Abstract
Blood cells of an adult vertebrate are continuously generated by hematopoietic stem cells (HSCs) that originate during embryonic life within the aorta-gonad-mesonephros region. There is now compelling in vivo evidence that HSCs are generated from aortic endothelial cells and that this process is critically regulated by the transcription factor Runx1. By time-lapse microscopy of Runx1-enhanced green fluorescent protein transgenic zebrafish embryos, we were able to capture a subset of cells within the ventral endothelium of the dorsal aorta, as they acquire hemogenic properties and directly emerge as presumptive HSCs. These nascent hematopoietic cells assume a rounded morphology, transiently occupy the subaortic space, and eventually enter the circulation via the caudal vein. Cell tracing showed that these cells subsequently populated the sites of definitive hematopoiesis (thymus and kidney), consistent with an HSC identity. HSC numbers depended on activity of the transcription factor Runx1, on blood flow, and on proper development of the dorsal aorta (features in common with mammals). This study captures the earliest events of the transition of endothelial cells to a hemogenic endothelium and demonstrates that embryonic hematopoietic progenitors directly differentiate from endothelial cells within a living organism.
Introduction
The first hematopoietic stem cells (HSCs) of vertebrates originate in an embryonic site called the aorta-gonad-mesonephros (AGM) region.1,2 Understanding the processes that initiate HSC generation in this niche is not only important for hematology but also has potential to advance our knowledge of tissue stem cell biology. Although it has been widely accepted that hemogenic endothelial cells of the dorsal aorta (DA) are the source of HSCs within the AGM region, in vivo evidence for this has only come forth very recently. An alternative view has been that HSCs differentiate from mesenchymal progenitors immediately underlying the DA.3,4 Histologic analyses have shown that embryonic blood always develops in intimate association with developing blood vessels.5 Blood and endothelial cells have overlapping expression of early gene markers.6-8 Cell and lineage tracing studies of endothelial cells in zebrafish, chick, and mouse embryos have shown that labeled cells are subsequently found in HSC sites.7,9-11 With advancements in live imaging technology, the fates of single mouse mesodermal cells were tracked by long-term microscopy to show that they can generate hemogenic endothelial cells that directly give rise to blood cells in culture.12 Live imaging of mouse DA explants has now shown that the process occurs in vivo.13 Real-time imaging of transparent transgenic zebrafish embryos has shown HSC emergence from the DA in an intact organism by a novel mechanism not requiring asymmetric cell division.14,15
In all vertebrates, the transcription factor Runx1 is absolutely essential for the development of definitive blood.4,16,17 Conditional deletion of Runx1 in mouse endothelial cells has demonstrated a requirement for Runx1 in the vasculature that is integral to its role in HSC generation.9 High-resolution microscopy within the zebrafish AGM revealed that the physical transition of hemogenic endothelial cells to nascent HSCs, by a process of bending and egress into the subaortic space, is strictly dependent on Runx1 activity.15 Expression of zebrafish runx1 is first detected in the AGM region in a subset of cells lining the floor of the DA at 22 hours postfertilization (hpf), before commencement of blood circulation.16,18 Soon after the onset of circulation at 25 hpf, runx1 also labels nascent embryonic blood progenitors, presumably including HSCs, borne in the AGM region.18,19 These cells located in the narrow region between the DA and caudal vein (CV) also begin to express cmyb and CD41 at approximately 27 and 35 hpf, respectively.19-22 The recent studies in zebrafish showing the transition of DA hemogenic endothelial cells to HSCs have taken advantage of the Tg(cmyb:EGFP) and Tg(CD41:EGFP) zebrafish transgenic lines where enhanced green fluorescent protein (EGFP) marks HSCs.14,15 Endothelial cells do not express cmyb or CD41, so that in these transgenics EGFP-positive HSCs were visualized when already occupying the space between the DA and CV.20,21 Confocal imaging detected low levels of cmyb and CD41 transgene expression in some hemogenic endothelial cells in the process of budding into the subaortic space from 33 hpf.14,15
In the work presented here, we have used runx1 as a marker of very early HSC commitment. By time-lapse imaging, we were able to directly track the dynamic transition of these vascular endothelial cells as they acquire EGFP expressed from a transgenic runx1 promoter. These EGFP-expressing endothelial cells later display HSC-like properties, strongly supporting the emergence of the earliest HSCs from hemogenic endothelium.
