DNA fragments were ligated in a second step to short “linker sequences,” amplified by PCR and analyzed on a custom-designed oligonucleotide microarray covering 25 626 HpaII-amplifiable fragments located at around 14 000 gene promoters. This analysis revealed significant overall methylation differences between MLL-rearranged (MLL-r), MLL-wild-type and normal samples. MLL-r ALL samples were characterized by global CpG island hypermethylation, while healthy controls and MLL-wild-type samples showed similar methylation patterns. To validate these findings, Schafer et al performed qRT-PCR on selected genes with a difference in promoter methylation in the previous analysis. Five of 7 genes with hypermethylated promoter regions had a significantly or at least markedly lower expression level. These qRT-PCR data were in accordance with previously published gene expression microarray data. In a further step, ALL cell lines with and without MLL fusion genes were investigated and treated with the demethylating agent decitabine. Genes previously found to be promoter-hypermethylated and silenced or underexpressed in MLL-r ALL were studied in these cell lines before and after decitabine treatment. Reversal of promoter methylation in MLL-r cell lines was revealed by sodium bisulfite treatment and methylation-specific PCR. Quick reexpression of these genes was observed, and this was associated with a cytotoxic effect on the MLL-r but not the MLL-wt cell lines.
In conclusion, the results reported by Schafer et al reveal interesting aspects of the biology of MLL-r infant ALL.1 However, they should be confirmed by studies involving larger numbers of patients. Several questions remain to be answered, especially concerning the molecular pathways leading to this global hypermethylation. The degree of methylation may also depend on the MLL fusion partner, as recently suggested.8 Nevertheless, there is increasing evidence that hypermethylation plays an important role in infants and perhaps also in older patients with MLL-r ALL. This should encourage the initiation of clinical studies investigating the potential benefit of demethylating agents in acute leukemias with MLL aberrations.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■
Acknowledgment
The author thanks Dr Joanne Weirowski for editing of the manuscript.