Abstract
Cooperating leukemogenic events in MLL-rearranged (MLL-r) infant acute lymphoblastic leukemia (ALL) are largely unknown. We explored the role of promoter CpG island hypermethylation in the biology and therapeutic targeting of MLL-r infant ALL. The HELP (HpaII tiny fragment enrichment by ligation-mediated polymerase chain reaction [PCR]) assay was used to examine genome-wide methylation of a cohort of MLL-r infant leukemia samples (n = 5), other common childhood ALLs (n = 5), and normals (n = 5). Unsupervised analysis showed tight clustering of samples into their known biologic groups, indicating large differences in methylation patterns. Global hypermethylation was seen in the MLL-r cohort compared with both the normals and the others, with ratios of significantly (P < .001) hypermethylated to hypomethylated CpGs of 1.7 and 2.9, respectively. A subset of 7 differentially hypermethylated genes was assayed by quantitative reverse-transcription (qRT)–PCR, confirming relative silencing in 5 of 7. In cell line treatment assays with the DNA methyltransferase inhibitor (DNMTi) decitabine, MLL-r (but not MLL wild-type cell lines) showed dose- and time-dependent cytotoxicity and re-expression of 4 of the 5 silenced genes. Methylation-specific PCR (MSP) confirmed promoter hypermethylation at baseline, and a relative decrease in methylation after treatment. DNMTi may represent a novel molecularly targeted therapy for MLL-r infant ALL.
Introduction
The 5-year event-free survival of childhood acute lymphocytic leukemia (ALL) now exceeds 80%.1 However, the outcome for infants with ALL is substantially worse. Despite intensified treatment, the event-free survival for children younger than 12 months with ALL is only 30% to 40%.2 Because of this, distinct mechanisms of leukemogenesis need to be explored to develop a rational basis for the design of novel therapies in this disease.
Infant ALL has long been known to be an outlier from other childhood lymphocytic leukemias. It is clinically distinct in presentation and is less sensitive to effective ALL chemotherapy.3-5 In addition, although morphologically and histochemically lymphoblastic, it is immunophenotypically unique, as it usually lacks the early lymphocyte antigen CD10 and has a propensity to coexpress myeloid antigens including CD15 and CD65.6 Finally, it is characterized by an extremely high incidence of reciprocal translocations involving the mixed-lineage leukemia (MLL, ALL1, HRX) gene.7 In comparison with ALL in older children, where MLL rearrangements (MLL-r's) are demonstrated in only 8% of cases,8 most studies demonstrate the rate in infants to be 70% to 80%, particularly for younger infants.8,9 Considered together, although there are now more than 60 characterized fusion partners of MLL,10 these findings have established MLL-r ALL to be a unique leukemia. This has been corroborated by gene expression profiling, which has shown MLL-r ALL to be easily differentiated from MLL wild-type (MLL-wt) ALL and acute myeloid leukemia (AML).11-14
The origin of MLL-r leukemia is an area of active investigation but several well-established mechanisms of leukemogenesis in other leukemias have been ruled out as contributing to the disease in infants. For example, a recent large genome-wide, high-density, single nucleotide polymorphism study of childhood ALL demonstrated a striking paucity of copy number alterations in MLL-r cases in comparison with other leukemias.15 In addition, although activating JAK mutations have been demonstrated in 10% of MLL-wt high-risk pediatric leukemias, they have not been discovered in MLL-r infant leukemia.16 Furthermore, a survey of the kinome in MLL-r infant leukemia samples found no activating tyrosine kinase mutations, other than a low rate of FLT3 mutations, in 30 samples (Melissa Wright, P.B., D.S., unpublished data, December 2008).
Several theories have been proposed to explain the leukemogenic potential of the MLL oncoprotein. These have included transcriptional activation, chromatin structure changes, association with signal transduction, dimerization or oligomerization leading to altered DNA binding, recruitment of transcriptional effector molecules, and sequestration of cofactors resulting in dominant-negative effects on target gene expression.17 Because an alteration of gene expression profiles is the common result of all of these hypothesized functions, it has been suggested that a unifying mechanism of MLL-r oncogenesis may be epigenetic regulation.17 Several groups have started examining the influence that histone modification may play in MLL-r oncogenesis.18 DNA methylation has yet to receive as much attention. However, it is known that 1 of the 2 essentially retained N-terminal domains necessary to produce an activated MLL oncoprotein with leukemic potential is a 100–amino acid area that displays homology to the regulatory portion of eukaryotic DNA methyltransferase 1 (DNMT1) and has been termed the MT domain.19 In addition, the MLL MT domain specifically recognizes unmethylated CpG dinucleotide sequences20 and has been shown to be transcriptionally repressive.21
The epigenetic phenomenon of CpG island hypermethylation in tumor suppressor gene promoters, leading to repression and silencing of expression, is an important contributor to oncogenesis.22 To test the hypothesis that CpG island hypermethylation may contribute to oncogenic transformation in MLL-r infant leukemia, we explored the role of global gene promoter hypermethylation in this disease. Here we show that MLL-r infant leukemia is globally hypermethylated in promoter CpG islands in comparison with other forms of childhood leukemia and normal controls, that the genes regulated by these promoters are largely repressed or silenced, and that in many cases these genes can be re-expressed when cells are treated with the demethylating agent decitabine. Thus, decitabine might be an effective antileukemic agent in infant ALL.
Methods
Cells
Primary patient samples were collected from patients treated at the Johns Hopkins Hospital, The Children's Hospital of Philadelphia, or Children's Mercy Hospital and healthy donors between 2004 and 2008. Samples were collected under the respective center's institutional review board–approved cell procurement protocols for newly diagnosed infants and children with ALL. Informed consent was obtained in accordance with the Helsinki protocol. Samples were enriched from diagnostic bone marrow collections by Ficoll-Hypaque centrifugation and stored within 5 hours of collection in 90% fetal bovine serum (FBS) and 10% dimethyl sulfoxide at −80°C until use. The diagnosis of ALL was based on morphology and flow cytometric analysis of immunophenotype. Cytogenetics were determined by standard procedures.
The NALM6, KOPN8, 380, and TANOUE cell lines were originally obtained from German Collection of Microorganisms and Cell Cultures (DSMZ), whereas the HB1119 cell line was provided from the laboratory of Dr Michael Cleary (Stanford University) and the SEMK2 cell line was provided from the laboratory of Dr Carolyn Felix (University of Pennsylvania). Cell lines were maintained in RPMI-1640 plus 10% FBS and 1% penicillin streptomycin at 37°C and 5% CO2.
CD34+-enriched and CD19+-enriched cord blood was obtained from StemCell Technologies, Inc. Cells used in this study are listed in Table 1.
