Abstract
Glucocorticoids play a critical role in the therapy of lymphoid malignancies, including pediatric acute lymphoblastic leukemia (ALL), although the mechanisms underlying cellular resistance remain unclear. We report glucocorticoid resistance attributable to epigenetic silencing of the BIM gene in pediatric ALL biopsies and xenografts established in immune-deficient mice from direct patient explants as well as a therapeutic approach to reverse resistance in vivo. Glucocorticoid resistance in ALL xenografts was consistently associated with failure to up-regulate BIM expression after dexamethasone exposure despite confirmation of a functional glucocorticoid receptor. Although a comprehensive assessment of BIM CpG island methylation revealed no consistent changes, glucocorticoid resistance in xenografts and patient biopsies significantly correlated with decreased histone H3 acetylation. Moreover, the histone deacetylase inhibitor vorinostat relieved BIM repression and exerted synergistic antileukemic efficacy with dexamethasone in vitro and in vivo. These findings provide a novel therapeutic strategy to reverse glucocorticoid resistance and improve outcome for high-risk pediatric ALL.
Introduction
Neoplasia is driven by a complex series of genetic lesions that regulate proliferation, apoptosis, response to cytokine signaling, invasion/metastasis, and angiogenesis. Compelling evidence now exists that epigenetic changes complement germline and somatic genetic lesions, resulting in aberrant gene expression in malignant cells.1 Such epigenetic alterations include methylation of DNA CpG dinucleotides; modification of nucleosome conformation by methylation, phosphorylation, acetylation, sumoylation, and ubiquitination of histone N-termini; and gene regulation by small noncoding RNAs termed microRNAs.2-4
The development of genome-wide analysis tools has highlighted the prevalence of epigenetic changes in tumor cells.1 A common feature of neoplasia is a global decrease, but frequent localized increase, in DNA methylation as well as altered patterns of chromatin and nucleosome structure that regulate accessibility of the transcriptional machinery to actively transcribed genes.5 This aberrant epigenetic regulation of gene expression has been implicated in multiple stages of cancer, from initiation through to the acquisition of drug resistance, leading to opportunities for the therapeutic use of epigenetic modifiers such as DNA methyltransferase inhibitors and histone deacetylase inhibitors (HDIs). For example, the HDI vorinostat (suberoylanilide hydroxamic acid) is currently approved in the United States for the third-line systemic treatment of cutaneous T-cell lymphoma and is in phase 1/2/3 clinical trials for a variety of hematologic malignancies and solid tumors.
The proapoptotic BH3-only bcl2 family member, BIM, plays a critical role in development and homeostasis of the lymphoid system6 and is a tumor suppressor in B lymphocytes.7 Moreover, loss of BIM confers resistance of normal and malignant B lymphocytes to glucocorticoids.8 Bim protein expression is regu-lated at the transcriptional level through transcription factors such as Foxo3a and E2F1,9,10 posttranscriptionally by microRNAs,11 and posttranslationally via phosphorylation.12 BIM is also reported to be the target of gene silencing in lymphoid malignancies via promoter methylation and biallelic deletion.13
The glucocorticoids dexamethasone and prednisolone are critical components of combination chemotherapy regimens used in the treatment of lymphoid malignancies, including pediatric acute lymphoblastic leukemia (ALL), which is known as the most common childhood malignancy. Several treatment protocols for childhood ALL now include an initial week of glucocorticoid monotherapy combined with a single intrathecal dose of methotrexate, and early response to treatment is one of the strongest predictors of outcome.14 Those children who fail to achieve a good early response (prednisolone poor responders, PPRs) are further stratified into treatment protocols for high-risk patients. A greater understanding of mechanisms associated with resistance to glucocorticoids in pediatric ALL would facilitate the design of treatment strategies to overcome resistance and improve outcome.
Several genome-wide expression analyses of glucocorticoid-induced genes, which included leukemic blasts harvested from pediatric ALL patients after treatment, have identified BIM as a frequently up-regulated Bcl-2 family member.15,16 Moreover, significantly lower BIM expression was detected in high-risk childhood ALL patients who exhibited slow early responses to a standard 4-drug induction regimen compared with those patients who responded rapidly.17
We previously showed that dexamethasone resistance in pediatric ALL xenografts established in immune-deficient (nonobese diabetic/severe combined immunodeficient [NOD/SCID]) mice directly from patient explants was associated with failure to induce Bim protein and mRNA expression in response to dexamethasone treatment.18,19 Moreover, dexamethasone resistance in these xenografts could not be attributed to dysfunctional glucocorticoid receptor (GR), in contrast to findings in which the authors used in vitro–cultured cell lines19,20 but consistent with reports that perturbations in expression and/or function of the GR are rare in primary ALL.21
In this study, we describe epigenetic repression of BIM in dexamethasone-resistant pediatric ALL xenografts and in leukemic blasts from PPRs compared with those from prednisolone good responders (PGRs). In contrast to previous reports, a rigorous analysis of the BIM promoter region revealed that increased DNA methylation was not the underlying mechanism of dexamethasone resistance. Instead, the BIM locus existed in a transcriptionally inert conformation as the result of decreased association of acetylated histone H3, which could be reversed with vorinostat leading to synergistic antileukemic effects in combination with dexamethasone both in vitro and in vivo. These findings present a paradigm for the rational use of epigenetic modifiers to improve the treatment of glucocorticoid-resistant pediatric ALL.
Methods
ALL xenograft model and primary patient samples
The process by which continuous xenografts from childhood ALL biopsies have been established in immunodeficient NOD/SCID mice has been previously described in detail.22 For in vitro experiments described in this study, xenograft cells harvested from mice at the quaternary serial passage were retrieved from cryostorage and cultured in QBSF-60 medium supplemented with Flt-3 ligand (20 ng/mL; kindly provided by Amgen), penicillin (100 U/mL), streptomycin (100 μg/mL), and l-glutamine (2mM). For drug treatments, dexamethasone (Sigma-Aldrich) was used at a final concentration of 1μM and vorinostat (Merck) at a final concentration of 100nM-5μM.
