Abstract
To develop safer and more effective vectors for gene therapy of X-linked severe combined immunodeficiency (SCID-X1), we have evaluated new self-inactivating lentiviral vectors based on the HIV virus. The CL20i4-hγc-Revgen vector contains the entire human common γ chain (γc) genomic sequence driven by the γc promoter. The CL20i4-EF1α-hγcOPT vector uses a promoter fragment from the eukaryotic elongation factor alpha (EF1α) gene to express a codon-optimized human γc cDNA. Both vectors contain a 400-bp insulator fragment from the chicken β-globin locus within the self-inactivating long-terminal repeat. Transduction of bone marrow cells using either of these vectors restored T, B, and natural killer lymphocyte development and function in a mouse SCID-X1 transplantation model. Transduction of human CD34+ bone marrow cells from SCID-X1 patients with either vector restored T-cell development in an in vitro assay. In safety studies using a Jurkat LMO2 activation assay, only the CL20i4-EF1α-hγcOPT vector lacked the ability to transactivate LMO2 protein expression, whereas the CL20i4-hγc-Revgen vector significantly activated LMO2 protein expression. In addition, the CL20i4-EF1α-hγcOPT vector has not caused any tumors in transplanted mice. We conclude that the CL20i4-EF1α-hγcOPT vector may be suitable for testing in a clinical trial based on these preclinical demonstrations of efficacy and safety.
Introduction
X-linked severe combined immunodeficiency (SCID-X1) is caused by loss-of-function mutations in the γc gene (also known as IL2RG),1,2 which encodes the common γ-chain (γc) subunit for the IL2, IL4, IL7, IL9, IL15, and IL21 receptors and is required for signal transduction with these cytokines.3 Human SCID-X1 is characterized by lack of T, natural killer (NK) cells, and nonfunctional B cells and is fatal early in life from progressive infections if left untreated. Allogeneic stem cell transplantation is the current standard of care and can be curative, particularly when a matched sibling donor is available.4 However, most patients lack a matched sibling donor and therefore receive transplants using haploidentical grafts obtained from a parent. These patients do significantly less well than those receiving matched sibling transplants, both in terms of overall survival and in terms of persistent immunodeficiency.5-7 Most experts agree that better treatment is desirable for SCID-X1 patients who lack a matched sibling donor.8-10
Gene therapy is one of several approaches that is being developed as an alternative to haploidentical allogeneic transplantation.11-15 Pioneering studies done in Europe using vectors derived from the mouse Moloney leukemia virus (MLV) have proven that this approach can be effective because of restoration of T-cell numbers and function as well as humoral immunity.11-14 However, the development of T-cell leukemia in 5 of 20 of these patients has uncovered a serious, unanticipated side effect associated with these gene therapy vectors.16-18 Comprehensive molecular analyses of the leukemia cases have identified vector insertions into a small number of T-cell proto-oncogenes, most particularly LMO2, leading to its aberrant and high-level expression.16-18 Further analysis of these integration sites has revealed that most, if not all, cases of gene activation were the result of the strong enhancer elements located in the U3 region of the MLV long-terminal repeat (LTR). Clearly, new vectors for SCID-X1 should not contain these MLV LTR enhancer elements to achieve better safety. This can be accomplished using self-inactivating (SIN) vectors that contain deletions in the U3 region of the LTR and is possible using either MLV or HIV-based vectors.
We chose to use lentiviral vectors because they are more efficient for transducing quiescent human hematopoietic stem cells (HSCs),19 and a recent study has suggested that, when an internal LTR enhancer is present, SIN lentiviral vectors require 10-fold higher copy numbers than an SIN-MLV vector to cause tumors in a tumor-prone mouse model,20 presumably because of the known differences in integration site profile between HIV versus MLV vectors.21 Another important consideration is the recent development of a stable packaging system for producing SIN lentiviral vectors.22 This eliminates the need for using transient transfection methods with multiple plasmid DNAs to produce lentiviral vector particles.
Whether one uses MLV or HIV SIN vectors, an important issue that has received relatively little study is the identification of suitable internal promoters to drive transcription of the transgene. Several recent studies have identified appropriate cellular promoters for this purpose, which include fragments from the EF1α gene,23 the phosphoglycerol kinase gene,24 and the bidirectional ubiquitously acting chromatin opening element.25 Although it appears that several of these promoters can achieve therapeutic levels of γc expression, little is known about the relative ability of these fragments to transactivate other cellular promoters.23,26 Another possibility is the use of the endogenous γc promoter itself, which would have the potential advantage of achieving a more physiologic expression pattern in hematopoietic cells derived from transduced HSCs.