Primary samples and cell lines
| Identifier . | Phenotype . | Cytogenetics . | Age . |
|---|---|---|---|
| Primary sample | |||
| M1 | Pro-B-cell ALL | t(4;11)(q23;21) → MLL/ AF4 | Infant |
| M2 | Pro-B-cell ALL | t(4;11)(q23;21) → MLL/AF4 | Infant |
| M3 | Pro-B-cell ALL | t(4;11)(q23;21) → MLL/AF4 | Infant |
| M4 | Pro-B-cell ALL | t(11;19)(q23;p13) → MLL/ENL | Infant |
| M5 | Pro-B-cell ALL | t(4;11)(q23;21) → MLL/AF4 | Infant |
| H1 | Pre-B-cell ALL | Hyperdiploid | Child |
| H2 | Pre-B-cell ALL | Hyperdiploid | Child |
| T1 | Pre-B-cell ALL | t(12;21) → TEL/AML | Child |
| T2 | Pre-B-cell ALL | t(12;21) → TEL/AML | Child |
| T3 | Pre-B-cell ALL | t(12;21) → TEL/AML | Child |
| N1 | CD19+ cord blood | Normal | Infant |
| N2 | CD34+ cord blood | Normal | Infant |
| N3 | CD19+ cord blood | Normal | Infant |
| N4 | CD19+ cord blood | Normal | Infant |
| N5 | CD34+ cord blood | Normal | Infant |
| Cell line | |||
| SEMK2 | Pro-B ALL | t(4;11)(q23;p21) → MLL/AF4 | Child |
| KOPN8 | Pre-B ALL | t(11;19)(q23;p13) → MLL/ENL | Infant |
| HB1119 | Pre-B ALL | t(11;19)(q23;p13) → MLL/ENL | Child |
| NALM6 | Pre-B ALL | t(5;12)(q33;p13) | Adult |
| 380 | Pre-B ALL | Near diploid with t(8;14) and t(14;18) | Child |
| TANOUE | Pre-B ALL | Hyperdiploid | Child |
| Identifier . | Phenotype . | Cytogenetics . | Age . |
|---|---|---|---|
| Primary sample | |||
| M1 | Pro-B-cell ALL | t(4;11)(q23;21) → MLL/ AF4 | Infant |
| M2 | Pro-B-cell ALL | t(4;11)(q23;21) → MLL/AF4 | Infant |
| M3 | Pro-B-cell ALL | t(4;11)(q23;21) → MLL/AF4 | Infant |
| M4 | Pro-B-cell ALL | t(11;19)(q23;p13) → MLL/ENL | Infant |
| M5 | Pro-B-cell ALL | t(4;11)(q23;21) → MLL/AF4 | Infant |
| H1 | Pre-B-cell ALL | Hyperdiploid | Child |
| H2 | Pre-B-cell ALL | Hyperdiploid | Child |
| T1 | Pre-B-cell ALL | t(12;21) → TEL/AML | Child |
| T2 | Pre-B-cell ALL | t(12;21) → TEL/AML | Child |
| T3 | Pre-B-cell ALL | t(12;21) → TEL/AML | Child |
| N1 | CD19+ cord blood | Normal | Infant |
| N2 | CD34+ cord blood | Normal | Infant |
| N3 | CD19+ cord blood | Normal | Infant |
| N4 | CD19+ cord blood | Normal | Infant |
| N5 | CD34+ cord blood | Normal | Infant |
| Cell line | |||
| SEMK2 | Pro-B ALL | t(4;11)(q23;p21) → MLL/AF4 | Child |
| KOPN8 | Pre-B ALL | t(11;19)(q23;p13) → MLL/ENL | Infant |
| HB1119 | Pre-B ALL | t(11;19)(q23;p13) → MLL/ENL | Child |
| NALM6 | Pre-B ALL | t(5;12)(q33;p13) | Adult |
| 380 | Pre-B ALL | Near diploid with t(8;14) and t(14;18) | Child |
| TANOUE | Pre-B ALL | Hyperdiploid | Child |
Infant indicates < 12 months old; child, 12 months old to 18 years old; and adult, > 18 years old.
DNA methylation analysis by HELP
High-molecular-weight DNA was isolated from Ficoll-Hypaque centrifugation–enriched diagnostic patient bone marrow samples using the PureGene kit (QIAGEN) and following the manufacturer's instructions and the HELP (HpaII tiny fragment enrichment by ligation-medicated polymerase chain reaction [PCR]) assay was carried out as previously described.23 All samples for microarray hybridization were processed by the Microarray Facility at the Cornell University Life Sciences Laboratories Center. Samples were labeled using cyanin-labeled random primers (9 mers) and then hybridized onto a human HG17 custom-designed oligonucleotide array (50-mer) covering 25 626 HpaII-amplifiable fragments located at gene promoters and imprinted regions. HpaII-amplifiable fragments are defined as genomic sequences contained between 2 flanking HpaII sites found within 200 to 2000 bp from each other. Annotation was performed using the University of California Santa Cruz (UCSC) Genome Browser24 2006 human assembly to identify genes located within 2000 bp 5′ to 2000 bp 3′ of the HpaII fragment. Each HpaII-amplifiable fragment on the array is represented by 15 individual probes distributed randomly across the microarray slide. Scanning was performed using a GenePix 4000B scanner (Molecular Devices) as previously described.25 Quality control was performed as previously described.26 DNA methylation was measured as the log(MspI/HpaII) ratio, where HpaII reflects the hypomethylated fraction of the genome (as HpaII is methylation-sensitive and therefore recognizes 5′-CCGG-3′ only when both cytosines are unmethylated) and MspI represents the whole genome reference (as MspI is methylation insensitive and recognizes 5′-CCGG-3′ regardless of cytosine methylation status). The raw data were processed using the Bioconductor oligo package (http://www.bioconductor.org/packages/release/bioc/html/oligo.html). The red and green channels were quantile normalized separately. For each channel, we summarized intensities in each probe set (representing HpaII fragments) using median polish as done by Robust Multi-array Average (RMA).27 Then, M values were calculated using the log (base 2) ratio of the green divided by the red channel intensity. Note that the higher values of M represent more methylation for the region associated with the respective probe set. For the unsupervised clustering, we first computed the standard deviation (SD) of the M values for each probe set. We filtered probe sets with low SD values. Specifically, we required the SD to be larger than the median SD across all probe sets. Note that this filtering step is necessary because most regions do not exhibit biologic variation, but rather, vary because of measurement error. Hierarchic clustering was applied to the M values of the probe sets surviving this filtering step. Note that class label information was not used in any of these steps. To find differentially methylated genes among the 3 groups (MLL-r, MLL-wild-type [MLL-wt], normal), we processed the raw data with RMA27 and computed a moderated t statistic and P value (adjusted for multiple comparison) using limma28 for each probe for each pairwise comparison (MLL-r vs MLL-wt, MLL-r vs normal, MLL-wt vs normal). All microarray data have been submitted to the Gene Expression Omnibus repository (accession no. GSE19671).29
Cytotoxicity assay
Cytotoxicity assays were performed on cell lines grown in RPMI-1640 plus 10% FBS plus 1% penicillin streptomycin with media refreshed for optimal survival. Each cell line was split into 5 flasks containing 250 mL of media and 150 million total cells just before treatment. 5-Aza-2′-deoxycytidine (decitabine; Sigma-Aldrich) was dissolved in diethylpyrocarbonate (DEPC)–treated water at a working stock concentration of 1mM. It was then filtered through a 0.22-μm syringe membrane before use. Decitabine was introduced into each culture flask within 1 hour of being dissolved, so that final concentrations of the drug were 0, 0.5, 1, 2, and 4μM on day 1 of the experiment. MTT (3-4,5-dimethylthiazol-2,5-diphenyltetrazolium) assays were performed on cells extracted under sterile conditions from the treatment flasks and introduced into a 96-well plate. Extractions for each drug concentration were performed in triplicate. The MTT plate was prepared from the flasks at 8, 24, 48, and 72 hours from introduction of drug, per the manufacturer's instructions (Roche) and read using a Bio-Rad model 680 plate reader.
DNA and RNA extraction
Ten million cells were removed from the cytotoxicity assay flasks at time points 0, 2, 4, 8, 24, 48, and 72 hours of treatment from which DNA and RNA were extracted for further analysis. DNA was extracted using the QIAamp DNA Mini Kit (QIAGEN) and RNA was extracted using the RNeasy Mini Kit (QIAGEN) per the manufacturer's instructions.
Gene expression assay
Gene expression assays were performed on both primary samples and cell lines treated in the cytotoxicity assay. Complementary DNA was produced from RNA recovered, as described in “DNA and RNA extraction,” by treatment with reverse transcriptase by standard procedures. Quantitative reverse-transcriptase PCR (qRT-PCR) was performed using materials from TaqMan Gene Expression Assays (Applied Biosystems) and carried out per the manufacturer's instructions. PCR was conducted using a Bio-Rad real-time iCycler and threshold cycle (Ct) results were calculated using the manufacturer's software. Single time-point results were reported as a ΔCt (gene Ct − GAPDH Ct), and single time-point gene expression data were reported as 2(−ΔCt). Comparative analysis was calculated using study group average gene expression data with statistical significance (P value) calculated using a 2-tailed distribution, 2-sample unequal variance Student t test. Copy number fold change was calculated by ΔΔCt (ΔCt at time 0 − ΔCt at time 0 + n).