Methods
Zebrafish (Danio rerio) maintenance and generation of zebrafish transgenics
The Tg(runx1P2:EGFP) line has been established and characterized previously.18 To generate the Tg(kdrl:nls-mCherry) line, the zebrafish kdrl promoter23 was first subcloned into the pCR II-TOPO vector (Invitrogen). The transgene plasmid was constructed and stable line established as described.18,24 Before cloning into the XhoI and SmaI sites of the Tol2 vector, the EGFP gene was replaced with the mCherry gene fused to a nuclear localization signal.25 The constructs were injected into embryos at the 1-cell stage and screened for germline integration by random intercrosses. Embryos were obtained from natural spawnings and raised in embryo medium at 28.5°C. All studies conducted for this manuscript have been reviewed and approved by the University of Auckland Animal Ethics Committee.
Morpholino injections
Morpholino (MO) antisense oligonucleotides were all purchased from GeneTools and injected at a dose of 0.25 to 0.5 pmol per embryo: Ctrl MO, 5′ CCTCTTACCTCAGTTACAATTTATA 3′ (standard control); Runx1 MO, 5′ TTTTCAGCATCTCACCTCGTCCGCT 3′; troponin T2 (Tnnt2) MO, 5′ CATGTTTGCTCTGATCTGACACGCA 3′; vascular endothelial growth factor a (Vegfa) MO, 5′ GTATCAAATAAACAACCAAGTTCAT 3′.
Statistical analysis
The number of runx1-EGFP/kdrl-mCherry-positive cells in the DA at 28, 35, and 48 hpf were scored as percentage of the total number of cells per field of view in the DA (n = 5; 1 field of view in 5 embryos). High-resolution images of the segments analyzed were captured by confocal microscopy to accurately count kdrl-mCherry nuclei. The range of DA cells counted per field of view ranged from 13 to 30 cells for 28 hpf embryos, 18 to 36 cells for 35 hpf embryos, and 25 to 49 cells for 48 hpf embryos. Bar graphs and statistical analyses were performed using Student t test (GraphPad Prism Version 5.0c).
Confocal imaging
Compound Tg(kdrl:nls-mCherry)/Tg(runx1P2:EGFP) embryos were characterized from crosses between the 2 transgenic parents. Time-lapse microscopy and image processing were performed as described.24 Embryos were anesthetized in tricaine, mounted in 1% (weight/volume) low-melt agarose in E3 and imaged with an Olympus FV1000 confocal microscope. Stacks of 15 to 30 images at 3 μm distance apart were taken at 7.5- to 10-minute intervals. The embryo was monitored regularly under brightfield and reembedded when the agarose was obstructing growth of the embryo. The resulting stack of images for each time point were processed with ImageJ software Version 1.43 (National Institutes of Health) and used to construct an image sequence. Time-lapse movies were performed for at least 5 embryos. Three-dimensional reconstructions of confocal images were generated using Volocity 5.0 image analysis software (Improvision/PerkinElmer Life and Analytical Sciences). Confocal imaging of double-transgenic (kdrl-mCherry/runx1P2-EGFP) embryos injected with MO was performed in a Nikon 90i/C1 plus confocal system using water immersion objectives. Still images were processed in Photoshop CS3 Extended (Adobe).
