Abstract
It is believed that hemopoietic stem cells (HSC), which colonize the fetal liver (FL) rapidly, expand to establish a supply of HSCs adequate for maintenance of hemopoiesis throughout life. Accordingly, FL HSCs are actively cycling as opposed to their predominantly quiescent bone marrow counterparts, suggesting that the FL microenvironment provides unique signals that support HSC proliferation and self-renewal. We now report the generation and characterization of mice with a mutant allele of Baf250a lacking exons 2 and 3. Baf250aE2E3/E2E3 mice are viable until E19.5, but do not survive beyond birth. Most interestingly, FL HSC numbers are markedly higher in these mice than in control littermates, thus raising the possibility that Baf250a determines the HSC pool size in vivo. Limit dilution experiments indicate that the activity of Baf250aE2E3/E2E3 HSC is equivalent to that of the wild-type counterparts. The Baf250aE2E3/E2E3 FL-derived stroma, in contrast, exhibits a hemopoiesis-supporting potential superior to the developmentally matched controls. To our knowledge, this demonstration is the first that a mechanism operating in a cell nonautonomous manner canexpand the pool size of the fetal HSC populations.
Introduction
During mouse ontogeny, adult-type hemopoietic stem cells (HSCs) emerge at around embryonic day 10 (E10) day postcoitus (dpc) and undergo a complex developmental processes occurring in the yolk sac, the aorta-gonad-mesonephros region, the placenta, and the fetal liver (FL). Before birth the HSC migrate to the bone marrow (BM), which remains the main site of hemopoiesis into adulthood.1 Between E12 and E16 dpc, the populations of FL HSCs expand nearly 40-fold to generate a near-sufficient supply of HSCs for the life of the animal.2 To achieve this population size, the majority of fetal HSC are cycling3 and likely undergo self-renewal divisions. In adult BM, in contrast, the majority of HSCs with long-term lympho-myeloid repopulation potential are quiescent, possibly dividing only 5 to 6 times during their lifetime.4,5
During homeostasis, the balance between HSC self-renewal and differentiation is regulated by the molecular crosstalk between HSCs and the various soluble, structural, and cellular constituents of BM microenvironment that they reside in known as the HSC niche.6 By inference, FL microenvironment may include temporary restricted niches that promote symmetrical self-renewing divisions of the fetal HSC, although the composition of these FL niches remains poorly defined. Experimental evidence indicates that the ancestors of the AFT024 cell line that support in vitro HSC self-renewal and differentiation7 may represent a key component of such niches. AFT024 cells are a clonal derivative of stromal cell culture established from E14.5 FL,8 and differentiate in response to appropriate stimuli into adipocytes, osteoblasts, chondrocytes, and vascular smooth muscle cells. These cells therefore share some characteristics with mesenchymal stem cells,9 which are present in all major hemopoietic sites throughout ontogeny,10 supporting the possibility that the putative fetal HSC niches comprise these multipotent cells and/or their differentiated progeny.
Conditions that favor self-renewal divisions of fetal HSC result from integration of HSC-specific intrinsic factors with extrinsic cues that originate from the corresponding microenvironment. The homeodomain protein MEIS1 is likely among the key intrinsic regulators of HSC activity in FL. MEIS1 deficiency is compatible with the establishment of definitive HSCs, but not with their expansion in FL,11,12 and similar functions were proposed for the core binding protein13 and the polycomb group gene mph1 (Rae-28).14 Interestingly, extrinsic regulators implicated in maintenance of adult stem cell identity, such as Angiopoietin 1 and c-Kit ligand, are dispensable for expansion of fetal HSC.15,16 Although requirement for Pitx2 activity for establishment and/or maintenance of the HSC-supportive ability of primary FL stromal cells has been identified,17 the identity of putative candidates that enhance the pool size of fetal HSC populations remains to be elucidated.
In a yeast 2-hybrid screen, we have recently identified BAF250a, a component of the Swi/Snf-BAF chromatin remodeling complex,18 as a putative MEIS1–interacting protein. Using a gene targeting approach, we generated a mutant Baf250a allele lacking exons 2 and 3 (Baf250aE2E3/+). Examination of Baf250aE2E3/E2E3 FL populations suggested that the Baf250aΔ2,3 allele enables normal establishment, maintenance, and differentiation of definitive HSC populations, but represents an important regulator of FL HSC populations.
Methods
Baf250a targeting vector and generation of Baf250aE2E3/E2E3 mice
The targeting vector (Figure 1A) comprised LoxP sites flanking approximately 11 kb of genomic Baf250a sequences (NM_0011080819) encompassing exons 2 and 3, and carried the PGK-neo cassette in reverse orientation inserted in intron 3. Detailed information about vector construction, strategy for selection of Baf250aE2E3 RI embryonic stem (ES) cells, and generation of chimeric mice is available upon request.
Animals and genotyping
The 129SvE Baf250aE2E3 chimeric mice were backcrossed at least 5 times into C57BL/6J (CD45.2+) background. Transplant recipients were C57BL/6Ly-Pep3b (CD45.1+) mice. Animals were housed in a specific pathogen-free animal facility and handled in accordance with the guidelines of the Animal Care and Use Committee at Université de Montréal, which also approved all experimental procedures. Genotyping of mice and embryos was performed using Sigma REDExtract-N-Amp Tissue PCR Kit following the recommendations of the manufacturer (Sigma-Aldrich) and the following primers: forward mutant, 5′-tacctattctctatcccctatt-3′; forward wild-type (WT), 5′-cctggttagttggttggtct-3′; and reverse 5′-ttacattcagcacctggcag-3′. For Southern blot analyses of Baf250a locus, the genomic DNA was digested as described in Figure 1. Probe used was the 400-bp fragment of genomic DNA located 3′ to the recombination site.