Data mining from previously published microarrays
Three large-sample gene expression arrays have been published comparing MLL-r ALL samples to MLL-wt ALL samples.11,13,14 Raw data from the arrays are available in public databases (Armstrong et al, http://www.broadinstitute.org/cgi-bin/cancer/publications/pub_paper.cgi?mode=view&paper_id=63; Yeoh et al, http://www.stjuderesearch.org/data/ALL1/index.html; and Ross et al, http://www.stjuderesearch.org/data/ALL3/dataFiles.html, all accessed in December 2009). Datasets were mined for our “genes of interest” using the Affymetrix probe numbers for the appropriate gene chip as published by http://www.genecards.org/. When greater than one probe was annotated to a particular gene of interest, all probes were given equal weight in our analysis. Average relative fluorescence was calculated for each relevant probe. For the Armstrong et al data, MLL-r ALL samples were compared with MLL-wt ALL samples. For the Yeoh et al and Ross et al data, MLL-r ALL samples were compared with a combination of TEL-AML1-r ALL and hyperdiploid ALL samples. Statistical significance for each gene was calculated using a 2-tailed distribution, 2-sample unequal Student t test.
MSP
Sodium bisulfite treatment of genomic DNA is known to convert unmethylated cytosines to uracil bases whereas methylated cytosines are protected from the conversion, thus enabling PCR amplification of CpG segments to be methylation specific.30 DNA was treated with sodium bisulfite using the EZ DNA Methylation Kit (Zymo Research) per the manufacturer's instructions. PCR was carried out following the protocol written for methylation-specific PCR (MSP) by Licchesi and Herman.31 MSP primers for DAPK1CONTROL and p73CONTROL,32 DAPK1,33 and HRK34 were taken from the published literature. MSP primers for CCR6 were designed using the UCSC Genome Browser human assembly (March 2006). The DNA sequence was explored from 1500 bp 5′ to 200 bp 3′ of the CCR6 start codon, assumed to be the gene's promoter region,35 for CpG islands and appropriate MSP primers using MethPrimer (University of California, San Francisco).36 MSP primers for CCR6 were as follows: forward methylated 5′-GTTTTTGTAGAAGTCGTTGGC-3′, reverse methylated 5′CATTTTCTACAATCTATAACCACGTA-3′, for ward unmethylated 5′-TTTGTTTTTGTAGAAGTTGTTGGTG-3′, and reverse unmethylated 5′-ATTTTCTACAATCTATAACCACATA-3′. Densitometry was performed using Bio-Rad Quantity One Software Version 4.6.5.
Results
Childhood ALL subtypes and MLL-r infant ALL samples have differential methylation patterns; MLL-r infant samples are characterized by several hypermethylated gene promoter regions
We hypothesized that gene promoter hypermethylation might be a characteristic of MLL-r infant ALL. Therefore, we carried out a genome-wide DNA methylation study using the HpaII tiny fragment enrichment by ligation-mediated PCR (HELP) assay. This technique has been shown to accurately identify the DNA methylation levels of CpG dinucleotides throughout the genome.23,37,38 We processed 15 samples, which included precursor B-cell ALL from infants carrying an MLL-r (n = 5), MLL-wild-type (wt) childhood leukemias including precursor B-cell ALL with a TEL-AML-r (n = 3), and precursor B-cell ALL with a hyperdiploid karyotype (n = 2) and normal controls of CD19+-enriched cord blood (n = 3) and CD34+-enriched cord blood (n = 2). The HELP assay was performed in this case using a long-oligonucleotide microarray representing the DNA methylation level of greater than 50 000 CpGs corresponding to 14 000 promoter regions.
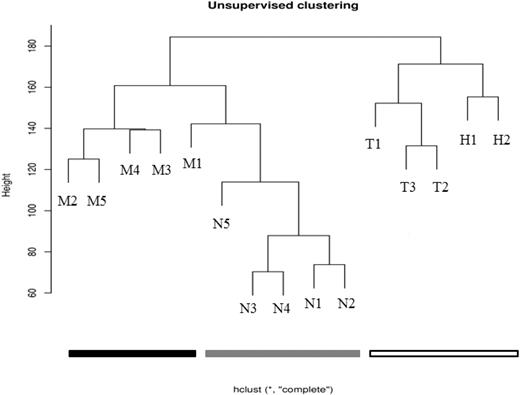
The HELP data were initially processed using an unsupervised analysis, in which the samples largely clustered into their biologic subtypes (Figure 1). All of the MLL-wt samples clustered together and separately from the MLL-r and normal samples. Furthermore, within the MLL-wt cohort, the TEL-AML-r samples and hyperdiploid samples further separated into subgroups. Although the MLL-r and normal samples were all contained within the same branch of the dendrogram, there was tight clustering of 4 of the 5 MLL-r samples and 4 of the 5 normal samples. This suggests that there are intragroup similarities and intergroup differences in the promoter methylation patterns of infant ALL and childhood ALL. These patterns may be reflective of each leukemia's unique underlying biology.
Unsupervised analysis shows clustering of primary samples into their biologic groups based on their HELP assay–determined methylome. Using raw data from the HELP assay, unsupervised analysis was performed on primary samples (Table 1). The MLL-r leukemia samples cluster together (black), the other (MLL-wt) ALL samples cluster together (white), and the normals cluster together (gray). This demonstrates that there are remarkable intragroup similarities and intergroup differences in promoter methylation patterns, and that these patterns are based on the underlying biology of the groups (determined primarily by recurrent cytogenetic abnormalities in the leukemias, or by lineage differentiation for the normals).
Unsupervised analysis shows clustering of primary samples into their biologic groups based on their HELP assay–determined methylome. Using raw data from the HELP assay, unsupervised analysis was performed on primary samples (Table 1). The MLL-r leukemia samples cluster together (black), the other (MLL-wt) ALL samples cluster together (white), and the normals cluster together (gray). This demonstrates that there are remarkable intragroup similarities and intergroup differences in promoter methylation patterns, and that these patterns are based on the underlying biology of the groups (determined primarily by recurrent cytogenetic abnormalities in the leukemias, or by lineage differentiation for the normals).
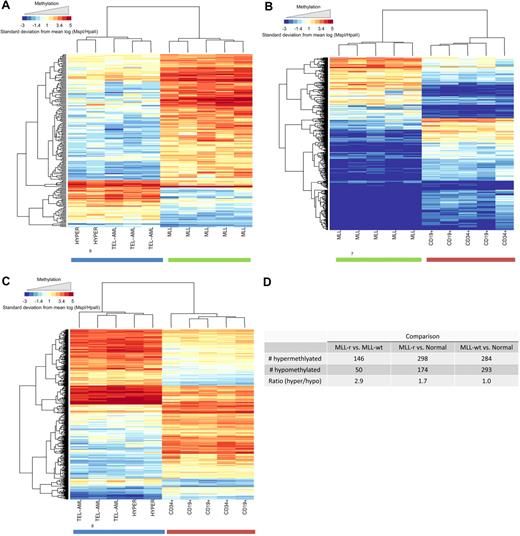
We extended the analysis of the HELP data by creating a supervised list of the most differentially (P < .001) methylated HpaII sites for each of 3 pairwise comparisons of the biologic groups (MLL-r vs MLL-wt, MLL-r vs normal, MLL-wt vs normal). The heat map for each of these comparisons is shown in Figure 2A, B, and C, respectively. The number of significantly hypermethlyated and significantly hypomethylated CpG sites for each comparison and the ratio of these numbers are shown in Figure 2D. It is evident that the MLL-r cohort is globally hypermethylated compared with both the MLL-wt leukemia cohort (ratio, 2.9) and the normal cohort (ratio, 1.7). The MLL-wt and the normal cohorts were similarly methylated (ratio, 1.0). Thus, global CpG island hypermethylation is comparatively specific to MLL-r infant ALL within this set of childhood lymphoid leukemias.