Primary patient samples were obtained from children presenting at Sydney Children's Hospital and at the Pediatric Clinic, Ospedale San Gerardo, with ALL at initial diagnosis or bone marrow relapse. Informed consent was obtained from each subject involved in the study. All human investigations were performed in accordance with the principles embodied in the declaration of Helsinki. Children presenting with ALL were enrolled in Australia and New Zealand Children's Cancer Study Group Study 8 and AIEOP-BFM ALL2000 protocol and were treated with an initial 7-day course of prednisolone and a single age-related dose of intrathecal methotrexate. Those patients with a day 8 peripheral blast count of > 109/L were classified as PPRs and those < 109/L as PGRs. Three sample groups were identified: (1) a group of PPRs; (2) a group of PGRs matched as closely as possible to PPRs in terms of age, sex, and leukemia phenotype; and (3) a group of samples from patients who relapsed within 2 years of initial diagnosis.
All animal studies had previous approval from the Animal Care and Ethics Committee of the University of NSW, whereas experiments that used patient biopsy material were approved by the Human Research Ethics Committees of the South East Sydney and Illawara Area Health Service, the University of New South Wales, San Gerardo Hospital, and the University of Milano-Bicocca.
In vitro cytotoxicity assays
In vitro drug sensitivity was assessed by use of the colorimetric methyl-thiazolyl-tetrazolium (MTT) assay, which measures a combination of inhibition of proliferation and cell death, and is described in detail elsewhere.18 The cytotoxic interactions between dexamethasone and vorinostat were evaluated by the use of the method of Chou and Talalay,23 as described in the supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Immunoblotting
Whole-cell protein extracts were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electro-transferred to PVDF membranes according to the manufacturer's instructions (Invitrogen). Membranes were probed with anti-Bim and anti-Actin polyclonal antibodies (Sigma-Aldrich) followed by horseradish peroxidase–conjugated donkey anti–rabbit secondary antibody (GE Healthcare). Protein binding was detected by chemiluminescence and visualized by autoradiography detection and phosphoimaging with a VersaDoc 5000 Imaging System (Bio-Rad). Data were analyzed by the use of QuantityOne software (Version 4.00; Bio-Rad).
Real-time RT-PCR
Real-time quantitative reverse transcription–polymerase chain reaction (RT-PCR) was carried out as previously described.19 A brief description and primer sequences are provided in the supplemental Methods.
ChIP and ChIP-chip arrays
Chromatin immunoprecipitation (ChIP) and ChIP-chip were carried out as previously described.24 Full details including primer sequences are provided in the supplemental Methods.
Methylated DNA immunoprecipitation assay
Genomic DNA was extracted from xenograft cells by the use of TRIzol reagent (Invitrogen), and 20 μg was sonicated by use of a Branson Sonoplus HD 2070. One microgram of sonicated DNA was immunoprecipitated overnight with 5-methyl-cytosine antibody (Abcam). Immunoprecipitated complexes were captured by incubation with salmon sperm DNA/protein A agarose (Upstate) for 1 hour and the immunocomplex washed to remove nonspecific binding in a series of buffers according to the manufacturer's recommendations (supplemental Methods). The immunoprecipitated DNA was eluted from the beads with freshly prepared 1% SDS, 0.1M NaHCO3 and purified by use of the QIAquick PCR purification kit according to the manufacturer's instructions. A total of 5 μL of immunoprecipitated DNA and a corresponding amount of input DNA was assayed by SYBR Green real-time PCR (Bio-Rad) according to the manufacturer's instructions (see supplemental Methods for primers).
Bisulfite sequencing
A total of 5 μL of bisulfite-treated DNA was amplified by nested PCR after a standard protocol (see supplemental Methods for preparation of bisulfite-treated DNA and primers). Amplicons were ligated into TOPO-TA pCR2.1 cloning vectors (Invitrogen) according to manufacturer's instructions and used to transform TOP10 Escherichia coli cells. One microgram of DNA from each clone was precipitated with ammonium acetate (0.7M final concentration) and sequenced (Eurofins MWG Operon). Ten clones from each amplicon were analyzed.
SEQUENOM methylation analysis
Genomic DNA extracted from xenograft cells was subjected to sodium bisulfite conversion by the use of the MethylEasy Xceed according to manufacturer's instructions (Human Genetic Signatures). Quantitative DNA methylation analysis was performed by the use of SEQUENOM EpiTYPER chemistry according to manufacturer's instructions (SEQUENOM Inc). PCR amplicons were derived from bisulfite-treated genomic DNA samples from each xenograft cell line (see supplemental Methods for primers and PCR conditions). Amplicons were subjected to SEQUENOM EpiTYPER chemistry consisting of shrimp alkaline phosphatase treatment, in vitro transcription, and RNA cleavage followed by analysis by matrix-assisted laser desorption-ionization time of flight mass spectrometry and SEQEUNOM EpiTYPER v1.05 software. Methylation ratios for each amplicon from each sample were plotted by the use of Heatmap.2 in R (http://www.r-project.org).
In vivo efficacy studies
Groups of 9 female NOD/SCID mice who were 6-8 weeks of age were inoculated with 5 × 106 xenograft cells and engraftment monitored by weekly enumeration of the proportion of human CD45+ cells in the peripheral blood (%huCD45+) as described previously.25 When the median %huCD45+ reached 1%, treatment was initiated after randomization: vorinostat was administered at 200 mg/kg for the first 7 days and then 100 mg/kg for 21 days26 ; dexamethasone was administered at 5 mg/kg Monday through Friday for 4 weeks; all via intraperitoneal injection. Leukemia progression was monitored throughout and after the course of drug treatment. An event was defined as the time taken from the initiation of treatment for the %huCD45+ to reach 25%, or when animals exhibited signs of morbidity associated with high leukemic infiltration of bone marrow and spleen. To allow comparisons between drug and vehicle treatment groups, event-free survival (EFS) was calculated for each mouse and represented by Kaplan-Meier analysis. The median EFS for the vehicle control group also was subtracted from that of the drug-treated groups to generate a leukemia growth delay.
Statistical analyses
Quantitative variables of normally distributed data were compared by the Student t test and nonnormally distributed data were compared by the Mann-Whitney U test, whereas categorical data were compared by the Fisher exact test. All statistical tests were 2-sided and the level of significance was set to .05.