In this study, we report new information on the use of the endogenous γc promoter in an SCID-X1 vector. We also describe the characteristics of an SCID-X1 vector that uses the EF1α promoter to express the γc gene and that is shown to be relatively less probable to cause transformation based on a variety of safety assays.
Methods
Animals
γc−/− mice were described before and used for bone marrow harvesting between 4 and 10 weeks.27 Female γc−/−Rag2−/− mice were purchased from Taconic Farms (#004111) and used as transplantation recipients at 4 to 10 weeks of age. All experimental procedures were approved by the Institutional Animal Care and Use Committee of St Jude Children's Research Hospital.
Vectors
The HIV-1 based pCL20 vector has been described.28 A 400-bp chicken β-globin chromatin insulator was inserted into the 3′ U3 of the LTR.29,30 The human γc genomic sequence, a 5341-bp fragment that contained regions 1205 bp upstream (proximal promoter) and 4136 bp downstream of the first base pair of the start codon (based on National Center for Biotechnology Information Build, Version 36.1), was amplified by polymerase chain reaction (PCR) using human BAC clone CTD-3090M11 (Invitrogen) as template and inserted into the pCL20 vector at the Mlu1 and Not1 restriction sites in a reverse genomic orientation (CL20i4-hγc-Revgen vector; Figure 1). In the second vector, a 233-bp human EF1α promoter31 and a codon optimized human γc cDNA, derived using a standard computational algorithm, were inserted into the same CL20 vector using standard subcloning techniques (CL20i4-EF1α-hγcOPT, or EF1α vector; Figure 1).22 Vesicular stomatitis virus G pseudotyped lentiviral vector particles were prepared by either transient transfection of 293T cells or using the stable inducible producer cell line.22
Bone marrow cell transduction and transplantation
Bone marrow cells from mice treated with 5-fluorouracil (150 mg/kg) 5 days before harvest were cultured in StepSpan serum-free expansion medium supplemented with recombinant growth factors, including murine stem cell factor, murine thrombopoietin, murine insulin-like growth factor II, and human fibroblast growth factor.32 At 24 hours later, cells were transduced with virus supernatant in tissue culture plates coated with RetroNectin.33 Another 24 hours later, cells were collected and 0.7 million to 1.5 million cells were injected intravenously into each γc−/−Rag2−/− mouse irradiated with 600 cGy using a 137Cs irradiator. At various time points after transplantation, peripheral blood samples were collected to perform complete blood count analysis using a FORCYTE Hematology System. Peripheral blood mononuclear cells were also stained for cell surface markers for flow cytometry analysis to evaluate lymphocyte reconstitution. The absolute number of each cell lineage is calculated by multiplying the complete blood count by the percentage of the lineage positivity obtained from flow cytometry. At 21 weeks, 3 mice from each group were killed, and cells of bone marrow, spleen, and thymus were analyzed for lymphoid reconstitution by flow cytometry. All the remaining mice were killed at 7 to 12 months for necropsy and immune reconstitution analysis. Bone marrow cells from some mice were used for secondary transplantation.
Transduction and in vitro differentiation of human SCID-X1 CD34+ hematopoietic cells
Bone marrow CD34+ cells from SCID-X1 patients were obtained with informed consent from parents in accordance with the Declaration of Helsinki and were cultured in X-VIVO 20 medium supplemented with recombinant human growth factors, including stem cell factor (rhSCF), thrombopoietin, FLT3 ligand, and interleukin-3 (IL-3) for 24 hours. A total of 1.2 × 105 cells were then transduced with vectors at a virus concentration of 9 × 107 tu/mL (multiplicity of infection [MOI] of 100) for 24 hours in RetroNectin-coated tissue culture plates. A total of 35 000 cells were then cocultured with an OP9 stromal layer expressing human Delta1 Notch ligand in α-minimum essential medium, 20% Hyclone fetal bovine serum, in the presence of recombinant human IL-7 (2 ng/mL), rhSCF (10 ng/mL), and recombinant human FLT3 ligand (5 ng/mL), in 1 well of a 6-well tissue culture plate, for T lymphocyte differentiation, or cultured in Myelocult in the presence of recombinant human IL-15 and rhSCF for NK-cell differentiation.34 Cells were replated on fresh OP9-DL1 cells every 7 days. At various time points, cells were collected to analyze for cell surface marker expression by flow cytometry.