Cell tracing
Tracing of cells expressing EGFP or EGFP/mCherry in transgenic embryos was performed using the 10 000 molecular weight dextran conjugate of CMNCBZ-caged carboxy-Q-rhodamine or 5-carboxymethoxy-2-nitrobenzyl (CMNB)–caged fluorescein, respectively (Invitrogen) as described.19 Exposure to light was minimized during injection and subsequent experimentation to prevent unintentional uncaging. The injected embryos were allowed to develop to the required age. Uncaging of the photoactivatable fluorophores (n = 10 embryos; 10 cells uncaged per embryo) was performed on an Olympus FV1000 confocal microscope. Cells containing uncaged rhodamine were imaged at a later time point by confocal microscopy as described in the previous paragraph. Detection of uncaged fluorescein was performed by antibody staining as previously described.19
Results
Analysis of compound runx1P2/kdrl fluorescent reporter embryos reveals that Runx1/EGFP-positive hematopoietic cells directly differentiate from a subset of Kdrl/mCherry endothelial cells
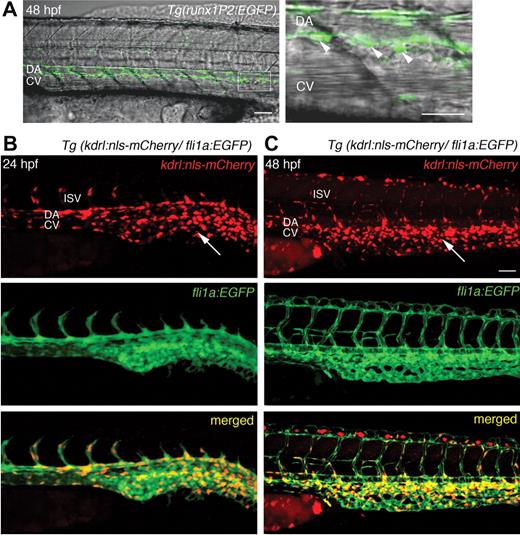
We have previously described the hematopoietic EGFP expression of the Tg(runx1P2:EGFP) line within the AGM region, and later in the thymus and kidney.18 The data corroborate that these Runx1/EGFP-positive cells have attributes consistent with an HSC identity. In the AGM region, rounded Runx1/EGFP-positive cells populate the subaortic space flanked by the main trunk vessels at 48 hpf, a finding consistent with the EGFP-expressing cells described within the cmyb and CD41 transgenics (Figure 1A). In the runx1P2 transgenic, EGFP expression within the AGM begins at 22 hpf, earlier than that demonstrated within the cmyb and CD41 reporter lines.
EGFP expression in HSCs of Tg(runx1P2:EGFP) and mCherry in the endothelium of Tg(kdrl:nls-mCherry) transgenic embryos. (A) Overlaid brightfield and fluorescent images of Tg(runx1P2:EGFP) at 48 hours postfertilization (hpf) displaying rounded HSCs (arrowheads in higher magnification of boxed area) just ventral to the DA. (B-C) Lateral views of Tg(kdrl:nls-mCherry)/Tg(fli1a:EGFP) compound transgenic embryos at 24 and 48 hpf. Red fluorescent mCherry was localized to the nuclei of all endothelial cells of the DA, CV, and intersegmental vessels (ISV) of the trunk, and numerous endothelial cells of the caudal vascular plexus (arrows). Scale bars represent 40 μm.
EGFP expression in HSCs of Tg(runx1P2:EGFP) and mCherry in the endothelium of Tg(kdrl:nls-mCherry) transgenic embryos. (A) Overlaid brightfield and fluorescent images of Tg(runx1P2:EGFP) at 48 hours postfertilization (hpf) displaying rounded HSCs (arrowheads in higher magnification of boxed area) just ventral to the DA. (B-C) Lateral views of Tg(kdrl:nls-mCherry)/Tg(fli1a:EGFP) compound transgenic embryos at 24 and 48 hpf. Red fluorescent mCherry was localized to the nuclei of all endothelial cells of the DA, CV, and intersegmental vessels (ISV) of the trunk, and numerous endothelial cells of the caudal vascular plexus (arrows). Scale bars represent 40 μm.
To examine whether there is early colocalization of runx1/EGFP expression in endothelial cells, mCherry, a red photostable fluorophore with relatively short maturation time, was chosen.26 Kdr is an early marker for both the hematopoietic and vascular lineages.6 A Tg(kdrl:EGFP) line has been made that recapitulates early kdrl expression.23 We generated the Tg(kdrl:nls-mcherry) line, where the transgene driven by the kdrl promoter was fused to a nuclear localization signal25 so that red fluorescence is restricted to the nuclei, allowing individual endothelial cells and their behavior to be easily distinguished. By 24 hpf, mCherry was expressed in endothelial cells of the DA, CV, and the intersegmental vessels (Figure 1B). Expression within the axial vessels became more prominent at 48 hpf (Figure 1C). To confirm nuclear mCherry expression within endothelial cells, Tg(kdrl:nls-mcherry) was crossed with the Tg(fli1a:EGFP) line.27 In the latter, the promoter of another vascular gene, fli1a, directs cytoplasmic EGFP expression within the developing vasculature27 (Figure 1B-C). In compound transgenics, nuclear mCherry expression was coincident with EGFP expression within endothelial cells (Figure 1B-C).