Results
Repression of BIM transcription in glucocorticoid-resistant pediatric ALL
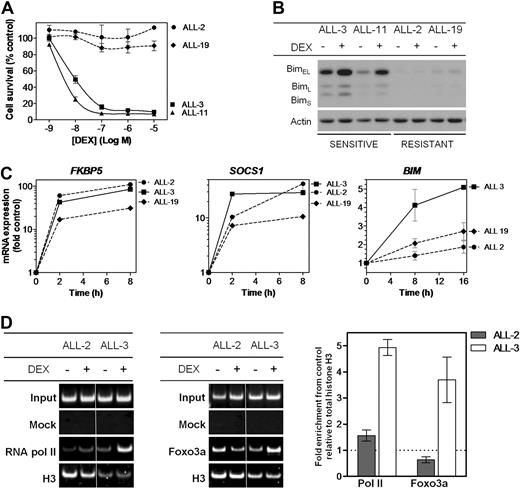
We have previously demonstrated that the in vivo and in vitro dexamethasone sensitivities of a panel of pediatric ALL xenografts closely reflected the clinical outcome of the patients from whom the xenografts were derived,18,22,25 indicating that the xenograft model system is likely to yield important insights into clinically relevant mechanisms of glucocorticoid resistance. The relative in vitro sensitivity of a subset of these xenografts is illustrated in Figure 1A, which shows that the dexamethasone half maximal inhibitory concentration (IC50) values of 2 resistant xenografts representative of the disease (ALL-2 and ALL-19, > 10μM) were > 1000-fold those of 2 sensitive xenografts (ALL-3 and ALL-11, 9.8nM and 3.7nM, respectively). The high level of dexamethasone resistance exhibited by ALL-2 and ALL-19 was also reflected in their failure to up-regulate Bim protein after exposure to dexamethasone (Figure 1B), consistent with previous reports.18,19 The in vitro and in vivo dexamethasone sensitivities of the entire xenograft panel used in this study are shown in supplemental Table 1.
Responses of ALL xenografts to dexamethasone in vitro. (A) Xenograft cells were retrieved from cryostorage, and metabolic activity was assessed after a 48-hour exposure to a serial dilution of dexamethasone (10−5-10−9) relative to vehicle-treated controls by MTT assay. (B) Protein expression of Bim was assessed by immunoblot analysis relative to Actin control in glucocorticoid-sensitive (ALL-3 and ALL-11) and -resistant (ALL-2 and ALL-19) xenograft cells after treatment with dexamethasone (1μM, 16 hours). (C) Gene expression of glucocorticoid responsive genes, FKBP5, SOCS1, and BIM, was analyzed by RT2-PCR and expressed as a fold of vehicle-treated controls after treatment with dexamethasone (1μM) for up to 16 hours. (D) ChIP was carried out to identify recruitment of RNA polymerase II to the TSS of the BIM locus, and Foxo3a to its binding site upstream of the TSS, with each PCR region evaluated relative to total histone H3 ChIP in the same sample. Results were visualized by polyacrylamide gel electrophoresis and quantified by SYBR-green PCR. Splicing within a representative gel is indicated by dividing spaces. Quantified results represent the mean ± SEM of 3 independent experiments.
Responses of ALL xenografts to dexamethasone in vitro. (A) Xenograft cells were retrieved from cryostorage, and metabolic activity was assessed after a 48-hour exposure to a serial dilution of dexamethasone (10−5-10−9) relative to vehicle-treated controls by MTT assay. (B) Protein expression of Bim was assessed by immunoblot analysis relative to Actin control in glucocorticoid-sensitive (ALL-3 and ALL-11) and -resistant (ALL-2 and ALL-19) xenograft cells after treatment with dexamethasone (1μM, 16 hours). (C) Gene expression of glucocorticoid responsive genes, FKBP5, SOCS1, and BIM, was analyzed by RT2-PCR and expressed as a fold of vehicle-treated controls after treatment with dexamethasone (1μM) for up to 16 hours. (D) ChIP was carried out to identify recruitment of RNA polymerase II to the TSS of the BIM locus, and Foxo3a to its binding site upstream of the TSS, with each PCR region evaluated relative to total histone H3 ChIP in the same sample. Results were visualized by polyacrylamide gel electrophoresis and quantified by SYBR-green PCR. Splicing within a representative gel is indicated by dividing spaces. Quantified results represent the mean ± SEM of 3 independent experiments.
We next compared BIM mRNA induction in ALL-2, -3, and -19 with reference to 2 genes (FKBP5 and SOCS1) previously identified to be up-regulated in leukemic blasts from patients treated with the glucocorticoid prednisolone.27 FKBP5 is a primary target of the GR and contains well-characterized glucocorticoid response elements (GREs),28,29 whereas SOCS1 is a secondary target gene for glucocorticoids, because protein synthesis has been shown to be required for its transcriptional activation.15 After exposure to dexamethasone, and regardless of their relative sensitivity to its cytotoxic effects (Figure 1A), all xenografts exhibited robust induction of both FKBP5 and SOCS1 (Figure 1C). In contrast, induction of BIM mRNA was repressed in ALL-2 and ALL-19 compared with the sensitive ALL-3, consistent with previous findings.19
To further characterize the mechanism of BIM transcriptional repression in dexamethasone-resistant xenografts, we assessed recruitment of RNA polymerase II and Foxo3a to a region just upstream of the BIM transcription start site (TSS, region 3, Figure 2A) by conventional and quantitative PCR (qPCR)-ChIP assays. After dexamethasone treatment of ALL-3, we observed a 4.9 ± 0.4- and 3.7 ± 1.2-fold increase in recruitment of RNA polymerase II and Foxo3a, respectively, to region 3 (Figure 1D; mean ± SEM). In contrast, there was a distinct lack of recruitment of both proteins in ALL-2. Moreover, conventional ChIP of histone H3 also indicated nucleosomal loss in this region in ALL-3 compared with ALL-2, consistent with active transcription.30
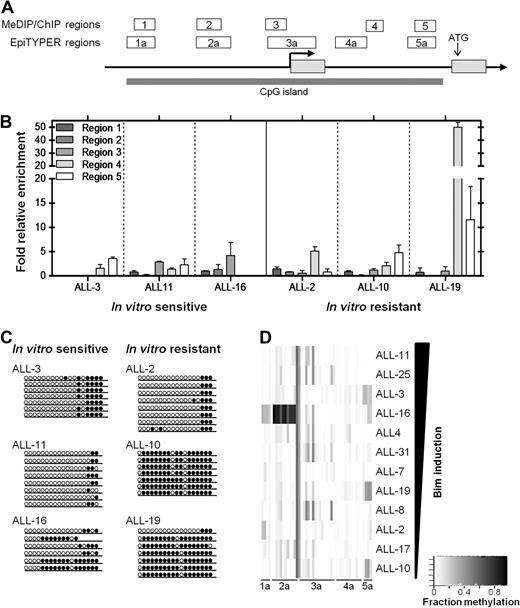
DNA methylation analysis of the CpG island identified across the BIM genomic locus. (A) The BIM genomic locus is represented by a schematic adapted from the UCSC genome browser (see supplemental Figure 3A), with regions analyzed in the separate assays (ChIP/MeDIP: regions 1-5; SEQUENOM: regions 1a-5a) indicated above the BIM gene, as well as the TSS (bold arrow), translation start site, and CpG island indicated. (B) MeDIP was carried out on 3 glucocorticoid-sensitive (ALL-3, -11, and -16) and 3 glucocorticoid-resistant (ALL-2, -10, -19) xenografts and enrichment for 5′-methyl-cytosine evaluated by qPCR across regions 1-5 indicated previously. (C) Bisulfite sequencing was carried out as described in “Bisulfite sequencing” to confirm MeDIP results for region 5. Open circles represent unmethylated sites, whereas closed circles represent methylated sites. At least 6 clones were sequenced for each xenograft. (D) SEQUENOM assays were carried out to measure methylation at individual CpG sites across BIM regions 1a-5a. Quantified methylation at each site is represented as a heat map ranging from white (0% methylation) to black (100% methylation). Xenografts are arranged vertically in decreasing order of Bim protein induced after treatment with dexamethasone (1μM, 16 hours).