Coimmunoprecipitation and Western blot analyses
Preparation of nuclear protein extracts,19 coimmunoprecipitations,20 and Western blot analyses21 were performed as previously described. Antibodies used were anti-BAF250a/PSG3 (Santa Cruz Biotechnology), anti-Brg1 (Abcam), and horseradish peroxidase-conjugated anti–rat and anti–rabbit antibodies (Santa Cruz Biotechnology).
Flow cytometry
To evaluate contribution of the transplanted test cells to repopulation of recipient mice, peripheral blood lymphocytes (PBLs) were incubated with fluorescein isothiocyanate-conjugated anti-Ly45.2, phycoerythrin (PE)–conjugated anti-CD45R and allophycocyanin (APC)–conjugated anti-Ly6G (BD Pharmingen). For evaluation of primitive hemopoietic cell populations in E14.5 dpc FL, the following antibodies were used: PE–cyanin 7 (PE-Cy7)–conjugated anti-CD150 (eBioscience), PE-conjugated anti-Ly6A/E (BD Pharmingen), and APC-conjugated anti-CD48, anti-TER119, anti-Ly6G, anti-CD45R, and anti-CD3 (eBioscience). To evaluate HSC populations in BM cells isolated from transplantation, chimeras cells were incubated with antibody mix including APC-conjugated anti-CD117 (BD Pharmingen), PE–cyanin 5.5 (PE-Cy5.5)–conjugated anti-Ly6A/E, PE-conjugated anti-CD135 (eBioscience), peridinin chlorophyll protein–Cy5.5-conjugated anti-CD45.2 (eBioscience), and biotinylated antibodies specific for Ly6G, CD3, CD4, CD8, CD11b, CD19, CD34, CD45R followed by incubation with streptavidin-conjugated APC-Cy7 (BD Pharmingen). To assess the proliferation activity of fetal progenitor/stem cell populations, pregnant females were injected intraperitoneally with 2 mg of 5-bromo-2′-deoxyuridine (BrdU) in phosphate-buffered saline (BD Pharmingen) 30 minutes beforesacrifice. FL cell populations were processed immediately as recommended by the manufacturer, and were stained with PE-Cy7–conjugated anti-CD150, fluorescein isothiocyanate–conjugated anti-CD48, anti-TER119, anti-Ly6G, anti-CD45R, and anti-CD3, Alexa Fluor 647–conjugated anti-BrdU (BD Pharmingen), and 4′-6-diamidino-2-phenylindole (Molecular Probes). Events were acquired using BD LSRII and BD FACSDiva 4.1 software (BD Pharmingen), which was also used for data analysis.
Cobblestone area–forming cells and clonogenic progenitor assays
Maintenance of AFT024 cells8 and cobblestone area–forming cell (CAFC) day-28 evaluation22 (CAFCday28) were performed as described. To establish primary stromal cell cultures, FL tissue was disrupted with gentle trituration in Dulbecco modified Eagle medium supplemented with 2% fetal bovine serum (FBS). The resulting cell suspension was pelleted, resuspended in long-term culture medium comprising alpha-minimum essential medium, 12.5% FBS, 12.5% horse serum (Invitrogen), 2 × 10−4 mol/L i-Inositol, 1.6 × 10−5 mol/L folic acid, 1 × 10−4 mol/L β-mercaptoethanol (Sigma-Aldrich), and 1 × 10−6 mol/L hydrocortisone hemisuccinate (StemCell Technologies), and plated at density 2 × 10−6 cells/cm2 on gelatin precoated tissue culture dishes or 96-well trays. After the 4-day incubation, half of the medium was changed. Confluent cell layers developing during the 8- to 10-day incubation were irradiated (20 cGy, 137Cs γ source; J. L. Shepherd) and used as feeder layers in experiments as described. All experiments were performed with primary stromal cell cultures. For evaluation of mesenchymal progenitor cell content (in colony-forming units–fibroblast [:CFU-F]:), 500 to 50 000 FL cells/cm2 in Dulbecco modified Eagle medium supplemented with 10% FBS were seeded in 6-well trays. After the 10-day incubation, the colonies of adherent cells were stained with 1% methylene blue solution in methanol. The CAFC and CFU-F frequencies were calculated using Limit Dilution Analysis software (StemCell Technologies). Clonogenic progenitor assays were performed as described.23
Competitive repopulation unit enumeration assays
For competitive repopulation assays, 2 × 105Baf250aE2E3/E2E3, Baf250aE2E3, or WT E14.5 dpc FL cells (CD45.2+) were mixed with 2 × 105 competitor E14.5 dpc FL cells (CD45.1+), and transplanted into irradiated (850 cGy, γCs source; J. L. Shepherd) C57BL/6Ly-Pep3b (CD45.1+) mice. Competitive repopulation unit (CRU) assays were performed as described.24 Briefly, various numbers of CD45.2+Baf250aE2E3/E2E3 or WT FL cells (200 cells-5 × 105 cells per recipient) were injected together with 105 competitor CD45.1+ BM cells into lethally irradiated CD45.1+ recipients (5-10 mice per group). Repopulation of the hematopoietic system in transplant recipients was evaluated by analysis of PBL for the presence of CD45.2+ (donor-derived) myeloid and B lymphoid cells at 20 weeks after transplantation. The CRU frequency in the transplanted FL populations was calculated using Limit Dilution Analysis software (StemCell Technologies).