MLL-r primary samples show global promoter hypermethylation compared with MLL-wt ALLs and normal controls in the HELP assay. Analysis of global methylation differences between the study groups is shown. Hierarchic clustering using a subset of probes, which demonstrated highly significant differences between groups (P < .001), was used to generate heat maps. Heat maps are shown for comparisons between (A) MLL-r ALL versus MLL-wt ALL, (B) MLL-r versus normal controls, and (C) MLL-wt ALL versus normal controls. Individual samples are represented on the heat map as columns, whereas individual probes sets are represented as rows. It is again evident that the groups cluster together and that within each group of samples there are certain probes that are differentially hypermethylated (red) or hypomethylated (blue). (D) Numeric representation of the heat maps shows that for the MLL-r group, there were 1.7- and 2.9-fold more probes, respectively, demonstrating relative hypermethylation than hypomethylation compared with either the other ALL samples or the normal marrow controls. This difference was not evident when MLL-wt ALL samples were compared with normal controls.
MLL-r primary samples show global promoter hypermethylation compared with MLL-wt ALLs and normal controls in the HELP assay. Analysis of global methylation differences between the study groups is shown. Hierarchic clustering using a subset of probes, which demonstrated highly significant differences between groups (P < .001), was used to generate heat maps. Heat maps are shown for comparisons between (A) MLL-r ALL versus MLL-wt ALL, (B) MLL-r versus normal controls, and (C) MLL-wt ALL versus normal controls. Individual samples are represented on the heat map as columns, whereas individual probes sets are represented as rows. It is again evident that the groups cluster together and that within each group of samples there are certain probes that are differentially hypermethylated (red) or hypomethylated (blue). (D) Numeric representation of the heat maps shows that for the MLL-r group, there were 1.7- and 2.9-fold more probes, respectively, demonstrating relative hypermethylation than hypomethylation compared with either the other ALL samples or the normal marrow controls. This difference was not evident when MLL-wt ALL samples were compared with normal controls.
Results of qRT-PCR assay of gene expression strongly correlate with HELP data and are consistent with known leukemia biology
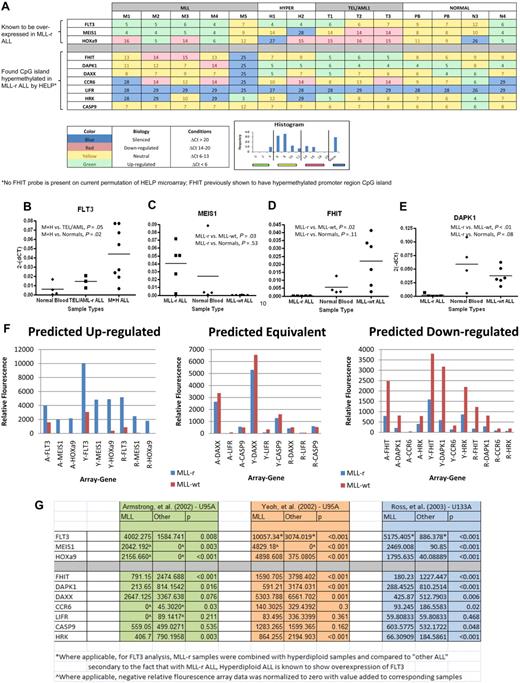
We selected a set of 12 genes to further analyze using qRT-PCR. Six genes were selected by correlating HpaII sites that were differentially hypermethlyated in MLL-r samples (P < .001) with their list of annotated genes (DAPK1, DAXX, CCR6, CASP9, LIFR, and HRK). We focused on these genes because they had previously been shown in the literature to be silenced, hypermethylated, epigenetically influenced, or related to tumor suppression or apoptosis in cancer. Four further genes were selected on the basis of known importance in MLL-r leukemia biology (FLT3, HoxA9, MEIS1, and FHIT). FLT3 is known to be highly expressed in MLL-r infant leukemia as well as precursor B-cell ALL with hyperdiploid cytogenetics.39 MEIS1 and HoxA9 have also been shown to be highly expressed in MLL-r leukemia.40 FHIT, on the other hand, has been demonstrated to be silenced due to promoter CpG methylation in MLL-r leukemia.41 Finally, 2 genes were selected as housekeeping genes (GAPDH and ABL; Tables 2–3). Reverse-transcriptase TaqMan qRT-PCR for all selected genes was run on each primary sample and normal controls. Results are shown in heat map format (Figure 3A). All 3 of the genes for which expression in MLL-r cases was expected to be relatively high were confirmed by qRT-PCR (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). This difference was particularly notable for FLT3 (Figure 3B) and MEIS1 (Figure 3C), in which these differences were highly significant. Of the 7 genes for which expression in MLL-r cases was expected to be relatively low because of differential promoter hypermethylation, 5 showed either statistical significance or a trend toward statistical significance in down-regulation or silencing. The only exceptions were LIFR, which was silenced in all, and CASP9, which was expressed at similar levels in all. The most striking differences were seen for FHIT (Figure 3D) and DAPK1 (Figure 3E).
Genes of interest from HELP analysis
| Gene . | Gene full name . | Location . | Comparison . | ΔM . | P . |
|---|---|---|---|---|---|
| LIFR | Leukemia inhibitory factor receptor-α | Chr 5p13-p12 | MLL vs hyperdiploid | 3.60 | < .001 |
| CCR6 | Chemokine (C-C motif) receptor 6 | Chr 6q27 | MLL vs normal controls | 3.76 | < .001 |
| DAXX | Death-associated protein 6 | Chr 6p21.3 | MLL vs normal controls | 3.43 | < .001 |
| DAPK1 | Death-associated protein kinase 1 | Chr 9q34.1 | MLL vs normal controls | 6.22 | < .001 |
| CASP9 | Caspase 9 | Chr 1p36.3-36.1 | MLL vs MLL-wt ALL | 3.84 | .005 |
| HRK | Harakiri | Chr 12q24.22 | MLL vs MLL-wt ALL | 4.17 | .005 |
| Gene . | Gene full name . | Location . | Comparison . | ΔM . | P . |
|---|---|---|---|---|---|
| LIFR | Leukemia inhibitory factor receptor-α | Chr 5p13-p12 | MLL vs hyperdiploid | 3.60 | < .001 |
| CCR6 | Chemokine (C-C motif) receptor 6 | Chr 6q27 | MLL vs normal controls | 3.76 | < .001 |
| DAXX | Death-associated protein 6 | Chr 6p21.3 | MLL vs normal controls | 3.43 | < .001 |
| DAPK1 | Death-associated protein kinase 1 | Chr 9q34.1 | MLL vs normal controls | 6.22 | < .001 |
| CASP9 | Caspase 9 | Chr 1p36.3-36.1 | MLL vs MLL-wt ALL | 3.84 | .005 |
| HRK | Harakiri | Chr 12q24.22 | MLL vs MLL-wt ALL | 4.17 | .005 |
Twelve genes of interest were selected for further analysis by TaqMan qRT-PCR. Six genes of interest were selected from the HELP analysis results. Probes that showed highly significant methylation differences between groups were matched with their corresponding gene based on location using the UCSC genome browser.24 This list of genes was further condensed by selecting genes that had previously been shown in the literature to be silenced, hypermethylated, epigenetically influenced, or related to tumor suppression or apoptosis. The other 6 genes appear in Table 3.
Genes of interest from literature
| Gene . | Gene full name . | Chromosome . | Rationale for selection . |
|---|---|---|---|
| FHIT | Fragile histidine triad | Chr 3p14.2 | Known to be hypermethylated in MLL-r infant ALL41 |
| FLT3 | FMS-like tyrosine kinase 3 | Chr 13q12.2 | Known to be up-regulated and of biologic importance in MLL-r infant ALL39 |
| HOXa9 | Homeobox A9 | Chr 7p15-p14 | Known to be up-regulated and of biologic importance in MLL-r infant ALL11 |
| MEIS1 | MEIS-homeobox 1 | Chr 2p14-p13 | Known to be up-regulated and of biologic importance in MLL-r infant ALL11 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Chr 12p13.31 | Housekeeping gene |
| ABL | C-ABL oncogene, receptor tyrosine kinase | Chr 9q34.1 | Housekeeping gene |
| Gene . | Gene full name . | Chromosome . | Rationale for selection . |
|---|---|---|---|
| FHIT | Fragile histidine triad | Chr 3p14.2 | Known to be hypermethylated in MLL-r infant ALL41 |
| FLT3 | FMS-like tyrosine kinase 3 | Chr 13q12.2 | Known to be up-regulated and of biologic importance in MLL-r infant ALL39 |
| HOXa9 | Homeobox A9 | Chr 7p15-p14 | Known to be up-regulated and of biologic importance in MLL-r infant ALL11 |
| MEIS1 | MEIS-homeobox 1 | Chr 2p14-p13 | Known to be up-regulated and of biologic importance in MLL-r infant ALL11 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Chr 12p13.31 | Housekeeping gene |
| ABL | C-ABL oncogene, receptor tyrosine kinase | Chr 9q34.1 | Housekeeping gene |
To complete the list of genes for qRT-PCR analysis from Table 2, 6 additional genes were selected. One gene, FHIT, was selected based on known reports of it being hypermethylated in MLL-r infant leukemia; 3 genes (FLT3, HOXa9, and MEIS1) were selected based on their known importance in MLL-r infant leukemia; and 2 housekeeping genes, ABL and GAPDH, were also selected.