DNA methylation analysis of the CpG island identified across the BIM genomic locus. (A) The BIM genomic locus is represented by a schematic adapted from the UCSC genome browser (see supplemental Figure 3A), with regions analyzed in the separate assays (ChIP/MeDIP: regions 1-5; SEQUENOM: regions 1a-5a) indicated above the BIM gene, as well as the TSS (bold arrow), translation start site, and CpG island indicated. (B) MeDIP was carried out on 3 glucocorticoid-sensitive (ALL-3, -11, and -16) and 3 glucocorticoid-resistant (ALL-2, -10, -19) xenografts and enrichment for 5′-methyl-cytosine evaluated by qPCR across regions 1-5 indicated previously. (C) Bisulfite sequencing was carried out as described in “Bisulfite sequencing” to confirm MeDIP results for region 5. Open circles represent unmethylated sites, whereas closed circles represent methylated sites. At least 6 clones were sequenced for each xenograft. (D) SEQUENOM assays were carried out to measure methylation at individual CpG sites across BIM regions 1a-5a. Quantified methylation at each site is represented as a heat map ranging from white (0% methylation) to black (100% methylation). Xenografts are arranged vertically in decreasing order of Bim protein induced after treatment with dexamethasone (1μM, 16 hours).
Epigenetic regulation of the BIM genomic locus
The BIM genomic locus on chromosome 2q13 includes a large (> 5 kb) CpG island spanning the promoter region, TSS and 5′-untranslated region (Figure 2A, supplemental Figure 3A). Because BIM silencing has been associated with DNA methylation in neoplastic lymphoid cell lines and primary biopsies,13 we used a multifaceted approach (methyl-DNA immunoprecipitation, or MeDIP,31 bisulfite sequencing,32 and SEQUENOM MassArray Epityper analysis33 ) to comprehensively assess the DNA methylation status at the BIM CpG island in dexamethasone-sensitive and -resistant ALL xenografts (see Figure 2A for regions analyzed by each technique, and supplemental Figure 3A for exact genomic alignments).
MeDIP analysis of 5 regions from 6 xenografts revealed enrichment in only one highly resistant xenograft, ALL-19, at regions 4 and 5, which span the 5′ untranslated region of the gene (Figure 2B). Two other dexamethasone-resistant xenografts, ALL-2 and ALL-10, showed only localized methylation with some enrichment evident in regions 4 and 5, respectively. Additional analysis by bisulfite sequencing confirmed increased region 5 DNA methylation in 2/3 resistant xenografts (ALL-10 and ALL-19 but not ALL-2) compared with 3 sensitive xenografts (Figure 2C; supplemental Figure 1). The high level of methylation exhibited by ALL-19 in the 5′-untranslated region was confirmed by bisulfite sequencing of region 4, indicating near-total DNA methylation (R.G.P., unpublished data, September 2006).
Because neither MeDIP nor bisulfite sequencing revealed a definite relationship between DNA methylation at the BIM locus and transcriptional silencing, we subjected all 12 xenografts to SEQUENOM MassArray EpiTYPER analysis. Figure 2D shows the level of DNA methylation at regions 1a-5a of the BIM locus (see Figure 2A, supplemental Figure 3A for schematic) in 12 xenografts arranged according to the extent of Bim induction after exposure to dexamethasone. These results confirmed no clear association between DNA methylation and BIM silencing across the panel of xenografts.
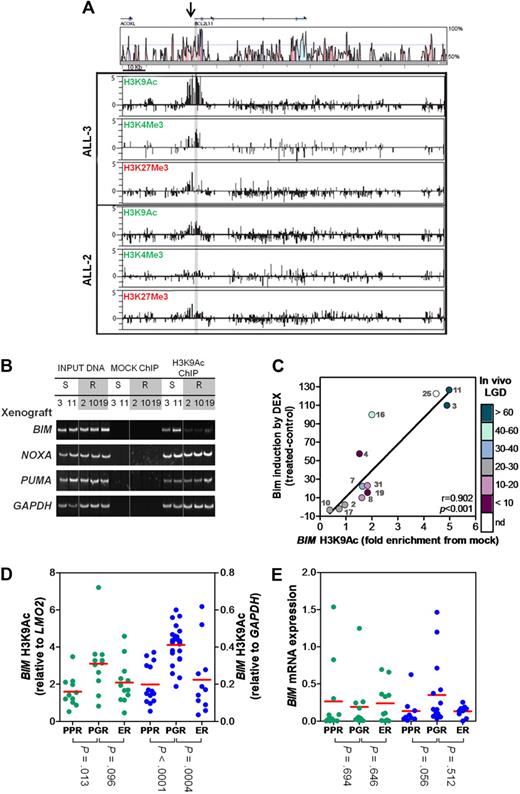
Because DNA methylation alone could not explain the lack of BIM induction, we investigated histone modification marks associated with actively transcribed (H3K9Ac, H3K4Me3) or inactive (H3K27Me3) genes at the BIM locus in ALL-2 (resistant) and ALL-3 (sensitive) by ChIP-chip analysis (Figure 3A). Although the majority of H3K9Ac and H3K4Me3 enrichment in ALL-3 was detected at the highly conserved 5′ region of the BIM gene, enrichment of both of these histone modification marks was dramatically decreased in ALL-2, consistent with a less transcriptionally accessible BIM promoter. In addition, despite some enrichment of H3K27Me3 at the BIM 5′ region, there was no noticeable difference between ALL-2 and ALL-3. These results were confirmed by qPCR-ChIP analysis of BIM region 3 (supplemental Figure 2).