Evaluation of the HSC activity in transplant recipients
Proportions of peripheral blood cells generated by the assay (CD45.2+) WT or Baf250aE2E3/E2E3 HSCs in transplant recipients at limit of dilution were used to determine the mean activity of stem cells (MAS) as described.25 Briefly, the proportion of chimerism shown by [:(% test donor cells) × 100 / (% test donor cells + % competitor cells)]: was calculated from the FACS data. The number of repopulating units (RU) in test cell populations was calculated as RU = (% chimerism) / (100 − % chimerism), and MAS was determined as RU divided by CRU. The CRU numbers in test cell populations were estimated as described in the previous paragraph. FACS data obtained from analyses of these mice were also used to evaluate the contribution of the transplanted test cells of WT or Baf250aE2E3/E2E3 (CD45.2+) or competitor WT (CD45.1+) to the regeneration of the HSC pool as defined by the CD117+Ly6A/E+ (CD135+Lin−) phenotype of recipient BM cells.
Microarray hybridization and analyses
A total of 10 μg of RNA was reverse transcribed using SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) according to the manufacturer's instructions. For the WT samples, 1 μg of purified cDNA was labeled with Cy5-labeled 9mers (Trilink Technologies) using 3′-5′ exo-Klenow fragment (New England Biolabs), whereas the Baf250aE2E3/E2E3 samples were labeled with Cy3-labeled 9mers. The 2-color hybridizations were carried out using 6 μg of Cy5-labeled WT and 6 μg of Cy3-labeled Baf250aE2E3/E2E3 cDNA using the NimbleGen mouse microarrays 2006-08-03_MM8_60mer_expr and hybridization kit as recommended by the manufacturer (Roche NimbleGen). Arrays were scanned at 5-μm resolution using a GenePix4000B scanner (Molecular Devices). Data from scanned images were extracted using NimbleScan 2.5 extraction software (Roche NimbleGen) and analyzed using Genespring.GX (Version 7.3; Agilent Technologies). All microarray data are available on ArrayExpress under accession number E-TABM-990.
Results
Generation of Baf250aE2E3/E2E3 mutant mice
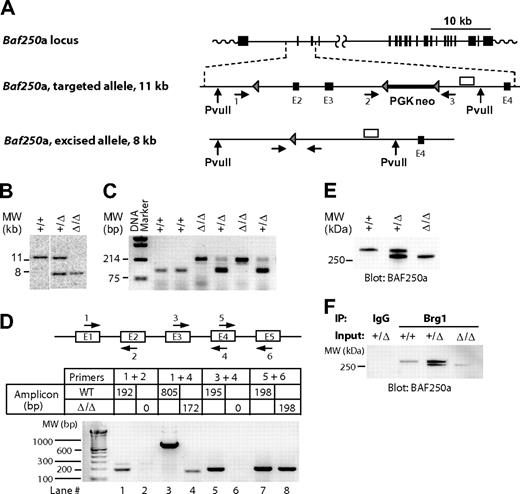
To examine the role of the Swi/Snf-BAF250a complexes in hemopoiesis, we generated a mutant mouse strain harboring a Baf250a allele (hereafter, Baf250aE2E3/E2E3) lacking exons 2 and 3 (Figure 1A). Southern blot analyses of the Baf250aE2E3/E2E3 locus (Figure 1B), or the polymerase chain reaction (PCR)–based genotyping (Figure 1C) confirmed deletion of the targeted region. Sequencing of the product generated by the reverse transcription (RT)–coupled PCR revealed that transcription of Baf250aE2E3/E2E3 allele proceeds from exon 1 directly into exon 4 (Figure 1D lanes 3 and 4, compare with lanes 1 and 2 or lanes 5 and 6) to generate mRNA coding in frame a BAF250aE2E3 protein lacking approximately 150 amino acids within the N-terminal region. This deletion had no effect on either expression levels of the mutated protein (Figure 1E), nor on its ability to interact with Brg1, the ATPase subunit of the Swi/Snf-BAF complex (Figure 1F).
Generation of Baf250aE2E3/E2E3 mouse strain. (A) Baf250a targeting strategy. Top, Baf250a locus (NM_001080819_MGI:1935147_Arid1a). Middle, the targeted allele before Cre-mediated excision. Bottom, the recombined Baf250aE2E3 allele. Exons (■), LoxP sites (▴), primers for PCR-based genotyping (numbered horizontal arrows), and 3′ external genomic probe (□) are shown. Positions of PvuII restriction sites are also denoted. (B) Southern blot analysis of Baf250a modification. DNA isolated from embryonic tissues was digested with PvuII, which cuts outside of the targeted region to generate a 11.6 kb WT and a 7.9 kb Baf250aE2E3 bands recognized by the 3′ probe depicted in Figure 1A. (C) PCR-based amplifications of Baf250aE2E3 allele. The primer pair 1 + 3 generates a 210-bp fragment identifying the excised Baf250aE2E3, and primers 2 + 3 pair generate the 90-bp WT fragment. Positions of primers are denoted in Figure 1A. (D) Quantitative RT-PCR–based analysis of Baf250aE2E3 mRNA. Note the absence of RT-PCR signal for homozygous Baf250aE2E3 cells when using primers positioned in exons 2 or 3, and the 172-bp segment amplified with primers in exons 1 and 4. Top, schema of primer positions. Bottom, amplicons generated by the denoted primer pairs. (E) Western blot analysis of BAF250a levels in WT, Baf250aE2E3, and Baf250aE2E3/E2E3 embryonic fibroblasts. The BAF250a antibody recognizes the approximately 280 kDa WT, and approximately 250 kDa mutated BAF250a proteins. (F) BAF250aE2E3 protein interacts with Brg1. Anti-Brg1 antibody coimmunoprecipitates WT and mutated BAF250a.