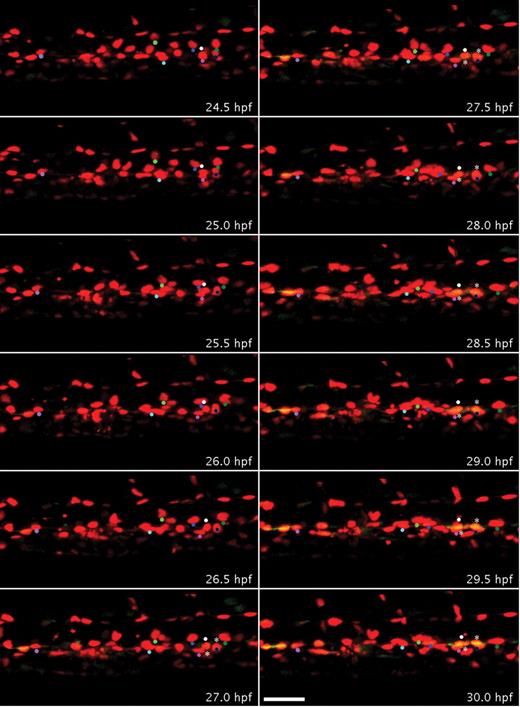
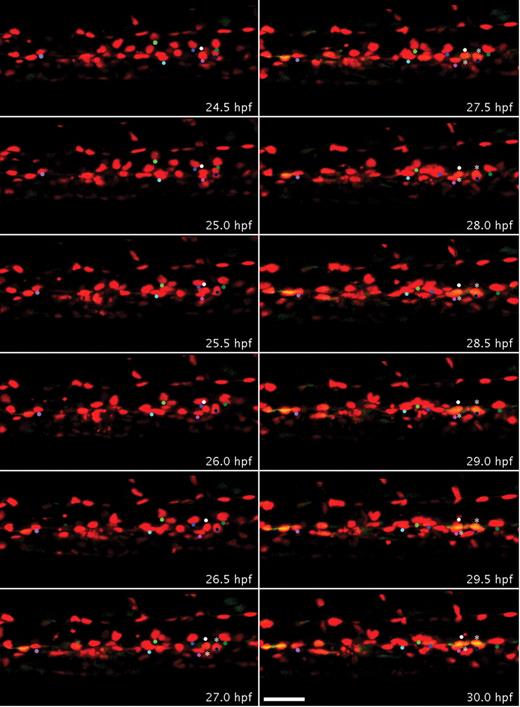
To determine whether HSCs develop by differentiation of endothelial cells in the AGM, the Tg(runx1P2:EGFP) line was crossed with the Tg(kdrl:nls-mCherry) line. To demonstrate that HSCs directly differentiate from the DA and later ingress to the subaortic space, time-lapse confocal imaging of these compound transgenic embryos was performed from 22 to 35 hpf. EGFP expression in some endothelial cells at 22 hpf was very weak,18 and overlapping green and red signals were first detectable at approximately 25 hpf (Figure 2; supplemental Video 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To clearly visualize colocalization of green and red signals within endothelial cells lining the ventral wall of the DA, x, y, and z slices from specific z-stack projections were analyzed in 3 dimensions (supplemental Figure 1). Time-lapse imaging showed that increasing green fluorescent signal in the DA always colocalized with nuclear red fluorescence (Figure 2), indicating that all Runx1/EGFP-positive hematopoietic progenitors directly emerged from endothelial cells expressing Kdrl/mCherry (11 EGFP+ cells/11 mCherry+ nuclei in Figure 2; supplemental Video 1). By 28 hpf, 33% (27 of 82) of ventral DA endothelial cells had become EGFP-positive (yellow). At approximately 32 hpf, some of these double-fluorescent cells began to form a rounder morphology compared with other endothelial cells (Figure 2). We observed a rare case where one of these double-positive cells, after an apparent cell division, migrated dorsally, also giving rise to an intersegmental vessel (33-34.5 hpf, supplemental Video 1). Overall, our analysis shows that the onset of overlapping runx1/EGFP and kdrl/mCherry expression is detected in a subset of endothelial cells at least 9 hours before these cells assume a rounded HSC-like morphology.
Endothelial cells of the DA transdifferentiate to form Runx1-positive blood progenitors. Embryos bred from a Tg(runx1P2:EGFP) and Tg (kdrl:nls-mCherry) cross were imaged by time-lapse confocal microscopy from 22 to 35 hpf, which captured the development of endothelial cells within a segment of the trunk DA (supplemental Video 1). Panels display frames every 30 minutes from 24.5 to 30 hpf. Colored dots were superimposed to track red to yellow transition of individual cells. At 24.5 hpf, all the nuclei of endothelial cells that line the DA only expressed mCherry. By 30 hpf, several red cells of the DA also expressed EGFP in the ventral aspect of the DA and appear yellow. More rounded double-fluorescent cells are marked by asterisks. Scale bar represents 40 μm.