Hematopoietic cell immortalization assay
Jurkat cell LMO2 activation assay
The provirus form of the CL20i4-hγc-Revgen vector and the CL2i4-EF1α-hγcOPT vector was generated by PCR amplification using genomic DNA from vector-transduced NIH3T3 cells and standard subcloning procedures. The provirus form of the 2 vectors was ligated into a plasmid containing LoxP511 and LoxP sites to make the cassette exchange plasmids, in which the provirus is flanked by LoxP511 and LoxP sites. A Jurkat cell clone (LTR) having an LTR–Green flourescent protein (GFP) insertion flanked by LoxP511 and LoxP sites in the first intron of LMO2 gene was used for Causes Recombination–mediated cassette exchange using the published procedures.37 The clones were subjected to a quantitative PCR screen first to select for vector positive clones, which include clones with random vector integrations and with targeted cassette-exchanged clones. The clones positive for vector integrations were then analyzed by Southern blot to select for clones that had successfully gone through cassette exchange. The LMO2 RNA and protein level in Jurkat cells and targeted clones were measured by quantitative reverse-transcription (RT)–PCR and Western blot assay.37
Results
Vector design and construction
All lentiviral vectors were constructed using the CL20 lentiviral backbone, a third-generation SIN HIV vector in which the viral enhancer/promoter region in the U3 region of the LTR was deleted to achieve an SIN design.38 In the first vector, we incorporated all 8 exons and 7 introns of the human γc gene along with the 1.2-kb proximal promoter into the CL20 vector in a reverse genomic orientation (CL20i4-hγc-Revgen, or Revgen vector; Figure 1A). This design is analogous to the design of current β-globin lentiviral vectors and requires reverse orientation to avoid loss of the introns during vector production.39
Schematic representation of the 2 novel lentiviral vectors for SCID-X1 gene therapy and their activity in various cell types. (A) Both vectors are based on third-generation lentiviral vector backbone. The enhancer/promoter in the long-terminal repeat region was deleted (ΔU3) and replaced with a 400-bp chromatin insulator element (Ins) from the chicken β-globin locus. The CL20i4-hγc-Revgen vector contains the entire human IL2RG gene (γc genomic), including all 8 exons, 7 introns, and the 1.2-kb promoter (γcPro), in reverse genomic orientation relative to the vector backbone. The CL20i4-EF1α-hγcOPT vector contains codon-optimized human γc cDNA (hγc), the expression of which is driven by a 233-bp human elongation factor α-promoter (EF1α). Ψ indicates viral packaging signal; cpPT, the central polypurine tract; RRE, lentiviral Rev Response Element; MSCV-hγc vector, γ-retrovirus vector expressing human γc cDNA and an IRES-GFP cassette; and IRES, internal ribosome entry site. (B) Epstein-Barr virus–transformed human B lymphoblast from an SCID-X1 patient (EBV-BSCID-X1 cells) and human epithelial cervical cancer cell line HeLa were transduced with either the Revgen vector or the EF1α vector. Two weeks later, cell-surface expressions of γc were measured by flow cytometry after staining with anti-CD132 monoclonal antibody. EBV-BWT cells indicates Epstein-Barr virus–transformed human B lymphoblasts from a healthy donor; MFI, mean fluorescence intensity of the gated subpopulation; and VCN, average vector copy number in the entire population.
Schematic representation of the 2 novel lentiviral vectors for SCID-X1 gene therapy and their activity in various cell types. (A) Both vectors are based on third-generation lentiviral vector backbone. The enhancer/promoter in the long-terminal repeat region was deleted (ΔU3) and replaced with a 400-bp chromatin insulator element (Ins) from the chicken β-globin locus. The CL20i4-hγc-Revgen vector contains the entire human IL2RG gene (γc genomic), including all 8 exons, 7 introns, and the 1.2-kb promoter (γcPro), in reverse genomic orientation relative to the vector backbone. The CL20i4-EF1α-hγcOPT vector contains codon-optimized human γc cDNA (hγc), the expression of which is driven by a 233-bp human elongation factor α-promoter (EF1α). Ψ indicates viral packaging signal; cpPT, the central polypurine tract; RRE, lentiviral Rev Response Element; MSCV-hγc vector, γ-retrovirus vector expressing human γc cDNA and an IRES-GFP cassette; and IRES, internal ribosome entry site. (B) Epstein-Barr virus–transformed human B lymphoblast from an SCID-X1 patient (EBV-BSCID-X1 cells) and human epithelial cervical cancer cell line HeLa were transduced with either the Revgen vector or the EF1α vector. Two weeks later, cell-surface expressions of γc were measured by flow cytometry after staining with anti-CD132 monoclonal antibody. EBV-BWT cells indicates Epstein-Barr virus–transformed human B lymphoblasts from a healthy donor; MFI, mean fluorescence intensity of the gated subpopulation; and VCN, average vector copy number in the entire population.