Generation of Baf250aE2E3/E2E3 mouse strain. (A) Baf250a targeting strategy. Top, Baf250a locus (NM_001080819_MGI:1935147_Arid1a). Middle, the targeted allele before Cre-mediated excision. Bottom, the recombined Baf250aE2E3 allele. Exons (■), LoxP sites (▴), primers for PCR-based genotyping (numbered horizontal arrows), and 3′ external genomic probe (□) are shown. Positions of PvuII restriction sites are also denoted. (B) Southern blot analysis of Baf250a modification. DNA isolated from embryonic tissues was digested with PvuII, which cuts outside of the targeted region to generate a 11.6 kb WT and a 7.9 kb Baf250aE2E3 bands recognized by the 3′ probe depicted in Figure 1A. (C) PCR-based amplifications of Baf250aE2E3 allele. The primer pair 1 + 3 generates a 210-bp fragment identifying the excised Baf250aE2E3, and primers 2 + 3 pair generate the 90-bp WT fragment. Positions of primers are denoted in Figure 1A. (D) Quantitative RT-PCR–based analysis of Baf250aE2E3 mRNA. Note the absence of RT-PCR signal for homozygous Baf250aE2E3 cells when using primers positioned in exons 2 or 3, and the 172-bp segment amplified with primers in exons 1 and 4. Top, schema of primer positions. Bottom, amplicons generated by the denoted primer pairs. (E) Western blot analysis of BAF250a levels in WT, Baf250aE2E3, and Baf250aE2E3/E2E3 embryonic fibroblasts. The BAF250a antibody recognizes the approximately 280 kDa WT, and approximately 250 kDa mutated BAF250a proteins. (F) BAF250aE2E3 protein interacts with Brg1. Anti-Brg1 antibody coimmunoprecipitates WT and mutated BAF250a.
Baf250asE2E3/+ mice were born at normal Mendelian ratios and remained healthy for a period of more than 12 months. Intercrossing of Baf250aE2E3/+ mice yielded expected proportions of correctly developed Baf250aE2E3/E2E3, Baf250aE2E3/+, and WT embryos up to E19.5 dpc. We obtained no living Baf250aE2E3/E2E3 offspring on either C57BL/6J (CD45.2+) or 129SvE genetic background. Homozygosity at Baf250aE2E3/+ locus thus likely resulted in a complex phenotype with perturbations possible in any of the processes essential for neonatal survival, namely normal parturition, breathing, suckling or maintenance of neonatal homeostasis.26 This study focused solely on the hemopoietic function of Baf250a as inferred from the activity of BAF250aE2E3/+, and characterization of other phenotypic characteristics of Baf250aE2E3/E2E3 will be reported separately.
Increased FL HSC pool size in Baf250aΔ2,3 mutant mice
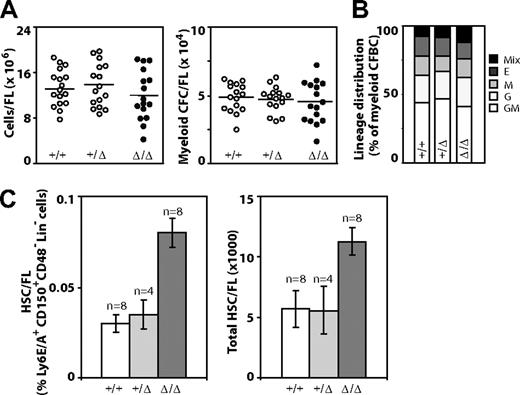
Examination of FL cell populations obtained from live and grossly normal E14.5 dpc WT, Baf250aE2E3/+, and Baf250aE2E3/E2E3 littermates revealed that erythroid, megakaryocytic, and myeloid cell lineages develop normally in mice from all 3 genotypes (data not shown). The total cellularity of Baf250aE2E3/E2E3 FL was comparable with that of the Baf250aE2E3/+ or WT littermate controls (Figure 2A left panel), and no quantitative (Figure 2A right panel) or qualitative differences (Figure 2B) in populations of myeloid clonogenic progenitors could be detected. Together, these observations implied that the WT allele of Baf250a is not essential for HSC specification, the onset of definitive hemopoiesis, or differentiation of blood cells.
Hemopoietic characterization of E14.5 dpc Baf250aE2E3/E2E3 FL cell populations. (A) Total number of FL cells (left) and number of myeloid clonogenic progenitors (right) in FL. (B) Frequency of distribution of different myeloid progenitors in FL. (C) The frequency of HSCs in E14.5 FL (left) as defined by the CD150+Ly6A/E+CD48−Lin− phenotype. The total number of HSCs in E14.5 FL (right). The HSC numbers were calculated from total mononuclear cell numbers and the HSC frequency determined for each FL population. Shown are mean (± SD). Genotypes of the examined FL populations are shown at with image. Mix indicates CFU-granulocyte, erythrocyte, monocyte/macrophage, megakaryocyte; E, CFU-erythroid; M, CFU-monocyte/macrophage; G, CFU-granulocyte; and GM, CFU-granulocyte-monocyte/macrophage.
Hemopoietic characterization of E14.5 dpc Baf250aE2E3/E2E3 FL cell populations. (A) Total number of FL cells (left) and number of myeloid clonogenic progenitors (right) in FL. (B) Frequency of distribution of different myeloid progenitors in FL. (C) The frequency of HSCs in E14.5 FL (left) as defined by the CD150+Ly6A/E+CD48−Lin− phenotype. The total number of HSCs in E14.5 FL (right). The HSC numbers were calculated from total mononuclear cell numbers and the HSC frequency determined for each FL population. Shown are mean (± SD). Genotypes of the examined FL populations are shown at with image. Mix indicates CFU-granulocyte, erythrocyte, monocyte/macrophage, megakaryocyte; E, CFU-erythroid; M, CFU-monocyte/macrophage; G, CFU-granulocyte; and GM, CFU-granulocyte-monocyte/macrophage.