Endothelial cells of the DA transdifferentiate to form Runx1-positive blood progenitors. Embryos bred from a Tg(runx1P2:EGFP) and Tg (kdrl:nls-mCherry) cross were imaged by time-lapse confocal microscopy from 22 to 35 hpf, which captured the development of endothelial cells within a segment of the trunk DA (supplemental Video 1). Panels display frames every 30 minutes from 24.5 to 30 hpf. Colored dots were superimposed to track red to yellow transition of individual cells. At 24.5 hpf, all the nuclei of endothelial cells that line the DA only expressed mCherry. By 30 hpf, several red cells of the DA also expressed EGFP in the ventral aspect of the DA and appear yellow. More rounded double-fluorescent cells are marked by asterisks. Scale bar represents 40 μm.
Runx1/Kdrl-positive HSCs migrate to the subaortic space and enter circulation through the CV to seed other hematopoietic sites
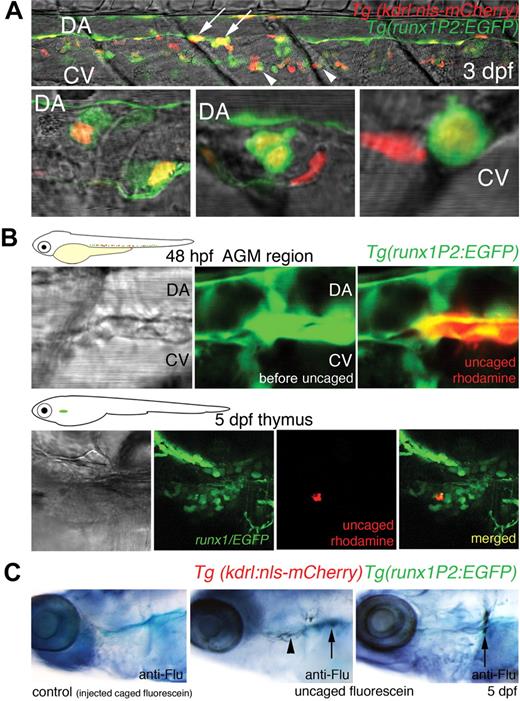
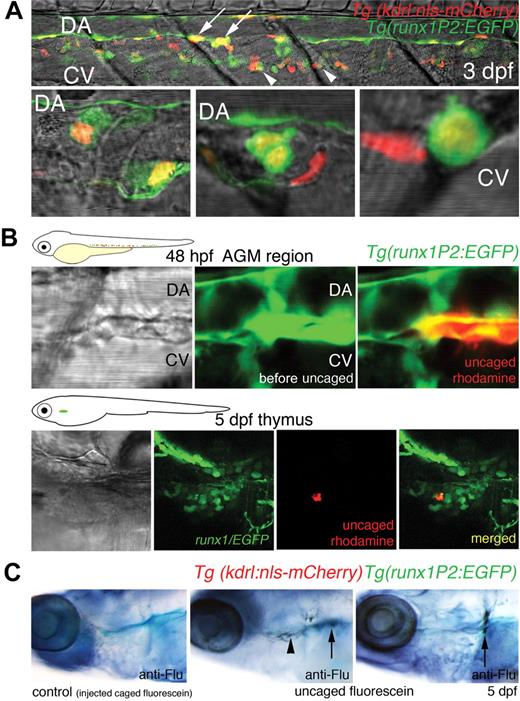
We have previously reported that HSCs become abundant in the region between the DA and CV from 48 hpf, suggesting that HSCs migrate to this site.18 To confirm that in the current system double-fluorescent cells arising from the DA populate the subaortic space, confocal images of a segment of the AGM within 3 days postfertilization (dpf) larvae were prepared. Several EGFP/mCherry-expressing cells with rounded morphology were shown to occupy the clearly defined subaortic space between the axial vessels at this stage. Some double-fluorescent cells were also imaged that appear to associate closely with the CV (Figure 3A). Zebrafish blood progenitors have been previously shown to migrate from the AGM to other hematopoietic niches, entering circulation by the CV.20 In an extended movie (until 43 hpf), we captured a double-fluorescent cell that became rounded and appeared to undergo cell division at 40 hpf. At 41 hpf, one of the daughter cells, after moving within the subaortic space, accessed the circulation through the CV (supplemental Video 2). This sequence of behavior of nascent blood progenitors, cell division, and heightened motility before entering the axial vein has been previously documented in zebrafish embryos.15,20 In at least 3 other time-lapse sequences performed, we have consistently observed that HSCs follow this venous route to enter circulation. We have observed continued EGFP expression in some elongated endothelial cells of the DA later than 35 hpf.18 As this pattern of expression had not been detected by in situ hybridization analysis for runx1,16 it may be the result of the absence of a silencer element in the transgene promoter or to residual EGFP signal after cell divisions. All double-positive cells in the subaortic space were rounded.