Histone H3 modifications at the BIM locus. (A) ChIP was carried out for the activating marks H3AcK9 and H3K4Me3 and the inactive mark H3K27Me3 on a representative sensitive (ALL-3) and resistant (ALL-2) xenograft. ChIP material was applied to customized tiling arrays to provide a comprehensive view of the BIM locus as described in “ChIP and ChIP-chip arrays.” (B) ChIP was carried out for H3K9Ac in 2 representative sensitive (ALL-3 and -11) and 3 representative resistant (ALL-2, -10, and -19) xenografts and visualized at the BIM, NOXA, PUMA and GAPDH genomic loci by PCR followed by polyacrylamide gel electrophoresis and SYBR staining. Splicing of samples within a single representative gel is indicated by dividing spaces. (C) ChIP was carried out for H3K9Ac at the BIM locus and quantified relative to the GAPDH locus. Relative BIM acetylation was plotted against the amount of Bim protein induced in each xenograft after treatment with dexamethasone (1μM, 16 hours) as previously published.18 The color-code alongside the y-axis designates the in vivo LGD in days for each xenograft (see supplemental Table 1); LGD indicates leukemia growth delay; nd, not determined. (D) H3K9 acetylation of BIM was evaluated by ChIP analysis of primary ALL samples in 2 independent patient cohorts (green, Australian; blue, Italian). Data from the Australian cohort were normalized to LMO2, whereas those from the Italian cohort to GAPDH. Individual results represent the mean of 4 experiments; red bar, mean of each subgroup. (E) Basal BIM mRNA expression was determined by RT2-PCR and grouped as for Figure 3D.
Histone H3 modifications at the BIM locus. (A) ChIP was carried out for the activating marks H3AcK9 and H3K4Me3 and the inactive mark H3K27Me3 on a representative sensitive (ALL-3) and resistant (ALL-2) xenograft. ChIP material was applied to customized tiling arrays to provide a comprehensive view of the BIM locus as described in “ChIP and ChIP-chip arrays.” (B) ChIP was carried out for H3K9Ac in 2 representative sensitive (ALL-3 and -11) and 3 representative resistant (ALL-2, -10, and -19) xenografts and visualized at the BIM, NOXA, PUMA and GAPDH genomic loci by PCR followed by polyacrylamide gel electrophoresis and SYBR staining. Splicing of samples within a single representative gel is indicated by dividing spaces. (C) ChIP was carried out for H3K9Ac at the BIM locus and quantified relative to the GAPDH locus. Relative BIM acetylation was plotted against the amount of Bim protein induced in each xenograft after treatment with dexamethasone (1μM, 16 hours) as previously published.18 The color-code alongside the y-axis designates the in vivo LGD in days for each xenograft (see supplemental Table 1); LGD indicates leukemia growth delay; nd, not determined. (D) H3K9 acetylation of BIM was evaluated by ChIP analysis of primary ALL samples in 2 independent patient cohorts (green, Australian; blue, Italian). Data from the Australian cohort were normalized to LMO2, whereas those from the Italian cohort to GAPDH. Individual results represent the mean of 4 experiments; red bar, mean of each subgroup. (E) Basal BIM mRNA expression was determined by RT2-PCR and grouped as for Figure 3D.
The ChIP-chip results prompted a more extensive analysis of H3K9Ac enrichment at region 3 in dexamethasone-sensitive and -resistant xenografts by conventional and qPCR-ChIP. Conventional ChIP analysis revealed a marked decrease in BIM region 3 H3K9Ac enrichment in 3/3 dexamethasone-resistant xenografts (ALL-2, ALL-10, and ALL-19) compared with 2 sensitive xenografts (ALL-3 and ALL-11), both of which exhibited robust enrichment (Figure 3B). In contrast, no differences in H3K9Ac associated with the promoter regions of 2 other proapoptotic BH3-only genes, PUMA and NOXA, were detected indicating specific histone deacetylation of the BIM locus (Figure 3B, see supplemental Figure 3B-C for schematics of genomic loci). Importantly, qPCR-ChIP analysis of all 12 xenografts revealed a strong correlation between BIM region 3 H3K9Ac and the extent of Bim protein induction after dexamethasone treatment (Figure 3C, R = 0.902, P < .001), indicating a significant role of the local chromatin environment at the BIM locus. Moreover, because early response to glucocorticoid monotherapy is used as a clinical surrogate for overall treatment response to multiagent chemotherapy, a critical finding was that the in vivo sensitivity of this panel of xenografts (supplemental Table 1) also significantly correlated with the extent of BIM region 3 H3K9Ac (R = 0.64, P = .03).
To verify that decreased H3K9Ac at the BIM promoter in glucocorticoid-resistant pediatric ALL was not an artifact of xenografting cells in immune-deficient mice we evaluated biopsy material from chemotherapy-naive patients before one week of prednisolone monotherapy (Table 1, supplemental Tables 2-3), as well as tissue harvested from patients when they experienced early relapse (supplemental Tables 2-3), in 2 independent cohorts of childhood ALL patients treated in either Australia or Italy. The Australian PPR and PGR patients were well matched in terms of sex, age at diagnosis, and ALL subtype, although the PPRs exhibited greater white cell counts, shorter length of CR1, and were stratified as being at greater risk than the PGRs (Table 1). Importantly, BIM region 3 H3K9Ac was significantly lower in the PPR group compared with PGRs in both independent cohorts (P = .013 and P < .0001 for the Australian and Italian cohorts, respectively; Figure 3D, Table 1, and supplemental Figure 4A,C). Moreover, the decrease in BIM H3K9Ac in patients at early relapse compared with PGRs was significant in the Italian cohort (P = .0004) and approached significance in the Australian cohort (P = .096; Figure 3D). Basal levels of BIM mRNA were determined in all patient samples (Figure 3E and supplemental Figure 4B,D) and failed to show a significant association with any particular subgroup. Similarly, evaluation of H3K9Ac of other BH3-only genes NOXA and PUMA revealed both genes to be in a mostly transcriptionally active conformation (> 3-fold relative to control gene), with no significant differences between patient subgroups (supplemental Figure 5). Therefore, the collective data from xenografts and patient biopsy material indicate that glucocorticoid resistance in pediatric ALL is associated with a significant and specific decrease in H3K9Ac at the BIM promoter, implying a transcriptionally inaccessible conformation.