The other vector we tested contains an internal 233-bp EF1α core promoter element to drive expression of human codon optimized γc cDNA (CL20i4-EF1α-hγcOPT or EF1α vector; Figure 1A).22 Our initial attempts at using the EF1α promoter to express a wild-type human γc cDNA did not lead to significant levels of B-lymphocyte reconstitution in the SCID-X1 mouse transplantation model, despite definitive evidence of vector transduction in secondary colony-forming unit–spleen (data not shown). A 400-bp chicken β-globin chromatin insulator element, which has been shown to have enhancer-blocking activity,30 was incorporated into the U3 region of the 3′ LTR of both the Revgen and the EF1α vectors and is copied into the 5′ LTR during reverse transcription to provide 2 copies flanking the transcription cassette (Figure 1A).
The Revgen vector and the EF1α vector were transiently produced in 293T cells with titers averaging approximately 7 × 106/mL and 1 × 107/mL, respectively, when measured on NIH3T3 cells by Southern blot. To assay for stability of the insulator fragment within the LTR, PCR analysis using primers flanking the 5′ and 3′ LTRs and genomic DNA from human CD34+ hematopoietic cells transduced with the EF1α vector showed the expected-sized LTR fragments (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), demonstrating that the 400-bp insulator element is relatively stable. Transduction of EBV−B cells from an SCID-X1 patient with either the Revgen vector or the EF1α vector led to similar and readily detectable levels of cell surface γc expression, whereas only the EF1α vector transduction in HeLa cells led to detectable level of γc (Figure 1B), consistent with the relative lymphoid specificity of the Revgen vector design and a broader expression spectrum of the EF1α promoter.
Both Revgen and EF1α vectors restored T-, B-, and NK-lymphocyte development in a murine SCID-X1 transplantation model
We carried out 2 separate transplantation experiments using SCID-X1 mice: one with the Revgen vector and the other with the EF1α vector. Each experiment included a control group that received mock-transduced cells (Mock) and, as a positive control, a murine stem cell virus (MSCV)–based γ-retroviral vector that expressed both γc and an Internal Ribosome entry site (IRES)–linked GFP cassette under control of the LTR promoter/enhancer (MSCV-hγc,Figure 1A).40 Transduced bone marrow cells from γc−/− mice were transplanted into sublethally irradiated γc−/−Rag2−/− recipient mice. The EF1α vectors were used at concentrations of 8.3 × 106 tu/mL (MOI = 3.3) and 2.3 to 4.6 × 107 tu/mL (MOI = 18-27). The Revgen transductions were done at a vector concentration of 1 × 107 tu/mL (MOI = 10).
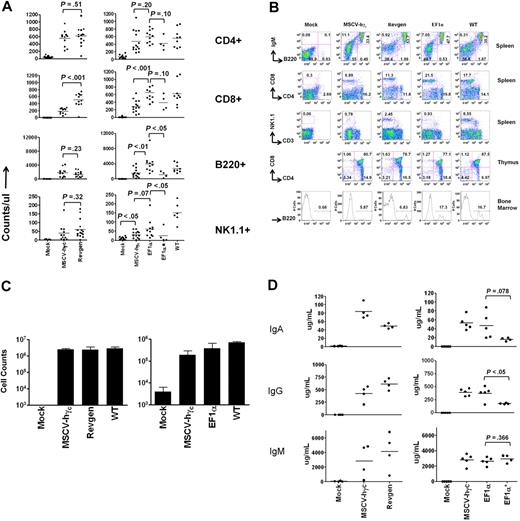
Both lentiviral vectors led to a significant number of B220+ B lymphocytes and CD4+ or CD8+ T lymphocytes as early as 5 to 6 weeks (data not shown). Lymphoid reconstitution data at 18 weeks after transplantation were shown in Figure 2A. Significant numbers of T and B cells were detected in recipient mice both with the Revgen vector (with 1 exception) and at both MOIs with the EF1α vector (P < .05 compared with mock), although lack of full reconstitution was seen in some cases at the low MOI. The level of T- and B-cell reconstitution was as good as or slightly better than that seen with the MSCV-hγc control vector. NK-cell development was partially restored with both lentiviral vectors (P < .05 compared with mock) but to levels less than that seen in wild-type mice. The numbers of NK cells obtained with either lentiviral vector appear to be higher than that obtained with the MSCV γ-retroviral control vector but did not reach statistical significance (P > .05; Figure 2A). Marginal NK reconstitution was seen in the small number of mice in the low MOI EF1α group. Three mice in each group were killed at 21 weeks, and spleen, thymus, and bone marrow cells were analyzed for immunophenotypes using flow cytometry. Both lentiviral vectors restored B-cell populations in the spleen and bone marrow. The spleen and thymus showed normal populations of T cells as defined by CD4 and 8 staining (Figure 2B and data not shown). When the remaining mice were killed at 7 to 8 months (EF1α group) and 11 to 12 months (Revgen group), significant levels of CD4+, CD8+, and B220+ cells persisted (Table 1).