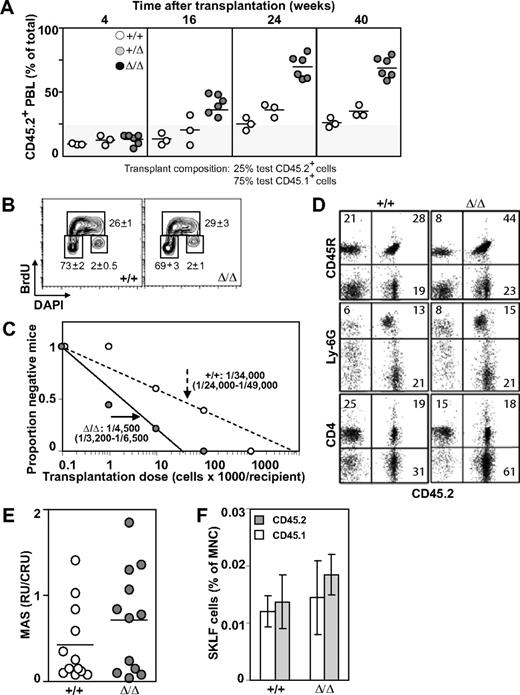
Baf250aE2E3/E2E3 FLs were characterized, however, by an approximately 2-times higher frequency and absolute number of phenotypically defined (CD150+Ly6A/E+CD48−Lin−) HSCs than WT or Baf250aE2E3/+ littermate controls (mean ± SD; P < .05, by t test; Figure 2C). To examine the competitive repopulation ability of the Baf250aE2E3/E2E3 FL cells, the test (CD45.2+) WT, Baf250aE2E3/+, and Baf250aE2E3/E2E3 cells were mixed with WT congenic (CD45.1+) competitor FL cells in a 1:4 ratio, and transplanted into irradiated (CD45.1+) recipients. At 24 weeks after transplantation, the peripheral blood leukocytes in recipients of WT and/or Baf250aE2E3/+ test cells retained the initial 1:4 test-to-competitor cell ratio (Figure 3A). In recipients of Baf250aE2E3/E2E3 cells, the progeny of mutant cells increased approximately 3-fold higher than the expected values, but no further increase was observed at 40 weeks after transplantation, suggesting that the repopulating activity of the transplanted mutant cells had reached the plateau levels. These observations suggested that homozygosity for Baf250aE2E3 correlated with the enhanced competitive repopulation ability of the mutant cell populations, but did not clarify whether this advantage occurred at the level of HSCs (ie, more HSCs per FL), or at the level of more mature progenitors (ie, higher proliferative potential per HSC).
Effect of Baf250aE2E3 on HSC activity. (A) Homozygosity at Baf250aE2E3 allele enhances the competitive repopulation ability of mutant FL cells. WT, Baf250aE2E3, or Baf250aE2E3/E2E3 (CD45.2+) FL cells were mixed in a 1:4 ratio with CD45.1+ WT FL cells, and transplanted into lethally irradiated CD45.1+ recipient mice. Contribution of donor cells in peripheral blood of recipient mice was assessed at different times after transplantation. (B) BrdU incorporation profile of the CD150+CD48−Lin− cell population was assessed after 30 minutes in vivo BrdU pulse. Cell numbers are mean (± SD) values obtained from 3 developmentally matched FL. (C) Limiting dilution analysis for estimation of CRU frequency in WT and mutant FL (2 FL populations and 2 cohorts of recipient mice for each genotype). (D) Lympho-myeloid repopulation of recipient mice transplanted 20 weeks before with 100 000 WT or 10 000 Baf250aE2E3/E2E3 FL cells. Representative FACS profiles demonstrate comparable contributions of the transplant cell populations to repopulation of myeloid (Ly6G+), B lymphoid (CD45R+), and T lymphoid (CD4+) PBL. Average value in each quadrant represents number determined for mice (n = 5 in each group). (E) Proliferation potential of the transplanted HSCs. MAS, or proportion of PBL generated by individual transplant test CRU, was determined for groups of mice (n = 12) transplanted with low numbers (1-3) of Ly45.2+ WT or Baf250aE2E3/E2E3 FL CRU, together with 10 BM-derived competitor Ly45.1+CRU. (F) Contributions of the transplanted WT and Baf250aE2E3/E2E3 CRUs to regeneration of adult HSC pool. Proportions of test CD45.2+ and competitor CD45.1+-derived CD117+Ly6A/E+ (CD135−Lin− cells) in the BM of recipients described for Figure 1E were determined at 24 weeks after transplantation. Results represent mean (± SD) determined for each group of recipients (n = 5).
Effect of Baf250aE2E3 on HSC activity. (A) Homozygosity at Baf250aE2E3 allele enhances the competitive repopulation ability of mutant FL cells. WT, Baf250aE2E3, or Baf250aE2E3/E2E3 (CD45.2+) FL cells were mixed in a 1:4 ratio with CD45.1+ WT FL cells, and transplanted into lethally irradiated CD45.1+ recipient mice. Contribution of donor cells in peripheral blood of recipient mice was assessed at different times after transplantation. (B) BrdU incorporation profile of the CD150+CD48−Lin− cell population was assessed after 30 minutes in vivo BrdU pulse. Cell numbers are mean (± SD) values obtained from 3 developmentally matched FL. (C) Limiting dilution analysis for estimation of CRU frequency in WT and mutant FL (2 FL populations and 2 cohorts of recipient mice for each genotype). (D) Lympho-myeloid repopulation of recipient mice transplanted 20 weeks before with 100 000 WT or 10 000 Baf250aE2E3/E2E3 FL cells. Representative FACS profiles demonstrate comparable contributions of the transplant cell populations to repopulation of myeloid (Ly6G+), B lymphoid (CD45R+), and T lymphoid (CD4+) PBL. Average value in each quadrant represents number determined for mice (n = 5 in each group). (E) Proliferation potential of the transplanted HSCs. MAS, or proportion of PBL generated by individual transplant test CRU, was determined for groups of mice (n = 12) transplanted with low numbers (1-3) of Ly45.2+ WT or Baf250aE2E3/E2E3 FL CRU, together with 10 BM-derived competitor Ly45.1+CRU. (F) Contributions of the transplanted WT and Baf250aE2E3/E2E3 CRUs to regeneration of adult HSC pool. Proportions of test CD45.2+ and competitor CD45.1+-derived CD117+Ly6A/E+ (CD135−Lin− cells) in the BM of recipients described for Figure 1E were determined at 24 weeks after transplantation. Results represent mean (± SD) determined for each group of recipients (n = 5).