TaqMan qRT-PCR on genes of interest shows preferentially silenced or decreased expression in MLL-r primary samples in comparison with MLL-wt ALL primary samples and in normal controls in comparison with published gene expression microarrays validating results. The 12 genes described in Tables 2 and 3 were analyzed by qRT-PCR on the HELP primary samples and 2 peripheral blood samples from healthy donors (PB). The results are summarized in the heat map (A), which has the genes of interest represented by rows and individual samples, by columns. Each box shows the ΔCt (gene Ct − GAPDH Ct), such that higher numbers indicate lower RNA expression. In addition, the heat map is color-coded based on its biology as described in the key. The color code was determined by natural peaks in a histogram of all Cts in the cohort. This histogram is also shown in the key. The MLL-r samples show a greater number of silenced genes and more underexpressed genes than do the other leukemias or normals. Ct values were then converted to relative gene expression using the 2(−ΔCt) method, and MLL-r expression levels were compared with MLL-wt and normal control levels (raw data in supplemental Table 1). By dot plot, many of the genes show statistically significant differences in gene expression between groups. In analysis, FLT3 (B) was statistically significantly up-regulated in the combination of MLL-r infant leukemia (M) and hyperdiploid (H) leukemia compared with normals; in addition, MEIS1 (C) was shown to be up-regulated in MLL-r leukemia; both of these phenomena are consistent with published literature. Finally, there was statistically significant down-regulation on FHIT (D) and DAPK1 (E) in MLL-r leukemia compared with “other leukemias” and normal controls. (F) To further validate our qRT-PCR results, we compared our data to 3 published gene expression microarrays. Based on our qRT-PCR results (A-E and supplemental Table 1), we predicted that the genes FLT3, MEIS1, and HOXa9 were up-regulated in MLL-r ALL in comparison with MLL-wt ALL, that the genes DAXX, LIFR, and CASPASE9 had equivalent expression in MLL-r ALL and MLL-wt ALL, and that the genes FHIT, DAPK1, CCR6, and HRK were down-regulated in MLL-r ALL in comparison with MLL-wt ALL. (G) Based on 3 independent gene expression microarrays using 2 different Affymetrix platforms (HG-U95A and HG-U133A) published by Armstrong et al (A),11 Yeoh et al (Y),14 and Ross et al (R),13 we confirmed the predictions.
TaqMan qRT-PCR on genes of interest shows preferentially silenced or decreased expression in MLL-r primary samples in comparison with MLL-wt ALL primary samples and in normal controls in comparison with published gene expression microarrays validating results. The 12 genes described in Tables 2 and 3 were analyzed by qRT-PCR on the HELP primary samples and 2 peripheral blood samples from healthy donors (PB). The results are summarized in the heat map (A), which has the genes of interest represented by rows and individual samples, by columns. Each box shows the ΔCt (gene Ct − GAPDH Ct), such that higher numbers indicate lower RNA expression. In addition, the heat map is color-coded based on its biology as described in the key. The color code was determined by natural peaks in a histogram of all Cts in the cohort. This histogram is also shown in the key. The MLL-r samples show a greater number of silenced genes and more underexpressed genes than do the other leukemias or normals. Ct values were then converted to relative gene expression using the 2(−ΔCt) method, and MLL-r expression levels were compared with MLL-wt and normal control levels (raw data in supplemental Table 1). By dot plot, many of the genes show statistically significant differences in gene expression between groups. In analysis, FLT3 (B) was statistically significantly up-regulated in the combination of MLL-r infant leukemia (M) and hyperdiploid (H) leukemia compared with normals; in addition, MEIS1 (C) was shown to be up-regulated in MLL-r leukemia; both of these phenomena are consistent with published literature. Finally, there was statistically significant down-regulation on FHIT (D) and DAPK1 (E) in MLL-r leukemia compared with “other leukemias” and normal controls. (F) To further validate our qRT-PCR results, we compared our data to 3 published gene expression microarrays. Based on our qRT-PCR results (A-E and supplemental Table 1), we predicted that the genes FLT3, MEIS1, and HOXa9 were up-regulated in MLL-r ALL in comparison with MLL-wt ALL, that the genes DAXX, LIFR, and CASPASE9 had equivalent expression in MLL-r ALL and MLL-wt ALL, and that the genes FHIT, DAPK1, CCR6, and HRK were down-regulated in MLL-r ALL in comparison with MLL-wt ALL. (G) Based on 3 independent gene expression microarrays using 2 different Affymetrix platforms (HG-U95A and HG-U133A) published by Armstrong et al (A),11 Yeoh et al (Y),14 and Ross et al (R),13 we confirmed the predictions.
Comparison with published gene expression microarrays validates TaqMan qRT-PCR results
To further validate that our TaqMan qRT-PCR results were generalizable to both the MLL-r and MLL-wt populations, we compared our gene expression data with 3 published large-sample arrays comparing MLL-r ALL to MLL-wt ALL.11,13,14 Based on our qRT-PCR results (Figure 3A-E and supplemental Table 1), we predicted that the genes FLT3, MEIS1, and HOXa9 were up-regulated in MLL-r ALL in comparison with MLL-wt ALL, that the genes DAXX, LIFR, and CASP9 had equivalent expression in MLL-r ALL and MLL-wt ALL, and that the genes FHIT, DAPK1, CCR6, and HRK were down-regulated in MLL-r ALL in comparison with MLL-wt ALL. When our results were compared with the 3 independent gene expression microarrays, which used 2 different Affymetrix platforms (HG-U95A and HG-U133A), we confirmed the predictions (Figure 3F). Both predicted up-regulated genes and predicted down-regulated genes were shown differentially expressed between MLL-r and MLL-wt ALL with robust statistical significance (Figure 3G).
Treatment with the demethylating agent 5-aza-2′-deoxycytidine (decitabine) selectively kills MLL-r precursor B-cell ALL cell lines and can be correlated with the re-expression of several silenced genes
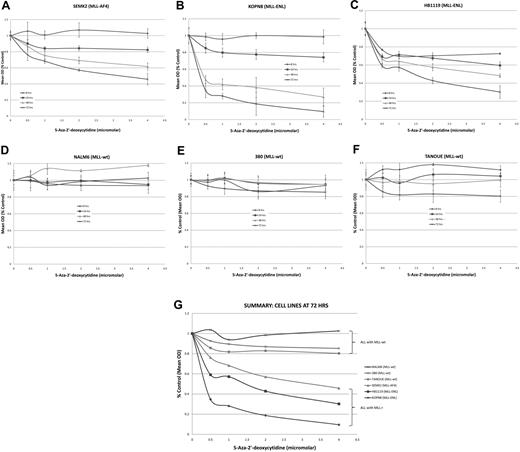
Having shown that the MLL-r infant ALL samples contained an increased number of hypermethylated gene promoters in comparison with childhood MLL-wt ALL and normal controls, and that 5 of the 7 genes with hypermethylated promoters were silenced or down-regulated, we investigated whether demethylating agents might serve to reverse aberrant methylation signaling and allow gene re-expression. In addition, if MLL-r infant ALL promoter hypermethylation follows the epigenetic paradigm of several other cancers,22 reversal of silencing of key tumor suppressor genes may lead to cytotoxicity of leukemia cells. Thus, ALL cell lines carrying the MLL-AF4 translocation (SEMK2), MLL-ENL translocation (KOPN8 and HB1119), and wild-type MLL (NALM6, 380 and TANOUE) were treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (decitabine) at concentrations of 0, 0.5, 1, 2, and 4 μM for 72 hours. We performed MTT assays at 8, 24, 48, and 72 hours and isolated RNA from each sample at 0, 2, 4, 8, 24, 48, and 72 hours. MLL-wt cell lines all showed robust time- and dose-dependent cell kill by 72 hours (Figure 4A-C,G). Although decitabine was clearly cytotoxic to the MLL-r cell lines, there was little response in the MLL-wt cell line (Figure 4D-G).