Reversal of BIM deacetylation and dexamethasone resistance with vorinostat
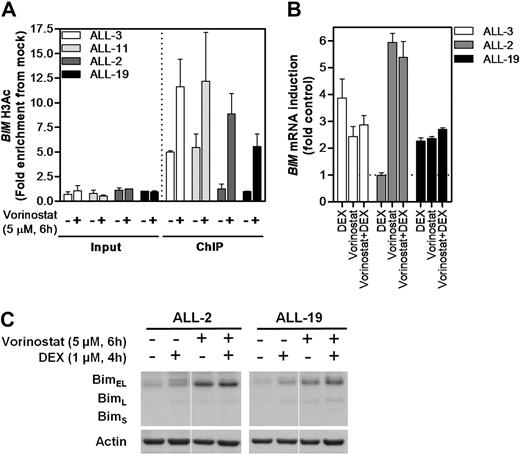
Vorinostat has been shown to reactivate expression of aberrantly silenced tumor-suppressor genes through its actions as a pan-inhibitor of class I and class II HDAC proteins.34 Therefore, we tested the ability of vorinostat to reverse BIM deacetylation and reactivate its expression in dexamethasone-resistant ALL xenograft cells. qPCR-ChIP of BIM region 3 H3K9Ac confirmed the differences in basal levels between sensitive (ALL-3 and ALL-11) and resistant (ALL-2 and ALL-19) xenografts (Figure 4A), and vorinostat exposure resulted in increased BIM H3K9Ac in all 4 xenografts tested. Notably, BIM H3K9Ac levels in the 2 resistant xenografts were increased by vorinostat treatment to levels at least comparable with basal acetylation in the 2 sensitive xenografts. Moreover, exposure to vorinostat resulted in reexpression of BIM mRNA and protein in ALL-2 (Figure 4B-C), although the addition of dexamethasone did not enhance this effect. Not unexpectedly, dexamethasone alone induced BIM expression in ALL-3 with no enhanced effects of vorinostat observed (Figure 4B), whereas vorinostat was relatively ineffective against ALL-19 (Figure 4B-C), consistent with the high level of CpG methylation detected in the promoter of this xenograft (Figure 2B-C, supplemental Figure 1).
Modulation of BIM acetylation and reexpression with vorinostat in vitro. (A) Two representative sensitive (ALL-3 and-11) and resistant (ALL-2 and -19) xenografts were treated with vorinostat (5μM, 6 hours) in vitro and subjected to H3AcK9 ChIP analysis. (B) BIM mRNA expression was determined by RT2-PCR after treatment with vorinostat (5μM, 6 hours), dexamethasone (1μM, 4 hours), or both in combination and expressed as a fold of untreated controls relative to EF-1α internal control. (C) Protein expression of Bim after treatment with vorinostat (5μM, 6 hours), dexamethasone (1μM, 4 hours), or both in combination was evaluated by immunoblot analysis and expressed relative to Actin in 2 dexamethasone-resistant xenografts (ALL-2 and ALL-19). Splicing within a representative gel is indicated by dividing spaces.
Modulation of BIM acetylation and reexpression with vorinostat in vitro. (A) Two representative sensitive (ALL-3 and-11) and resistant (ALL-2 and -19) xenografts were treated with vorinostat (5μM, 6 hours) in vitro and subjected to H3AcK9 ChIP analysis. (B) BIM mRNA expression was determined by RT2-PCR after treatment with vorinostat (5μM, 6 hours), dexamethasone (1μM, 4 hours), or both in combination and expressed as a fold of untreated controls relative to EF-1α internal control. (C) Protein expression of Bim after treatment with vorinostat (5μM, 6 hours), dexamethasone (1μM, 4 hours), or both in combination was evaluated by immunoblot analysis and expressed relative to Actin in 2 dexamethasone-resistant xenografts (ALL-2 and ALL-19). Splicing within a representative gel is indicated by dividing spaces.
We next tested the in vitro and in vivo antileukemic effects of the vorinostat/dexamethasone combination. Fixed-ratio in vitro cytotoxicity assays showed that the vorinostat/dexamethasone combination was strongly synergistic against ALL-2 across the entire range of vorinostat concentrations (Figure 5A and Table 2). In contrast, vorinostat only enhanced the cytotoxic effects of dexamethasone against ALL-19 at the 2 greatest concentrations (2 and 4μM; Figure 5B and Table 2), which are greater than those that are clinically achievable.35
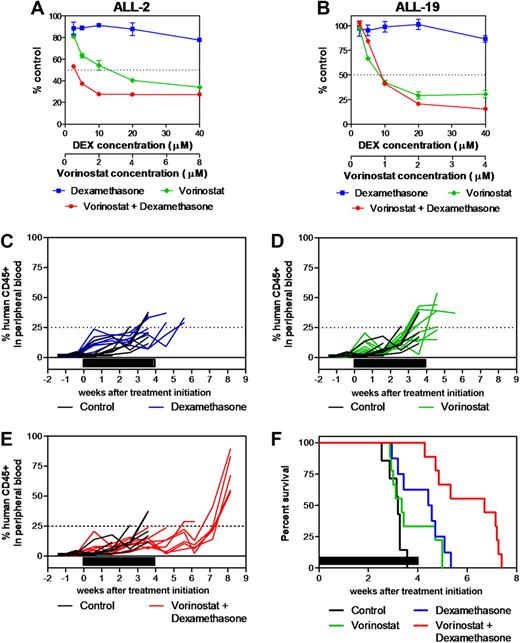
Synergistic antileukemia effects of vorinostat and dexamethasone in vitro and in vivo. (A) ALL-2 and (B) ALL-19 xenograft cells were treated with vorinostat (green), dexamethasone (blue), or both (red) in vitro at a fixed ratio of concentrations as described in “In vitro cytotoxicity assays” and mitochondrial activity was assessed by MTT assay. Data represent the mean ± SEM of 3 independent experiments. (C-F) The in vivo antileukemia effects of dexamethasone (C, blue; 5 mg/kg Monday through Friday for 4 weeks; n = 9) and vorinostat (D, green; 200 mg/kg for 7 days then 100 mg/kg for 21 days; n = 9) individually and in combination (E, red; n = 9) were evaluated against ALL-2 inoculated in NOD/SCID mice in vivo. Colored lines indicate the %huCD45+ in each mouse within the drug-treated groups, while black lines show the progression in vehicle-treated controls (n = 9). (F) Kaplan-Meier graphs indicate the EFS of each cohort in the weeks post treatment initiation. Solid bars in panels C-F represent treatment periods.