To help clarify this observation, we first performed an in vivo BrdU incorporation assay combined with intracellular staining for DNA content (4′-6-diamidino-2-phenylindole). These experiments revealed no noticeable difference between the cell-cycle progression of WT and Baf250aE2E3/E2E3 CD150−CD48−Lin− cells (mean ± SD, n = 3; Figure 3B). We also observed that mutant and WT populations displayed similar proportions of apoptotic cells as determined by analyses of Annexin V stained cells (data not shown). We next determined functional HSC numbers in E14.5 Baf250aE2E3/E2E3 and WT FL cell populations using the CRU assay. Estimation of CRU numbers was performed using the maximum likelihood method previously described.24 At 18 weeks after transplantation, the frequency of Baf250aE2E3/E2E3 CRU was 1/4500 (n = 2; 95% confidence interval [:CI]:; range 1/3200-1/6500; Figure 3C), or approximately 4550 CRU per Baf250aE2E3/E2E3 FL. In comparison, the CRU frequency in the WT controls was estimated at 1/34 000 cells (2 independent experiments; 95% CI; range 1/24 000-1/49 000 cells). Mutant cells retained their differentiation ability and contributed to repopulation of B lymphoid (CD45R+), myeloid (Ly6G−), and T lymphoid (CD4+) PBL (Figure 3D). Recipient mice remained healthy during an 8-month observation period (n = 25), exhibited a sustained balanced production of both the test cell-derived erythroid, myeloid, and lymphoid cells in their BM, spleen, and thymus (Figure 3D, and data not shown), and the total production of functional blood cells (data not shown). Together, these observations indicated that the competitive repopulation advantage of Baf250aE2E3/E2E3 FL cells resulted from a higher number of Baf250aE2E3/E2E3 HSCs in the transplantation inocula, implying that Baf250a activity may limit the size of the HSC pool in FL.
Baf250aE2E3/E2E3 HSCs show normal proliferation and self-renewal activity
As a strategy to compare the intrinsic proliferation potential of our Baf250aE2E3/E2E3 mutant and WT CRU, we measured the number of mature blood cells produced by individual test CD45.2+ CRU transplanted in irradiated recipients along with 10 CD45.1+ BM-derived competitor CRU. At 21 weeks after transplantation, analysis of MAS25 showed that the proportion of hemopoietic progeny produced by individual Baf250aE2E3/E2E3 CRU was slightly higher, but not statistically different from controls (Figure 3E). Using phenotypical assays defined by Adolfsson et al,27 we next compared the size of the regenerated HSC pool with the relative contribution of mutant (CD45.2+) and WT (CD45.1+) HSCs to this pool. Results of these analyses revealed at best a minor, but not statistically significant difference between the levels of HSC pool size regenerated by the transplanted Baf250aE2E3/E2E3 or WT HSCs (Figure 3F). Together, these results suggested that cell nonautonomous mechanisms are responsible, at least in part, for the observed increase in HSC pool size in the FL of our homozygous Baf250aE2E3/E2E3 mice.
Baf250aE2E3 enhances the HSC supporting ability of the FL microenvironment
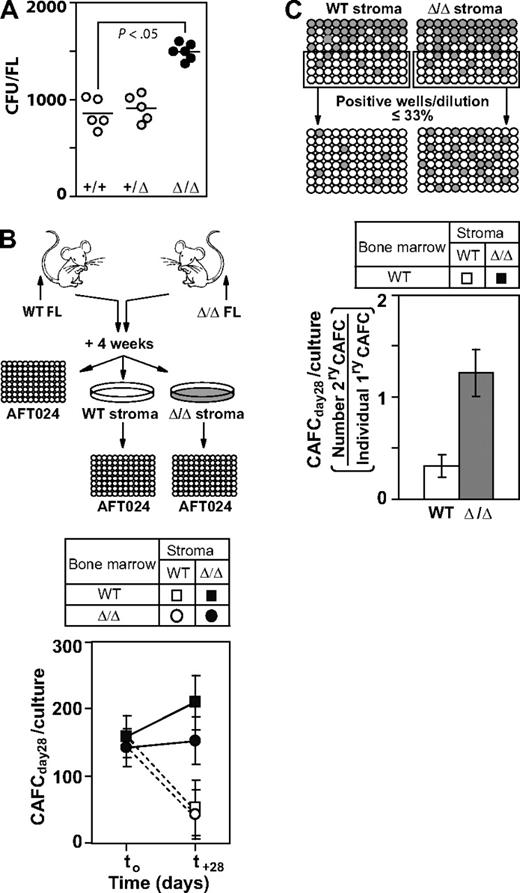
Given that mesenchymal cell populations were proposed to represent an important cellular component of the hemopoiesis-supporting FL microenvironment9 we first analyzed the mesenchymal progenitor cell (CFU-F) content in FL derived from the WT, Baf250aE2E3/+, and Baf250aE2E3/E2E3 mice. We found that these cells were consistently present at higher numbers in the mutant animals compared with controls (Baf250aE2E3/E2E3: mean ± SEM, 1451 ± 313; WT: 797 ± 74, P < .05; Figure 4A). Note that FL cellularity of both genotypes was similar (Figure 2A), suggesting that the increase in the mesenchymal cell content may have contributed to creation of microenvironment that enabled the Baf250aE2E3/E2E3–associated expansion of fetal HSC popbulations.