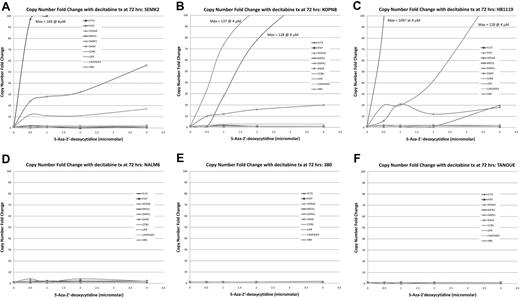
MTT assays on cell lines treated with the demethylating agent 5-Aza-2′-deoxycytidine (decitabine) show preferential cytotoxicity in MLL-r cell lines. ALL cell lines were treated with decitabine at 0-, 0.5-, 1-, 2-, and 4-μM concentrations over 72 hours. Cell lines with an MLL-r (A-C) showed robust dose- and time-dependent cell kill. However, the MLL-wt cell lines (D-F) showed no response to decitabine. A summary graph representing the 72-hour time point for all cell lines treated is presented as panel G.
MTT assays on cell lines treated with the demethylating agent 5-Aza-2′-deoxycytidine (decitabine) show preferential cytotoxicity in MLL-r cell lines. ALL cell lines were treated with decitabine at 0-, 0.5-, 1-, 2-, and 4-μM concentrations over 72 hours. Cell lines with an MLL-r (A-C) showed robust dose- and time-dependent cell kill. However, the MLL-wt cell lines (D-F) showed no response to decitabine. A summary graph representing the 72-hour time point for all cell lines treated is presented as panel G.
Biologic correlates to the MTT assay results were seen in a parallel examination of the treated cells using TaqMan gene expression assays. As noted, the genes FHIT, DAPK1, CCR6, and HRK were shown to be underexpressed or silent in our primary samples. This was also seen in our MLL-r cell lines (supplemental Figure 1). When treated with decitabine, there was re-expression of FHIT, DAPK1, and CCR6 in the SEMK2 cells (Figure 5A), of DAPK1 and CCR6 in the KOPN8 cells (Figure 5B), and of FHIT, DAPK1, CCR6, and HRK in the HB1119 cells (Figure 5C) but no change in expression in the NALM6, 380, and TANOUE cells (Figure 5D-F). The re-expression occurred relatively quickly, with several genes responding in MLL-r cell lines treated with 4μM decitabine within 24 hours (supplemental Figure 2).
TaqMan qRT-PCR on MLL-r cell lines shows dose-dependent re-expression of genes of interest when treated with decitabine. TaqMan qRT-PCR was performed on cell lines treated with decitabine at concentrations of 0, 0.5, 1, 2, and 4μM. Copy number fold change was estimated using the ΔΔCt method. (A-C) In cell lines carrying MLL-r several of the genes of interest showed robust dose-dependent increase in copy number, whereas this phenomenon was not seen in the MLL-wt cell lines (D-F). This demonstrates biologic correlation with the MTT assays.
TaqMan qRT-PCR on MLL-r cell lines shows dose-dependent re-expression of genes of interest when treated with decitabine. TaqMan qRT-PCR was performed on cell lines treated with decitabine at concentrations of 0, 0.5, 1, 2, and 4μM. Copy number fold change was estimated using the ΔΔCt method. (A-C) In cell lines carrying MLL-r several of the genes of interest showed robust dose-dependent increase in copy number, whereas this phenomenon was not seen in the MLL-wt cell lines (D-F). This demonstrates biologic correlation with the MTT assays.
MSP validates HELP assay results and provides evidence that the mechanism of gene re-expression with decitabine treatment is reversal of promoter methylation
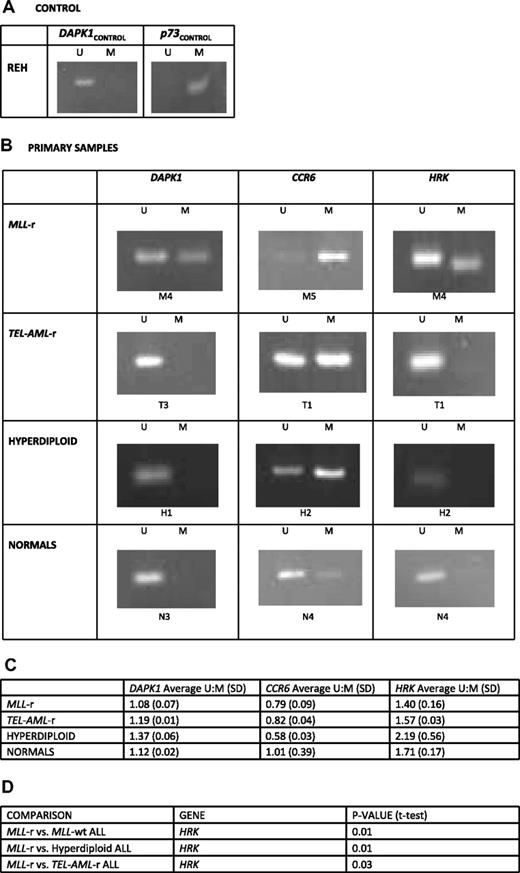
To both validate the findings from the HELP assay and verify that the re-expression of the genes DAPK1, HRK, and CCR6 was linked to gene promoter hypermethylation and not a secondary effect of decitabine, we performed methylation-specific PCR (MSP) on primary samples and cell lines both before and after treatment with decitabine. Post–bisulfite-treated methylation-specific primers for the promoter regions of the genes DAPK1 and HRK were based on previously published MSP data, whereas primers for CCR6 were custom designed. Complete bisulfite conversion was assured by treating REH genomic DNA (gDNA) under the same conditions and running MSP for the gene promoters DAPK1 and p73, as it is known that the DAPK1 promoter region is completely unmethylated whereas the p73 promoter is completely methylated in REH cells (Figure 6A).
Methylation-specific PCR demonstrates preferential promoter methylation of genes of interest in MLL-r ALL compared with MLL-wt ALL and normal controls. DNA from the REH cell line and HELP primary samples were treated with bisulfite to specifically convert unmethylated cytosines to uracil. PCR primers for anticipated post–bisulfite-methylated (M) and unmethylated (U) promoter sequences for the genes DAPK1, p73, CCR6, and HRK were used to determine their methylation status. (A) REH cells were used as control to assure complete bisulfite conversion because in REH cells the DAPK1 promoter is completely unmethylated whereas the p73 promoter is completely methylated. (B) Examples are shown from treatment of the HELP assay primary samples. In DAPK1 and HRK, only the MLL-r sample shows a methylated amplicon. In CCR6, although all samples show methylated amplicons, the MLL-r sample is clearly preferentially methylated as it shows only a dim unmethylated amplicon. (C) Densitometry was performed on all amplicons for all primary samples in each group and average unmethylated amplicon–methylated amplicon (U:M) densitometry ratios and their SDs are shown. The DAPK1 and HRK promoters are preferentially methylated in MLL-r ALL cells in comparison with other leukemias and normals; CCR6 promoters have some methylation in all leukemias, whereas in MLL-r ALL it is preferentially methylated and preferentially unmethylated in normals. (D) With the exception of, in the CCR6 gene, MLL-r versus other leukemias and MLL-r versus hyperdiploid ALL (where hyperdiploid leukemia showed greater methylation than MLL-r) all genes and groups compared trended toward statistical significance, with MLL-r leukemia having a lower U:M ratio (more methylation); despite small numbers, several comparisons reached statistical significance and are listed.