Cells of the DA expressing both Runx1/EGFP and Kdrl/mCherry migrate to the DA/CV subaortic region, thymus, and kidney. (A) Confocal images of a segment of the trunk of a larvae 3 days postfertilization (dpf) showing the AGM region. (Top panel) A 6-somite span of the subaortic space flanked by the DA and CV. (Bottom 3 panels) High-resolution images of double-positive cells (not from the top panel) occupying the subaortic space. Yellow cells with rounded morphology expressing both Runx1/EGFP and Kdrl/mCherry were seen intimately associated with the floor of the DA (arrows), within the subaortic space, or close to the CV (arrowheads). EGFP expression was also observed within elongated endothelial cells of the DA, distinct from the round presumptive HSCs. Some mCherry-positive cells within the region were part of intersegmental vessels. (B) Photoactivatable rhodamine-dextran uncaged in Runx1/EGFP-expressing cells of the AGM region in 48-hpf embryos was traced to the thymus at 5 dpf. (C) Photoactivatable CMNB-fluorescein uncaged in double-fluorescent (Runx1/EGFP, Kdrl/mCherry) AGM cells at 48 hpf was detected in the thymus ( ) and kidney (↑) at 5 dpf by fluorescein antibody staining. No antibody staining was observed in control embryos injected with caged fluorescein (n = 10).
) and kidney (↑) at 5 dpf by fluorescein antibody staining. No antibody staining was observed in control embryos injected with caged fluorescein (n = 10).
Cells of the DA expressing both Runx1/EGFP and Kdrl/mCherry migrate to the DA/CV subaortic region, thymus, and kidney. (A) Confocal images of a segment of the trunk of a larvae 3 days postfertilization (dpf) showing the AGM region. (Top panel) A 6-somite span of the subaortic space flanked by the DA and CV. (Bottom 3 panels) High-resolution images of double-positive cells (not from the top panel) occupying the subaortic space. Yellow cells with rounded morphology expressing both Runx1/EGFP and Kdrl/mCherry were seen intimately associated with the floor of the DA (arrows), within the subaortic space, or close to the CV (arrowheads). EGFP expression was also observed within elongated endothelial cells of the DA, distinct from the round presumptive HSCs. Some mCherry-positive cells within the region were part of intersegmental vessels. (B) Photoactivatable rhodamine-dextran uncaged in Runx1/EGFP-expressing cells of the AGM region in 48-hpf embryos was traced to the thymus at 5 dpf. (C) Photoactivatable CMNB-fluorescein uncaged in double-fluorescent (Runx1/EGFP, Kdrl/mCherry) AGM cells at 48 hpf was detected in the thymus ( ) and kidney (↑) at 5 dpf by fluorescein antibody staining. No antibody staining was observed in control embryos injected with caged fluorescein (n = 10).
) and kidney (↑) at 5 dpf by fluorescein antibody staining. No antibody staining was observed in control embryos injected with caged fluorescein (n = 10).
To confirm that rounded runx1-positive cells function as HSCs and migrated to other hematopoietic sites, such as the thymus and kidney (the zebrafish equivalent of mammalian bone marrow), cell tracing using caged fluorophores was performed.19 Uncaged rhodamine dextran in rounded runx1/EGFP cells (10 cells per embryo) within the AGM region at 48 hpf was later traced to the thymus at 5 dpf (Figure 3B). For uncaging experiments of double-fluorescent cells, CMNB-caged fluorescein was used, and uncaged fluorescein was detected at 5 dpf in the thymus (in 5 of 7 embryos) and kidney (in 4 of 7 embryos) by antifluorescein antibody staining (Figure 3C). We have previously shown that, in runx1P2:EGFP transgenics, EGFP-expressing cells localized to the larval thymus and to the larval, juvenile, and adult kidney.18 Similar cell tracing analysis has been previously performed on rounded cells between the DA and CV that express runx1 and cmyb, and a population of uncaged cells also later homed to the thymus and kidney.19 Taken together, we have shown that runx1-positive cells within the AGM region, consistent with an HSC identity, are able to seed other hematopoietic sites.