Ex vivo gene therapy of an SCID-X1 mouse model with either the Revgen vector or the EF1α vector potently restores lymphocyte development. (A) Counts of peripheral blood lymphocyte subpopulations (CD4+ or CD8+ T lymphocytes and B220+ B lymphocytes) and natural killer (NK1.1+) cells at 18 weeks after transplantation. The Revgen vector and the EF1α vector were evaluated in 2 separate transplantation experiments with their own negative mock and positive MSCV-hγc vector controls (experiment 1 on the left and experiment 2 on the right). Each dot represents data from a single mouse. EF1α* indicates 4 of 10 mice transplanted with cells transduced with the EF1α vector at a lower MOI of 3.3 showed significant reconstitution; and WT, data from nontransplanted wild-type C57BL/6J mice to serve as reference. (B) Relative proportions (percentage) of lymphoid subpopulations in spleen, bone marrow, and thymus of transplanted mice analyzed at 21 weeks. (C) Proliferation of peripheral blood T lymphocytes taken from recipient mice at 14 weeks in response to concanavalin A and interleukin-2 stimulation. The number of cells was counted on day 6 for the left panel (n = 3 for WT, n = 4 for the other groups) and on day 5 for the right panel (n = 3 for WT, n = 5 for other groups). (D) Plasma immunoglobulin levels of recipient mice at 14 weeks after transplantation. Each dot represents data for a single mouse. The 3 panels on the left and the 3 panels on the right represent data from transplantation experiments 1 and 2, respectively. P values were obtained by t test.
Ex vivo gene therapy of an SCID-X1 mouse model with either the Revgen vector or the EF1α vector potently restores lymphocyte development. (A) Counts of peripheral blood lymphocyte subpopulations (CD4+ or CD8+ T lymphocytes and B220+ B lymphocytes) and natural killer (NK1.1+) cells at 18 weeks after transplantation. The Revgen vector and the EF1α vector were evaluated in 2 separate transplantation experiments with their own negative mock and positive MSCV-hγc vector controls (experiment 1 on the left and experiment 2 on the right). Each dot represents data from a single mouse. EF1α* indicates 4 of 10 mice transplanted with cells transduced with the EF1α vector at a lower MOI of 3.3 showed significant reconstitution; and WT, data from nontransplanted wild-type C57BL/6J mice to serve as reference. (B) Relative proportions (percentage) of lymphoid subpopulations in spleen, bone marrow, and thymus of transplanted mice analyzed at 21 weeks. (C) Proliferation of peripheral blood T lymphocytes taken from recipient mice at 14 weeks in response to concanavalin A and interleukin-2 stimulation. The number of cells was counted on day 6 for the left panel (n = 3 for WT, n = 4 for the other groups) and on day 5 for the right panel (n = 3 for WT, n = 5 for other groups). (D) Plasma immunoglobulin levels of recipient mice at 14 weeks after transplantation. Each dot represents data for a single mouse. The 3 panels on the left and the 3 panels on the right represent data from transplantation experiments 1 and 2, respectively. P values were obtained by t test.