Methylation-specific PCR demonstrates preferential promoter methylation of genes of interest in MLL-r ALL compared with MLL-wt ALL and normal controls. DNA from the REH cell line and HELP primary samples were treated with bisulfite to specifically convert unmethylated cytosines to uracil. PCR primers for anticipated post–bisulfite-methylated (M) and unmethylated (U) promoter sequences for the genes DAPK1, p73, CCR6, and HRK were used to determine their methylation status. (A) REH cells were used as control to assure complete bisulfite conversion because in REH cells the DAPK1 promoter is completely unmethylated whereas the p73 promoter is completely methylated. (B) Examples are shown from treatment of the HELP assay primary samples. In DAPK1 and HRK, only the MLL-r sample shows a methylated amplicon. In CCR6, although all samples show methylated amplicons, the MLL-r sample is clearly preferentially methylated as it shows only a dim unmethylated amplicon. (C) Densitometry was performed on all amplicons for all primary samples in each group and average unmethylated amplicon–methylated amplicon (U:M) densitometry ratios and their SDs are shown. The DAPK1 and HRK promoters are preferentially methylated in MLL-r ALL cells in comparison with other leukemias and normals; CCR6 promoters have some methylation in all leukemias, whereas in MLL-r ALL it is preferentially methylated and preferentially unmethylated in normals. (D) With the exception of, in the CCR6 gene, MLL-r versus other leukemias and MLL-r versus hyperdiploid ALL (where hyperdiploid leukemia showed greater methylation than MLL-r) all genes and groups compared trended toward statistical significance, with MLL-r leukemia having a lower U:M ratio (more methylation); despite small numbers, several comparisons reached statistical significance and are listed.
After assurance of complete unmethylated cytosine to uracil conversion, MSP was carried out on gDNA from all primary samples (Figure 6B) and cell lines (supplemental Figure 3) and densitometry was performed on each amplicon. From densitometry results, the average unmethylated amplicon–methylated amplicon (U:M) ratios were calculated for each biologic subtype, with lower ratios indicating a higher degree of methylation. The DAPK1 (1.08 in MLL-r vs 1.19 in TEL-AML-r, 1.37 in hyperdiploid, and 1.12 in normal controls) and HRK (1.40 in MLL-r vs 1.57 in TEL-AML-r, 2.19 in hyperdiploid, and 1.71 in normal controls) promoters were preferentially methylated in MLL-r ALL cells in comparison with other leukemias and normals. CCR6 promoters had some methylation in all leukemias, but were preferentially methylated in MLL-r ALL and preferentially unmethylated in normals. With the exception of the CCR6 gene, where hyperdiploid leukemias showed greater methylation than MLL-r leukemias, all genes and groups compared either reached or trended toward statistical significance, with MLL-r leukemias having a lower U:M ratio (Figure 6C-D). In cell line analysis, MSP U:M amplicon ratios of genes of interest increases during treatment with decitabine in MLL-r cell lines, indicating that methylation was reversed during the course of treatment (Figure 7). In summary, we found that for the genes identified by the HELP assay and shown in the cell lines to be re-expressed after decitabine treatment, the promoter regions were reversibly methylated. This serves both as a validation of the HELP data and implies that reversal of methylation is the likely mechanism of gene re-expression and cell kill during treatment with decitabine.
MSP unmethylated–methylated amplicon ratio of genes of interest increases during treatment with decitabine in MLL-r cell lines. PCR primers for anticipated post–bisulfite-methylated (M) and unmethylated (U) promoter sequences for the genes DAPK1, CCR6, and HRK were used to determine their methylation status. MSP was performed on DNA from each cell line before treatment (time 0) with decitabine and again at 72 hours of treatment with 2 and 4μM decitabine. In addition, densitometry was performed on all amplicons and the unmethylated amplicon–methylated amplicon (U:M) densitometry ratios were calculated, with a smaller ratio indicating a lower degree of methylation. The graph demonstrates that for DAPK1, CCR6, and HRK the time 0 U:M ratios are generally smaller for the MLL-r cell lines ([A] SEMK2, [B] KOPN8, and [C] HB1119) than for the MLL-wt cell lines ([D] NALM6, [E] 380, and [F] TANOUE) and there is a posttreatment trend toward increased U:M ratios in the MLL-wt cell lines, indicating a demethylating effect of decitabine treatment. This phenomenon was not seen in the MLL-wt cell lines.
MSP unmethylated–methylated amplicon ratio of genes of interest increases during treatment with decitabine in MLL-r cell lines. PCR primers for anticipated post–bisulfite-methylated (M) and unmethylated (U) promoter sequences for the genes DAPK1, CCR6, and HRK were used to determine their methylation status. MSP was performed on DNA from each cell line before treatment (time 0) with decitabine and again at 72 hours of treatment with 2 and 4μM decitabine. In addition, densitometry was performed on all amplicons and the unmethylated amplicon–methylated amplicon (U:M) densitometry ratios were calculated, with a smaller ratio indicating a lower degree of methylation. The graph demonstrates that for DAPK1, CCR6, and HRK the time 0 U:M ratios are generally smaller for the MLL-r cell lines ([A] SEMK2, [B] KOPN8, and [C] HB1119) than for the MLL-wt cell lines ([D] NALM6, [E] 380, and [F] TANOUE) and there is a posttreatment trend toward increased U:M ratios in the MLL-wt cell lines, indicating a demethylating effect of decitabine treatment. This phenomenon was not seen in the MLL-wt cell lines.
Discussion
This study revealed several interesting findings. First, in an unsupervised analysis using HELP data, each of the 3 sample cohorts (MLL-r leukemias, MLL-wt leukemias, and normal controls) tended to cluster together. Genome-wide methylation studies have been previously used to classify cancers,42,43 but unique genome-wide “methylome signatures” have not been previously described in pediatric leukemias. The findings suggest that there are remarkable intragroup similarities and intergroup differences in promoter methylation patterns among the study populations and that these patterns are driven by the underlying biology of each group (primarily determined by recurrent cytogenetic abnormalities in the leukemias). In addition, this lends further evidence suggesting that infant MLL-r ALL is a unique leukemia.
Second, it also demonstrated that MLL-r infant ALL demonstrates global promoter CpG island hypermethylation in comparison with the groups studied here, which included other common childhood ALLs and normal controls. This shows that promoter CpG island hypermethylation is comparatively specific to infant ALL within the subset of childhood lymphoid leukemias, suggesting that therapeutic agents that reverse DNA hypermethylation may have selective antileukemic efficacy in infants with MLL-r ALL. Further studies will be needed to determine whether this finding is specific to infants with MLL-r ALL, or whether it also applies to older children and adults with MLL-r ALL or to patients of any age with either de novo or treatment-related MLL-r acute myeloid leukemia (AML).
Third, the DNA methyltransferase inhibitor (DNMTi) 5-aza-2′-deoxycytidine (decitabine) was preferentially cytotoxic to MLL-r ALL cell lines, and the mechanism of action of this selective cytotoxicity is likely reversal of promoter methylation and up-regulation of silenced tumor suppressor genes. It is known that DNA methyltransferase (DNMT) is required for the maintenance of promoter hypermethylation and several DNA methyltransferase inhibitors (DNMTis) have attained Food and Drug Administration approval for myelodysplastic syndromes and are undergoing phase 2 to 3 trials in adult AML and chronic myeloid leukemia.44,45 We selected decitabine for this study not only because it is a powerful, well-described, and Food and Drug Administration–approved DNMTi, but also because it can be dissolved in water, thus avoiding the known effect dimethyl sulfoxide has on the methylation profile, even at low concentrations.46 Of 7 genes expected to be relatively hypermethylated based on HELP assay results or prior data, 5 (DAPK1, LIFR, HRK, FHIT, and CCR6) were shown to be biologically silenced and 4 (DAPK1, HRK, FHIT, and CCR6) showed reactivation after treatment with decitabine. To both validate the HELP assay results and show that the silencing and subsequent reactivation after decitabine treatment were secondary to promoter CpG island hypermethylation, we performed methylation-specific PCR (MSP) on primary cells. The DAPK1, HRK, and CCR6 genes clearly show reversible promoter hypermethylation by MSP in the MLL-r samples. Although there is mounting clinical evidence of the efficacy of DNMTis in patients with diseases known to have global hypermethylation of promoter CpG islands, the exact in vivo mechanism of action of these drugs is still a subject of debate.47 However, Gore et al showed that the methylated p15 and CDH-1 promoters were unmethylated in 6 of 6 responders and not unmethylated in 0 of 6 nonresponders in myelodysplastic syndrome/AML patients receiving combination DNMTi and histone deacetylase inhibitor therapy.48 Although the subject of DNMTi in vivo activity continues to be studied, we have provided further in vitro evidence that DNMTi may be a rational therapeutic strategy to explore in the high-risk MLL-r infant ALL patient population.