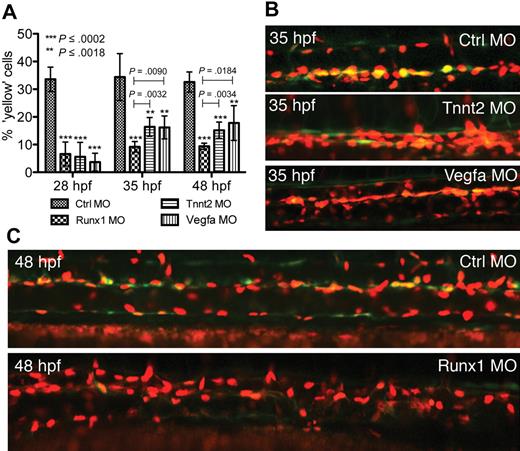
Emergence of EGFP/mCherry-positive cells in the AGM region is dependent on transcriptional regulation by Runx1, blood flow, and development of the DA
It is known that Runx1-dependent formation of HSCs is modulated by blood flow and normal development of the DA.15,28-31 As further support that the double-fluorescent cells in the DA in our compound transgenics represent HSCs, we examined the contributions of Runx1 activity, blood flow, and DA development to their emergence. Disruption of either cardiac function or aortic development resulted in a significant decrease in the numbers of EGFP/mCherry-positive cells in the AGM region (Figure 4). MO knockdown of cardiac Tnnt2 or Vegfa resulted in embryos that failed to develop a heartbeat or a functional DA, respectively, with the consequent absence of blood flow. Similar to uninjected double-transgenic embryos, 33% of ventral DA endothelial cells in embryos injected with control MO had become EGFP-positive by 28 hpf. By depleting Tnnt2 or Vegfa, the numbers of double-fluorescent cells in the AGM region compared with those injected with control MO were initially reduced 6- to 9-fold (28 hpf; ctrlMO 33%, Tnnt2MO 6%, VegfaMO 4%; Figure 4A). By 35 hpf, the population of EGFP/mCherry-positive cells in the AGM region of embryos injected with Tnnt2MO or VegfaMO were half those injected with control MO (ctrlMO 33%, TnntMO 16%, VegfaMO 16%; Figure 4A-B). Runx1 morphants, while maintaining normal circulation (primitive erythrocytes), exhibited a more marked diminution of HSC numbers at 35 hpf and 48 hpf (ctrlMO 33%, Runx1MO, 9%; Figure 4A,C). This highlights a more stringent requirement for cell-autonomous Runx1 activity in HSC formation and is consistent with data showing that hematopoietic defects resulting from abnormal DA formation, as a consequence of impaired Vegf signaling, can be rescued by runx1 overexpression.28
Depletion of Runx1, Vegfa, or Tnnt2 impairs development of double-fluorescent (Runx1/EGFP, Kdrl/mCherry) HSCs in the AGM region. After MO injection, double-fluorescent cells (yellow) in the DA were scored as a percentage of total cells in the DA per field of view in a total of 5 embryos. (A) Quantification and (B-C) lateral views of representative embryos. Error bars represent SD. Statistical significance was measured by P value (Student t test). For each test bar (Runx1 MO, Tnnt2 MO, Vegfa MO), asterisks denote P values for comparisons with Ctrl MO for that developmental stage. Capped lines with P values denote comparisons between Runx1 MO with either Tnnt2 MO or Vegfa MO.
Depletion of Runx1, Vegfa, or Tnnt2 impairs development of double-fluorescent (Runx1/EGFP, Kdrl/mCherry) HSCs in the AGM region. After MO injection, double-fluorescent cells (yellow) in the DA were scored as a percentage of total cells in the DA per field of view in a total of 5 embryos. (A) Quantification and (B-C) lateral views of representative embryos. Error bars represent SD. Statistical significance was measured by P value (Student t test). For each test bar (Runx1 MO, Tnnt2 MO, Vegfa MO), asterisks denote P values for comparisons with Ctrl MO for that developmental stage. Capped lines with P values denote comparisons between Runx1 MO with either Tnnt2 MO or Vegfa MO.