Long-term splenic lymphoid reconstitution in transplanted mice
| . | CD4+, % . | CD8+, % . | B220+IgM+, % . | NK1.1+, % . |
|---|---|---|---|---|
| Transplantation experiment 1 (11-12 mo) | ||||
| Mock (n = 7) | 8.1 ± 5.3 | 0.5 ± 0.8 | 0.2 ± 0.2 | 0.1 ± 0.2 |
| MSCV-hγc (n = 4) | 12.8 ± 10.5 | 4.2 ± 2.9* | 20.6 ± 13.8* | 1.6 ± 2.8 |
| Revgen (n = 10) | 12.7 ± 5.8 | 10.2 ± 4.9* | 16.1 ± 9.2* | 1.1 ± 1.8 |
| Transplantation experiment 2 (7-8 mo) | ||||
| Mock (n = 8) | 8.4 ± 3.9 | 1.2 ± 1.2 | 0.1 ± 0.1 | 0.1 ± 0.1 |
| MSCV-hγc (n = 11) | 20.6 ± 6.2* | 13.8 ± 5.4* | 29.3 ± 12.9* | 0.6 ± 0.7* |
| EF1α (n = 8) | 16.9 ± 5.5* | 16.8 ± 7.7* | 44.4 ± 11.3* | 1.0 ± 1.1* |
| EF1α* (n = 4) | 18.1 ± 1.7* | 16.5 ± 6.3* | 30.7 ± 13.9* | 0.2 ± 0.2 |
| . | CD4+, % . | CD8+, % . | B220+IgM+, % . | NK1.1+, % . |
|---|---|---|---|---|
| Transplantation experiment 1 (11-12 mo) | ||||
| Mock (n = 7) | 8.1 ± 5.3 | 0.5 ± 0.8 | 0.2 ± 0.2 | 0.1 ± 0.2 |
| MSCV-hγc (n = 4) | 12.8 ± 10.5 | 4.2 ± 2.9* | 20.6 ± 13.8* | 1.6 ± 2.8 |
| Revgen (n = 10) | 12.7 ± 5.8 | 10.2 ± 4.9* | 16.1 ± 9.2* | 1.1 ± 1.8 |
| Transplantation experiment 2 (7-8 mo) | ||||
| Mock (n = 8) | 8.4 ± 3.9 | 1.2 ± 1.2 | 0.1 ± 0.1 | 0.1 ± 0.1 |
| MSCV-hγc (n = 11) | 20.6 ± 6.2* | 13.8 ± 5.4* | 29.3 ± 12.9* | 0.6 ± 0.7* |
| EF1α (n = 8) | 16.9 ± 5.5* | 16.8 ± 7.7* | 44.4 ± 11.3* | 1.0 ± 1.1* |
| EF1α* (n = 4) | 18.1 ± 1.7* | 16.5 ± 6.3* | 30.7 ± 13.9* | 0.2 ± 0.2 |
P < .05 versus mock (t test).
We also tested whether the vector-encoded γc transgene expression was functional by measuring T-cell proliferation responses. Mononuclear cells were collected either from peripheral blood at 14 weeks or from spleen at 7 months after transplantation and assayed in vitro for proliferative responses to concanavalin A and IL-2 stimulation (Figure 2C), or anti-CD3/CD28 stimulation (supplemental Figure 2). These studies showed restoration of proliferative responses in T cells from both lentiviral vectors and from the MSCV retrovirus control vector. The reconstituted B lymphocytes were also functional as evidenced by detection of normal levels of immunoglobulins A, G, and M in the peripheral blood (Figure 2D). In contrast, mice in the mock group showed undetectable levels of circulating immunoglobulin.
Vector copy number was analyzed by quantitative PCR in peripheral blood mononuclear cells collected at 14 weeks after transplantation (Figure 3). The average vector copy number per cell was 3 vector genomes/cell or less in most mice. The exception was 4 lentiviral-treated mice with a copy number of approximately 4 to 5 vector genomes per cell (Figure 3). The mice in the low MOI EF1α group averaged only a single copy per cell. Taken together with the reconstitution data, these results suggest that the lentiviral vectors are relatively potent for restoring T- and B-cell development in this model and, in at least some cases, can reconstitute T and B lymphopoiesis with 1 or 2 vector genomes on average. NK-cell development may require higher copy numbers, or higher number of vector transduced hematopoietic stem cells, at least with EF1α vector.
Average vector copy number in peripheral blood mononuclear cells at 14 weeks after transplantation. Genomic DNA from peripheral blood mononuclear cells of transplanted mice were extracted and used for average vector copy number (VCN) measurement using quantitative PCR assay. Each dot represents data for a single mouse. The left panel and the right panel represent data from transplantation experiments 1 and 2, respectively.
Average vector copy number in peripheral blood mononuclear cells at 14 weeks after transplantation. Genomic DNA from peripheral blood mononuclear cells of transplanted mice were extracted and used for average vector copy number (VCN) measurement using quantitative PCR assay. Each dot represents data for a single mouse. The left panel and the right panel represent data from transplantation experiments 1 and 2, respectively.