Baf250aE2E3/E2E3 stromal cell cultures support the in vitro self-renewal of progenitor/stem cell populations. (A) Enumeration of mesenchymal stem/progenitor cells in WT, Baf250aE2E3/+, and Baf250aE2E3/E2E3 FL cell populations. Each circular symbol represents individual FL population. Note the statistically significant difference between the WT, or Baf250aE2E3/+, and the Baf250aE2E3/E2E3 FL (unpaired t test, and 2-tailed P value determined). (B) Maintenance of CAFCday28 in the presence of WT or Baf250aE2E3/E2E3 stromal cells. Primary stromal cell cultures (4-cm - 10-cm dishes for each genotype) were established using pools of 4 FL cell populations. BM cells isolated from recipients of WT or Baf250aE2E3/E2E3 FL (2 recipients per genotype, > 85% of donor-derived PBL) were cocultured with stromal cell cultures for 28 days. CAFCday28 frequency at the beginning of experiment (t0), or after 28-day maintenance in stromal cocultures, were determined by plating an aliquot of the input BM cells, or entire cellular content of the stromal cocultures, at limit dilution on pre-established AFT024 cell layers. Results are mean (± SD), of 2 independent experiments performed in duplicate and are presented as the number of CAFC per 3 × 107 input BM cells. (C) Baf250aE2E3/E2E3 stromal cell cultures support self-renewal of progenitor/stem cell populations. WT BM cells were seeded at limit dilution on pre-established WT or Baf250aE2E3/E2E3 stromal cell layers in 96-well trays (4 independent stromal cell cultures for each genotype). After 4-week coculture, all wells estimated to comprise a single cobblestone area were replated on freshly established WT or Baf250aE2E3/E2E3 stromal cell layers, and the frequency of secondary CAFCday28 was determined after additional 4-week maintenance.
Baf250aE2E3/E2E3 stromal cell cultures support the in vitro self-renewal of progenitor/stem cell populations. (A) Enumeration of mesenchymal stem/progenitor cells in WT, Baf250aE2E3/+, and Baf250aE2E3/E2E3 FL cell populations. Each circular symbol represents individual FL population. Note the statistically significant difference between the WT, or Baf250aE2E3/+, and the Baf250aE2E3/E2E3 FL (unpaired t test, and 2-tailed P value determined). (B) Maintenance of CAFCday28 in the presence of WT or Baf250aE2E3/E2E3 stromal cells. Primary stromal cell cultures (4-cm - 10-cm dishes for each genotype) were established using pools of 4 FL cell populations. BM cells isolated from recipients of WT or Baf250aE2E3/E2E3 FL (2 recipients per genotype, > 85% of donor-derived PBL) were cocultured with stromal cell cultures for 28 days. CAFCday28 frequency at the beginning of experiment (t0), or after 28-day maintenance in stromal cocultures, were determined by plating an aliquot of the input BM cells, or entire cellular content of the stromal cocultures, at limit dilution on pre-established AFT024 cell layers. Results are mean (± SD), of 2 independent experiments performed in duplicate and are presented as the number of CAFC per 3 × 107 input BM cells. (C) Baf250aE2E3/E2E3 stromal cell cultures support self-renewal of progenitor/stem cell populations. WT BM cells were seeded at limit dilution on pre-established WT or Baf250aE2E3/E2E3 stromal cell layers in 96-well trays (4 independent stromal cell cultures for each genotype). After 4-week coculture, all wells estimated to comprise a single cobblestone area were replated on freshly established WT or Baf250aE2E3/E2E3 stromal cell layers, and the frequency of secondary CAFCday28 was determined after additional 4-week maintenance.
Using procedures shown in Figure 4B and the CAFC assay,7,22 we next assessed the ability of WT and Baf250aE2E3/E2E3 FL-derived stromal cell cultures to support stem/primitive progenitor cell populations in vitro. Through the design of limiting dilution experiments, we found that CAFCday28 represented approximately 1/200 000 of freshly isolated, unfractionated BM cells isolated from recipients of either Baf250aE2E3/E2E3 or WT FL (> 85% donor-derived PBL), for a total of approximately 150 input CAFCday28 (Figure 4B). After a 28-day maintenance on the WT stroma, the number of WT, and Baf250aE2E3/E2E3 CAFCday28 decreased to approximately one-third of their input values. Baf250aE2E3/E2E3 stroma, in contrast, supported maintenance and/or expansion of CAFCday28 for both genotypes of the input cell populations (Figure 4B). The ability of WT and Baf250aE2E3/E2E3 stroma to support the self-renewal divisions of CAFCday28 was evaluated by replating individual cobblestones onto freshly established WT or Baf250aE2E3/E2E3 stromal cell layers (Figure 4C). In the presence WT stroma, only approximately 1/3 of the CAFCday28 generated a daughter cell capable of forming secondary late cobblestone areas. Baf250aE2E3/E2E3 stroma, in contrast, enabled self-renewal divisions for a majority of CAFCday28, and a noticeable proportion (mean ± SEM, 18% ± 5%, n = 53) generated more than 1 daughter colony. Given that the estimated t½ of cobblestone areas is only 4 to 5 days22 these observations strongly suggested that Baf250aE2E3/E2E3 stroma surpassed the developmentally matched WT controls in its ability to support and/or promote the self-renewal divisions in stem/progenitor cell populations.