Synergistic antileukemia effects of vorinostat and dexamethasone in vitro and in vivo. (A) ALL-2 and (B) ALL-19 xenograft cells were treated with vorinostat (green), dexamethasone (blue), or both (red) in vitro at a fixed ratio of concentrations as described in “In vitro cytotoxicity assays” and mitochondrial activity was assessed by MTT assay. Data represent the mean ± SEM of 3 independent experiments. (C-F) The in vivo antileukemia effects of dexamethasone (C, blue; 5 mg/kg Monday through Friday for 4 weeks; n = 9) and vorinostat (D, green; 200 mg/kg for 7 days then 100 mg/kg for 21 days; n = 9) individually and in combination (E, red; n = 9) were evaluated against ALL-2 inoculated in NOD/SCID mice in vivo. Colored lines indicate the %huCD45+ in each mouse within the drug-treated groups, while black lines show the progression in vehicle-treated controls (n = 9). (F) Kaplan-Meier graphs indicate the EFS of each cohort in the weeks post treatment initiation. Solid bars in panels C-F represent treatment periods.
The combination of both drugs was then evaluated in vivo against ALL-2. Dexamethasone alone significantly delayed ALL-2 progression by 9.1 days compared with vehicle-treated control mice (P = .012, Figure 5C-F, and Table 3), whereas vorinostat was ineffective (P = .18, Figure 5D-F, and Table 3) despite increasing histone H4 acetylation in leukemic blasts in vivo (C.D. and R.B.L., unpublished data, April 2008). Importantly, the combination of dexamethasone and vorinostat delayed the progression of ALL-2 by 24.5 days (P = .0006; Figure 5E-F and Table 3), which was 14.3 days greater than additive compared with each drug alone (P = .005 relative to dexamethasone alone, P = .001 relative to vorinostat alone).
Discussion
This report has shown that glucocorticoid resistance in pediatric ALL xenografts and primary biopsy specimens involves epigenetic transcriptional repression of BIM, reflected in a reduction of acetylated histone H3 associated with the BIM promoter region leading to a transcriptionally inert chromatin conformation. BIM repression was specific compared with the histone acetylation status of the promoter regions of 2 other BH3-only genes, PUMA and NOXA. Furthermore, notwithstanding the possible effects on the expression of other genes, pharmacologic “reacetylation” of the BIM locus with vorinostat was associated with synergistic antileukemic efficacy with dexamethasone both in vitro and in vivo. These results support the clinical testing of epigenetic modifiers in glucocorticoid-resistant pediatric ALL.
A surprising finding of our study was that high level dexamethasone resistance was not associated with dysfunctional GR, because known glucocorticoid-induced genes (FKBP5 and SOCS1) were highly up-regulated in all xenografts tested, and we have previously shown intact GR signaling in all xenografts.19 The general lack of dysfunctional GR in glucocorticoid-resistant ALL xenografts or primary biopsy specimens19,21,36 indicates that pharmacologic restoration of sensitivity is more likely than in instances where the GR is dysfunctional, which is the finding in the majority of studies in which the authors used leukemia cell lines.19,20
Silencing of BIM has been reported to occur via mechanisms, including promoter methylation in Burkitt lymphoma and diffuse large B-cell lymphomas,13 and biallelic deletion in mantle cell lymphomas.37 Nevertheless, cytogenetic abnormalities at the BIM locus (on chromosome 2q13) were not detected in a large cohort of pediatric ALL cases analyzed by genome-wide single nucleotide polymorphism arrays,38 nor in any of the xenografts tested in this panel,39 suggesting that if BIM repression were to occur in ALL it would be via mechanisms other than gene deletion, such as DNA methylation. However, in contrast to a previous report in lymphoma,13 but in agreement with a recent study in multiple myeloma,40 a comprehensive analysis of the BIM promoter region using 3 complementary techniques revealed no overall association between methylation of the CpG island and BIM repression in pediatric ALL. The reasons for this apparent discrepancy are not immediately clear, but may relate to differences in methodology or disease subtype.
On the basis of these findings, BIM CpG methylation as a silencing mechanism was not studied in the patient samples, although its implications cannot be discounted. Increased BIM CpG methylation was identified in only one highly resistant xenograft (ALL-19). Interestingly, this xenograft was intractable to reversal of dexamethasone resistance by vorinostat, in contrast to ALL-2 in which BIM CpG methylation was not observed. These findings further support a significant role for BIM repression in glucocorticoid resistance of pediatric ALL, but also indicate that selection of patients with the appropriate “epigenotype” is likely to be a critical factor when attempting to reverse glucocorticoid resistance with HDIs in the clinic.