Restoration of T-cell development in primary human CD34+ cells from SCID-X1 patients
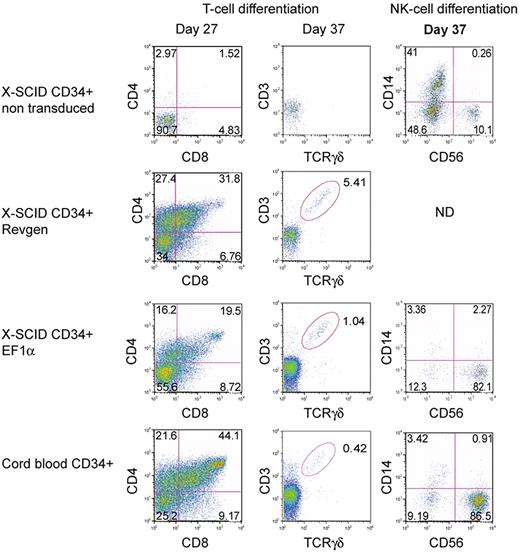
To determine whether these vectors could correct the phenotype in human SCID-X1 hematopoietic cells, we used an in vitro T-cell development assay based on OP9/DL-1 stromal cells that supports T-cell development and differentiation from human CD34+ hematopoietic cells.41 Both vectors were used to transduce CD34+ cells from an SCID-X1 patient at an MOI of 100 before culture on OP9/DL-1 cells. Human CD34+ cells from a healthy donor were used as positive control, whereas nontransduced SCID-X1 CD34+ cells were used as negative control. Ten days after transduction, the vector copy number in bulk cultured cells as assessed by real-time PCR was 1.5 and 3.1 copies per cell for the Revgen and EF1α vector, respectively. At 27 days of culture in the presence of IL7, SCF, and Flt3L, significant levels of double-positive CD4+/CD8+ T cells were detected in the Revgen-transduced and the EF1α-transduced groups as well as in the normal CD34+ control (Figure 4). T-cell maturation occurred in both vector-transduced populations as demonstrated by the presence of a rearranged γδ T-cell receptor in CD3+ cells. In contrast, there were essentially no CD4+/CD8+ T cells detected in the mock-transduced SCID-X1 cultures, nor were there any significant levels of CD3+/TCRγδ+ cells in the mock group (Figure 4). This result was repeated with cells from a second patient (data not shown). These results demonstrate that both the Revgen and the EF1α vector were effective in human SCID-X1 hematopoietic cells.
Restoration of T- and NK-cell development in human SCID-X1 cells by the Revgen and the EF1α vectors. For T-cell differentiation, CD34+ cells from an SCID-X1 patient were transduced with either the Revgen or the EF1α vector and cultured on an OP9-DL1 stromal cell layer. Double-positive (DP, CD4+CD8+) as well as CD3+TCRγσ+ cells were observed on day 27 and day 37, respectively, using flow cytometric analysis. For NK-cell differentiation, CD34+ cells from a separate SCID-X1 patient were transduced with the EF1α vector and then cultured in Myelocult. Cells were taken on day 37, and expression of NK-cell marker (CD56) was analyzed by flow cytometry. ND indicates not done.
Restoration of T- and NK-cell development in human SCID-X1 cells by the Revgen and the EF1α vectors. For T-cell differentiation, CD34+ cells from an SCID-X1 patient were transduced with either the Revgen or the EF1α vector and cultured on an OP9-DL1 stromal cell layer. Double-positive (DP, CD4+CD8+) as well as CD3+TCRγσ+ cells were observed on day 27 and day 37, respectively, using flow cytometric analysis. For NK-cell differentiation, CD34+ cells from a separate SCID-X1 patient were transduced with the EF1α vector and then cultured in Myelocult. Cells were taken on day 37, and expression of NK-cell marker (CD56) was analyzed by flow cytometry. ND indicates not done.
The EF1α vector was also used to transduce CD34+ cells from a different SCID-X1 donor, who had a Val-130-Gly γc mutation that was confirmed in his brother and found in their mother in a heterozygous state. This γc mutation was associated with a low number of CD3+ T cells and NK cells and is presumed to be hypomorphic on this basis. These cells were transduced and then incubated in an NK-cell differentiation culture that contained IL-15 and rhSCF.34 After 37 days of culture, 82% of the cells in the culture displayed the CD56+/CD14− NK-cell phenotype, similar to that seen with the wild-type CD34+ cells (87% CD56+/CD14−). In contrast, only 10% of the cells from the mock-transduced group expressed this NK-cell phenotype (Figure 4).