To examine whether HSC-stimulating activity of Baf250aE2E3/E2E3 stroma correlates with a distinct molecular signature, we also analyzed the global gene expression profiles of primary Baf250aE2E3/E2E3 and WT FL-derived stromal cell cultures (n = 3), and identified 26 differentially expressed transcripts (Table 1). Baf250aE2E3/E2E3 stromal cells consistently expressed more than 2-fold higher levels of transcripts coding for soluble factors that promote HSC self-renewal (ie, insulin-like growth factor binding protein 2 [:Igfbp2]:),28,29 or survival [:Tip-1b]:)30 compared with controls, and exhibited a more than 2-fold lower activity of genes implicated in induction of senescence (ie, [:Igfbp5]:31 and oxidative stress-induced apoptosis [:Stc1]:).32
Together, the results of our experiments suggest that homozygosity at the Baf250aE2E3 allele promotes the expansion HSC populations by increasing the potential of the FL environment to support HSC self-renewal, and implicate Baf250a as a cell nonautonomous negative regulator of fetal, but not adult HSC populations.
Discussion
Results of studies presented in this report indicate that activity of a mutant Baf250a allele lacking the amino acids encoded by exon 2 and 3 results in increase in the number of FL HSCs that retain their full competitive reconstitution and differentiation potential. We also show that this Baf250a mutation provides no advantage to fetal HSCs in regeneration of the adult BM HSC pool, but markedly enhances the ability of FL-derived stromal cells to support the in vitro maintenance of primitive hemopoietic cells. To our knowledge, this is the first demonstration that a mechanism operating in a cell nonautonomous manner can modulate self-renewal of the fetal HSCs.
In contrast to early developmental arrest because of the absence of mesoderm formation reported for Baf250a deficiency,33 the Baf250aE2E3/E2E3 embryos develop without any detectable abnormality, suggesting that the deleted region of protein is dispensable for developmental function(s) of BAF250a. This region codes for less than 10% of amino acids comprising BAF250a. It is highly unstructured and predicted to comprise no distinct domains associated with protein functions (UniProt A2BH40 and http://elm.eu.org./), and is not essential for interaction with Brg1 (Figure 1F), or with other components of the Swi/Snf-BAF complex (data not shown). It harbors clusters of a predicted WW and Stat3 interaction motifs and putative mitogen-activated protein kinase, glycogen synthase kinase 3, and phosphatidylinositol-3-kinase-related phosphorylation sites (UniProt A2BH40 and http://elm.eu.org./), the deletion of which could have affected the ability of BAF250aE2E3 to participate in a subset of BAF250a-dependent functions. The BAF250aE2E3 thus could act as an intrinsic regulator of HSC behavior. Our observations that after transplantation into adult recipients, the Baf250aE2E3/E2E3 FL-derived HSCs exhibit proliferation and differentiation behavior indistinguishable from their WT counterparts suggest, however, that the potential cell autonomous BAF250aE2E3 activity could be restricted to fetal HSC populations, but would play no major role in regulation of adult HSC behavior.
Results of experiments presented in this report suggest that this mutant Baf250a allele regulates the size of the fetal HSC pool indirectly, through control of the FL microenvironment. Examination of histologic sections revealed no noticeable differences between the WT and Baf250aE2E3/E2E3 FL in vivo (L. Gaboury, IRIC Histological Facility, oral communication, June 2008, and data not shown), but we show that stromal cell layers established from Baf250aE2E3/E2E3 FL are noticeably more efficient than their WT counterparts in the in vitro maintenance/expansion of primitive hemopoietic cells. Moreover, Baf250aE2E3/E2E3 stroma induced equivalent responses from Baf250aE2E3/E2E3 and WT stem/progenitor cell populations, suggesting that the efficiency of this maintenance/expansion was determined solely by the origin of stromal cell layers, and not by the genotype of the input BM cells. One possible explanation for the observed differences between the supportive ability of Baf250aE2E3/E2E3 and WT stromal cultures is the increase in proportion of mesenchymal stem/progenitor cells observed in mutant stroma. This cell type has been recognized for its ability to support self-renewal and differentiation of hemopoietic cells.9 Enhanced in vitro survival and/or expansion of these cells could thus at least partly recapitulate the stimulatory environment of Baf250aE2E3/E2E3 FL. The Baf250aE2E3/E2E3 stromal cell populations also expressed elevated levels of transcripts coding for Igfbp2, a soluble protein capable of promoting ex vivo expansion of mouse28 and human HSCs,29 which may have contributed to the HSC expanding ability of Baf250aE2E3/E2E3 FL.
Results of studies presented are mostly consistent with a model predicting that Baf250aE2E3 controls the size of the fetal HSC pool indirectly, through regulation of the FL microenvironment, and thereby identify a novel cell nonautonomous mechanism that promotes expansion of fetal HSC populations.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Special thanks are owed to Melanie Frechette and Angele Fournier for assistance with the animal care and transplantation experiments, Nadine Fradette for selection of Baf250aE2E3/+ ES clones, and Sébastien Harton from the Transgenic Animal Facility of the Institute for Research in Immunology and Cancer (IRIC) for generation of Baf250aE2E3/+ chimeras. We also thank Josée Hébert from Molecular Cytogenetics Facility at the IRIC-Maisonneuve-Rosemont Hospital for karyotyping the Baf250aE2E3/+ ES clones, Louis A. Gaboury from the Histological Facility of IRIC for histologic evaluations of tissue sections, Danièle Gagné from the Flow Cytometry Facility of IRIC for sorting and assistance in FACS analysis, and Josette-Renée Landry for microarray hybridization and scanning.
This work was supported by a grant from the Canadian Cancer Society (G.S.). G.S. is a recipient of a Canadian Research Chair in Molecular Genetics of Stem Cells.
Authorship
Contribution: J.K. and J.C. conceived, designed, and performed the experiments; A.M., B.T.W., S.G., I.L., and J.L. performed and analyzed experiments; C.P. and G.S. conceived and designed the experiments; and J.K. and G.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guy Sauvageau, Faculty of Medicine, Université de Montreal, CP 6128, Succursale Centre-Ville, Montreal, QC, H3C 3J7 Canada; e-mail: guy.sauvageau@umontreal.ca.