Discussion
A central challenge in hematology research is to recapitulate the generation and expansion of HSCs in vitro for clinical applications. Although differentiated blood cells can be readily produced in culture, it is still not possible to generate HSCs that are able to retain engraftment and self-renewing properties when transplanted in vivo. The last decade has seen great advances in our understanding of the molecular programs that regulate key properties of HSC specification. Because vertebrate HSCs are generated during embryogenesis, it has become clear that a precise understanding of the developmental processes within successive niches, from HSC emergence to their maturation, is also paramount for simulation of the formation of HSCs. An important step in this direction is the recent confirmation in vivo that HSCs are directly derived from hemogenic endothelial cells of the embryonic DA.
In the current study, we captured in real-time in a living organism the transdifferentiation of endothelial cells located in the ventral wall of the DA to hemogenic endothelial cells, and their subsequent transition to hematopoietic cells with attributes consistent with an HSC identity. Critical to this analysis is the use of a specific zebrafish Runx1-EGFP transgenic line. Vertebrate Runx1 is transcribed from 2 alternative promoters.32 We have previously described the establishment of 2 transgenic zebrafish lines, with EGFP driven by the distal promoter, runx1P1, or the proximal promoter, runx1P2.18 In the latter transgenic line, Tg(runx1P2:EGFP), we detected the earliest hematopoietic EGFP expression within hemogenic endothelial cells of the AGM region. EGFP expression within the hemogenic endothelium was not observed with the distal runx1 promoter line, Tg(runx1P1:EGFP).18 A mouse line with a reporter knocked into the Runx1P2 promoter also displayed expression within hemogenic endothelial cells.33 This level of conservation between zebrafish and mammals highlights a unique role for Runx1 transcribed from the proximal promoter for HSC generation.
In the transparent Tg(kdrl:nls-mCherry/runx1P2:EGFP) zebrafish embryo, the potential hemogenic endothelial cells are initially indistinguishable from the other cells lining the DA. This vessel becomes patent and functional at 25 hpf, soon after the heart starts beating, mainly for circulation of primitive erythrocytes. As visualized in the double-fluorescent transgenic embryos, initiation of runx1 expression in a subset of aortic endothelial cells distinguishes those with hemogenic potential. Because the earliest runx1 expression (EGFP) in the AGM region was strictly observed to colocalize with nuclear mCherry expression in endothelial cells, mesenchymal cells can be ruled out as a source of embryonic HSCs. Only Runx1-EGFP/Kdrl-mCherry-positive cells proceeded to assume a rounded morphology characteristic of hematopoietic cells and subsequently populated the subaortic space. An important research focus that comes from this work is to determine the detailed molecular mechanisms and interactions that dictate how an apparently functional and differentiated subpopulation of endothelial cells can give rise to HSCs. This could open up the exciting possibility of generating hematopoietic cells from endothelial tissue cultures for therapeutic use.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Alhad Mahagaonkar for managing the fish facility, Annie Chien and the University of Auckland Biomedical Imaging Research Unit for technical support, and J. Chen, B. Weinstein, and K. Kawakami for sharing their reagents.
This work was supported by the Auckland Medical Research Foundation and the Foundation for Research, Science and Technology, New Zealand. E.Y.N.L. was supported by a Bright Futures Top Achievers Doctoral Scholarship.
Authorship
Contribution: E.Y.N.L. and M.V.F. designed and performed the experimental work, analyzed the data, and wrote the paper; C.J.H. performed experimental work and analyzed the data; and P.S.C. and K.E.C. planned the project, analyzed the data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Maria Vega Flores, Department of Molecular Medicine & Pathology, School of Medical Sciences, University of Auckland, Private Bag 92019, Auckland, New Zealand; e-mail: m.flores@auckland.ac.nz; and Kathryn E. Crosier, Department of Molecular Medicine & Pathology, School of Medical Sciences, University of Auckland, Private Bag 92019, Auckland, New Zealand; e-mail: ke.crosier@auckland.ac.nz.




 ) and kidney (↑) at 5 dpf by fluorescein antibody staining. No antibody staining was observed in control embryos injected with caged fluorescein (n = 10).
) and kidney (↑) at 5 dpf by fluorescein antibody staining. No antibody staining was observed in control embryos injected with caged fluorescein (n = 10).