Suppression of DAPK1, CCR6, and HRK expression has been previously correlated with cancer and tumor progression. However, only DAPK1 has been specifically linked to MLL-r ALL in past studies. The death-associated protein kinase 1 (DAPK1) is part of a 5-member family of proapoptotic serine/threonine kinases that are ubiquitously expressed and are capable of inducing apoptosis.49 DAPK1 hypermethylation has been specifically linked to pediatric leukemias carrying the MLL-AF4 oncoprotein,32 the development of chronic lymphocytic leukemia,50 progression of chronic myeloid leukemia to blast crisis,33 and several solid tumors.51,52 This study confirms the previous findings and also suggests that DAPK1 promoter hypermethylation may be a phenomenon of several different MLL translocations.
Chemokines are a superfamily consisting of small chemotactic cytokines that interact with G-protein–coupled receptors and CCR6 is the receptor for the inflammatory and homeostatic chemokine CCL20.53 Interestingly, CCR6 is up-regulated in several solid tumors,54,55 although gene and protein expression studies have found it absent or greatly down-regulated in a host of hematologic malignancies including precursor B-cell ALL,56 chronic lymphocytic leukemia,57 and adult T-cell leukemia/lymphoma.58 This is the first study to suggest that CCR6 expression may be regulated by promoter region hypermethylation or other epigenetic phenomena. This study should spur further investigation into the possible epigenetic control of CCR6 and its potential role in leukemogenesis.
HRK (Harakiri) is a well-described member of the BH3 family of proteins known to selectively interact with the prosurvival proteins Bcl-2 and Bcl-xL to promote apoptosis.59 Its promoter hypermethylation and silencing have been described previously in solid tumors but not in leukemia.34,60 However, in a recent gene expression array, it was shown to be down-regulated in l-asparaginase resistant B-lineage ALL cells.61 This finding correlates well with the studies cited in this section, noting that infant leukemia blasts in vitro are l-asparaginase resistant.3,4 It is not yet clear how the epigenetic modifications of DAPK1, CCR6, and HRK might influence infant ALL. However, their known role in other cancers and their known importance in cell survival, along with the results from our study, support the concept that their silencing, and reactivation after treatment with decitabine, may well play a role in the biology of MLL-r leukemias.
Limitations of the study include the small number of samples analyzed within each group and the fact that several important subsets of childhood ALL were not represented (eg, Philadelphia chromosome–positive ALL). Definitive conclusions will require further studies with larger samples sizes. It will be of interest, for example, to determine methylation patterns in a wider spectrum of MLL-r infant ALL cases, including samples with other MLL fusion partners (eg, AF9, AF6) and samples from various age groups (eg, < 90 days vs > 90 days). It will also be of considerable interest to extend this analysis to cohorts of other MLL-r leukemias such as MLL-r ALL in older children and adults and MLL-r AML.
The epigenetic phenomenon of CpG island hypermethylation in tumor suppressor gene promoters is known to be an important contributor to oncogenesis.22 The MLL gene, which is rearranged in 80% of infant leukemia cases, is known to harbor domains with epigenetic activity, and silencing of several tumor suppressor genes in MLL-r infant leukemia has been described in the literature.32,41 In this study, we have demonstrated that silencing of many genes through global promoter region CpG island hypermethylation is a characteristic of MLL-r infant leukemia samples compared with other childhood leukemias and normal controls. In addition, we have demonstrated that the HELP assay can identify individual genes whose promoters are hypermethylated in MLL-r infant ALL and that many of the genes identified are transcriptionally silenced. Finally, we showed not only that the demethylating agent decitabine preferentially kills MLL-r lymphoblastic leukemia cell lines but also that this response correlates with the up-regulation of several of the identified silenced genes. Future studies using increased numbers of primary samples will be important to generalize these findings and to better correlate methylated promoter CpG islands with genome-wide gene expression. This should enable us to better identify a set of biologically important genes that are both hypermethylated and down-regulated in an effort to increase our understanding of leukemogenesis in infant ALL and to determine the ultimate gene set whose re-expression can be targeted with new treatment strategies.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the National Cancer Institute (K23 CA111728 [P.B.] and T32 CA060441 [E.S.]), the Damon Runyon-Lilly Clinical Investigator Award (30-06 [P.B.]), the Leukemia & Lymphoma Society (SCOR 7372-07 [P.B. and D.S.]), the Children's Cancer Foundation (P.B.), the Gabrielle's Angel Foundation (P.B.), and the Optimist International Research Fellowship (E.S.).
Authorship
Contribution: E.S. designed and performed research, performed data analysis, and wrote the paper; R.I. performed data analysis and edited the paper; S.N. and E.M. performed research; D.S. contributed to data analysis and edited the paper; M.E.F. designed research, contributed to data analysis, and edited the paper; A.M. designed research, contributed to data analysis, and edited the paper; and P.B. designed research, performed data analysis, and wrote and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Patrick Brown, Johns Hopkins Oncology, CRB1 2M49, 1650 Orleans St, Baltimore, MD 21231; e-mail: pbrown2@jhmi.edu.

![Figure 7. MSP unmethylated–methylated amplicon ratio of genes of interest increases during treatment with decitabine in MLL-r cell lines. PCR primers for anticipated post–bisulfite-methylated (M) and unmethylated (U) promoter sequences for the genes DAPK1, CCR6, and HRK were used to determine their methylation status. MSP was performed on DNA from each cell line before treatment (time 0) with decitabine and again at 72 hours of treatment with 2 and 4μM decitabine. In addition, densitometry was performed on all amplicons and the unmethylated amplicon–methylated amplicon (U:M) densitometry ratios were calculated, with a smaller ratio indicating a lower degree of methylation. The graph demonstrates that for DAPK1, CCR6, and HRK the time 0 U:M ratios are generally smaller for the MLL-r cell lines ([A] SEMK2, [B] KOPN8, and [C] HB1119) than for the MLL-wt cell lines ([D] NALM6, [E] 380, and [F] TANOUE) and there is a posttreatment trend toward increased U:M ratios in the MLL-wt cell lines, indicating a demethylating effect of decitabine treatment. This phenomenon was not seen in the MLL-wt cell lines.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/23/10.1182_blood-2009-09-243634/4/m_zh89991052360007.jpeg?Expires=1769129882&Signature=uNT9UbjcqO4hwCyZSP-nzHGEMTkhm8w9QHxLSBhpMJD3cOgRoYNJSXzIFNLla5JR~poQC7X7LbpE2imIOr0~KH6Ui5ICDgh2lza5hW1Kagfa-F-82-U6Lf3Nxhqu7i4YsV~vFYCfi2W8T9CRRmlrJ2KDgmhJsTDqnAs-JBwSzUsOeodbopN4MQhuRNl88oy14GnAq3Mvf-jARWjZ0SvbvZ07bMH~6GCCEXmW4DbWKhd3lx03JZDc5MKNov8KKucz4XYjPD~fcV7QUqC8p0Ro83L4ww7L56worwxQyOevOLZ3YgsOBwrjAFnq~5ObB~G0X4B9bV1YL9RibJRM1e0xWg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)