Glucocorticoids are known to regulate more than 200 genes,41 although the mechanisms by which they induce BIM expression are poorly defined because there are no conserved GREs in the BIM promoter region. Foxo3a represents a candidate transcription factor likely to be centrally involved in BIM induction,42 and, indeed, our data demonstrate differential recruitment of Foxo3a to the BIM promoter after dexamethasone treatment of sensitive and resistant xenografts. Nevertheless, the underlying mechanism for reduced transcriptionally accessible BIM in dexamethasone-resistant ALL remains to be defined, both in the presence or absence of concomitant DNA methylation. HDACs are recruited to DNA indirectly via interaction with multiprotein transcriptional repressor complexes, and their dysregulated expression occurs in many malignancies.43 Further studies in which the authors seek to identify repressor complexes bound to the aberrantly silenced BIM locus may shed light on the mechanism of BIM silencing and afford the opportunity for more selective epigenetic therapy with compounds targeting a particular HDAC. Alternative mechanisms resulting in chromatin remodeling include changes in nucleosomal structure mediated by ATP-hydrolyzing enzymes, of which the SWI/SNF type have been demonstrated to be involved in the steps leading to the formation of transcriptionally inactive heterochromatin.44 Intriguing findings determined on the basis of genome-wide studies of primary samples have implicated differential expression of catalytic components of the SWI/SNF complex in glucocorticoid resistance in pediatric ALL cells. The implication of these findings, though, is that such differences will affect the global transcriptional activity of the GR rather than having specific effects on singular genes.45
The mechanism of BIM repression does not appear to involve global repression of other BH3-only genes because neither PUMA, a gene required for optimal glucocorticoid-induced apoptosis of T-lymphocytes,8 nor NOXA loci were in a transcriptionally inaccessible chromatin conformation in glucocorticoid-resistant ALL xenografts or biopsy specimens compared with their sensitive counterparts. Because BIM-induced apoptosis is critical for development and homeostasis of the lymphoid system,46 and BIM is a tumor suppressor gene in lymphoid malignancy,7 one component of ALL etiology might be to specifically silence BIM, which in turn would lead to a glucocorticoid-resistant phenotype even in cells naive of prior exposure to pharmacologic concentrations of glucocorticoids. Alternatively, BIM silencing may represent only one component of a broader mechanism leading to poor treatment outcome, indicating that a common pattern of gene repression might be revealed by genome-wide analysis, for example by ChIP-chip or ChIP-Seq.
The question remains why differential BIM expression has not been consistently identified in genome-wide screens of glucocorticoid-resistant ALL,47,48 despite being up-regulated in leukemic blasts from children treated with prednisolone.15,49 In fact, the only study that we are aware of in which basal levels of BIM expression significantly correlated with treatment response stratified patients according to rapid or slow early response to a standard 4-drug regimen, which included a glucocorticoid.17 Despite the strong association between BIM promoter acetylation status and up-regulation of Bim protein in ALL xenografts (Figure 3C) basal BIM mRNA and protein expression levels exhibited no such correlation (P.S.B., Rosemary C. O'Brien, and R.B.L., unpublished data, June 2008). Moreover, the significant difference in BIM H3K9Ac between leukemic blasts from PPRs and PGRs (Figure 3D) was also not reflected in a similar difference in basal BIM expression (Figure 3E). Although in current studies, authors focus on delineating the intricate “Bcl-2 rheostat” of gene and protein expression in determining tumor response to chemotherapy, in this study we indicate that the epigenomic status may play an important, and currently ill-defined, role that encompasses not only expression levels of pro- and antiapoptotic proteins, but their ability to be activated by apoptotic stimuli, ultimately determining cellular response to chemotherapy. In this study, the inherent ability of ALL cells to up-regulate BIM after dexamethasone exposure proved to be an important, but not sole, determinant of treatment response. Future studies may provide further insight into the role of individual members of the Bcl-2 family of proteins acting in concert with BIM to commit dexamethasone-sensitive cells to apoptosis.
Another surprising finding of this study was the extremely high levels of dexamethasone resistance exhibited by the xenografts (> 1000-fold), despite them expressing a functional GR and up-regulating known GR-responsive genes after exposure to dexamethasone.18,19 Nevertheless, the mechanisms associated with glucocorticoid resistance are likely to be clinically relevant, because both in vitro and in vivo dexamethasone sensitivity correlates with treatment outcome of the patients from whom the xenografts were derived.25 Moreover, the candidate resistance mechanism identified in the xenografts, that of epigenetic BIM repression, was confirmed in leukemic blasts from independent cohorts of ALL patients. Despite this strong evidence, it is unlikely that epigenetic BIM repression is the sole mechanism of glucocorticoid resistance in pediatric ALL. For example, increased expression of the antiapoptotic MCL1 gene has been strongly implicated.48 Therefore, BIM repression is likely to be only one component of a multifactorial drug resistance mechanism.
The use of epigenetic modifiers, such as DNA demethylating agents or HDIs, is a burgeoning area of cancer treatment.43 Nevertheless, for a disease such as pediatric ALL where cure rates now approach 80% it is essential to prove therapeutic concepts using in vivo preclinical models before advancement into clinical trials. Despite vorinostat demonstrating unimpressive in vivo single-agent activity against many of the xenografts used in this study,50 our findings support the strategic use of HDIs to promote improved response to glucocorticoid therapy in patients whose lymphoid malignancies exhibit BIM repression associated with decreased H3K9Ac but not increased CpG methylation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We gratefully acknowledge the staff and patients of the Center for Children's Cancer and Blood Disorders, Sydney Children's Hospital, for provision of primary ALL samples; Rachael Papa and Clare Boland for technical assistance; and Dr Rosemary Sutton and Nicola Venn for assistance in selecting patient biopsy specimens. Vorinostat was generously provided by Merck, Sharp and Dohme and the National Cancer Institute, National Institutes of Health.
Children's Cancer Institute Australia for Medical Research is affiliated with The University of New South Wales and Sydney Children's Hospital.
This work was supported by the Children's Cancer Institute Australia for Medical Research, a fellowship (R.B.L.) and grants from The Australian National Health and Medical Research Council and the Anthony Rothe Memorial Trust, a University Postgraduate Award from the University of New South Wales (P.S.B.), an Australian Postgraduate Award (C.D.), the Center for Children's Cancer and Blood Disorders, Sydney Children's Hospital (V.A.B.), by a Postgraduate Scholarship from the Leukemia Foundation of Australia (V.A.B.), by the Associazione Italiana Ricerca sul Cancro (AIRC, IG-4637). and by the Italian Prin program–Ministry of University and Research (PRIN-MIUR).
Authorship
Contribution: P.S.B., R.G.P., N.C.W., C.D., V.A.B., G.C., A.B., C.G.-P., D.M.-S., B.G., R.S., J.M.C., G.M.M., J.E.P., and R.B.L. designed the study; P.S.B., R.G.P., M.E.J., N.C.W., C.D., B.S., G.G., A.M., and V.M. performed experiments; and P.S.B. and R.B.L. contributed to writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Associate Professor Richard B. Lock, Children's Cancer Institute Australia for Medical Research, Lowy Cancer Research Centre, University of New South Wales, PO Box 81, Randwick NSW 2031, Australia; e-mail: rlock@ccia.unsw.edu.au.
REFERENCES
Author notes
P.S.B. and R.G.P. contributed equally to this work.