Only the EF1α vector does not activate LMO2 expression when inserted into intron 1 of LMO2 in Jurkat T cells
We directly compared the potential safety of the Revgen and the EF1α vector using an LMO2 activation assay in human Jurkat T cells. In this assay, Cre-Lox recombination is used to insert vector constructs in an antisense orientation into the exact location within intron 1 of LMO2 that was seen in patient 4 of the French trial.37 Unmodified Jurkat T cells show no detectable expression of LMO2 protein by Western blot analysis; but when a murine retrovirus (MFG) vector was inserted in an antisense orientation into this position, large amounts of LMO2 protein expression were detected, recapitulating what was seen in the patient's tumor cells.37
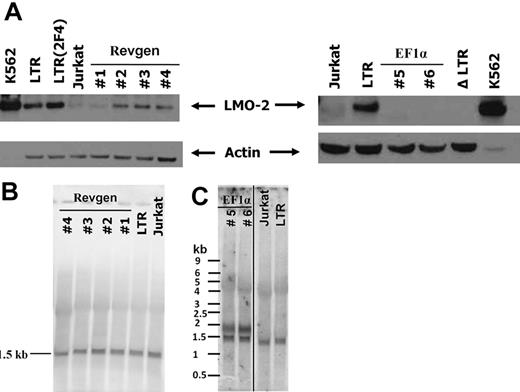
Using this assay, we inserted the Revgen and EF1α proviral genomes into the LMO2 locus in an antisense vector orientation. The orientation of the γc expression cassette in the Revgen vector is in the same orientation as the LMO2 gene, whereas the γc gene in the EF1α vector is in the opposite orientation. We were able to successfully obtain 2 targeted clones for the EF1α vector and 4 targeted clones for the Revgen vector as demonstrated by Southern blot analysis (supplemental Figure 3). LMO2 protein expression was clearly detectable in all of the 4 Revgen clones, although at levels less than that seen with the MLV LTR (Figure 5A). Quantitative PCR for LMO2 transcript was also consistent with protein data and directly showed that transcriptional activation of LMO2 occurred as the result of Revgen insertion (supplemental Figure 4). Northern blot analysis of RNA from these clones using the full-length γc cDNA as probe showed a single 1.5-kb band, which corresponds to both the vector-derived and the endogenous γc allele transcripts (Figure 5B). Lack of any other aberrantly sized transcripts suggests that transcriptional read-through or aberrant splicing of the Revgen transcripts had not occurred.
The EF1α vector lacks LMO2 activation activity, whereas the Revgen vector activates LMO2 expression. (A) The provirus form of the Revgen and the EF1α vector was targeted to the first intron of the LMO2 gene locus in Jurkat T lymphocytes by Cre-mediated homologous recombination. The expression of LMO2 in the targeted Jurkat cell clones was analyzed by immunoblot. Clones 1 to 4 are 4 Jurkat clones targeted with the Revgen vector and clones 5 and 6 are 2 clones targeted with the EF1α vector. Human erythroid leukemic cell line K562 cells constitutively express high level of LMO2 and is used as positive control. Jurkat cell itself does not express detectable level of LMO2. LTR indicates Jurkat clone with a γ-retrovirus LTR-GFP cassette targeted into the first intron of the LMO2 gene in Jurkat cells; LTR(2F4), a subclone of the LTR clone that was culture for equivalent time as the Revgen and EF1α clones, showing the stability of LMO2 activation by the LTR enhancer; and ΔLTR, a Jurkat clone with the LTR-GFP cassette deleted by Cre-mediated recombination, showing that LMO2 expression is turned off. (B) Northern blot analysis of RNA preparations from the 4 Jurkat clones with targeted Revgen vector insertion. The full-length γc cDNA was used as probe. A single band of 1472 bp demonstrates that the γc transcript derived from the Revgen vector is indistinguishable from that of the endogenous γc allele of Jurkat cells. (C) Northern blot analysis of RNA preparations from the 2 Jurkat clones with targeted EF1α vector insertion. The full-length codon-optimized γc cDNA was used as probe. This probe cross-reacts with the endogenous γc RNA, which is expressed in Jurkat cells and is shown as a 1472-bp band. The RNA species transcribed from the EF1α vector is shown as another predicted main band of approximately 1.9 kb. A vertical line has been inserted between the EF1α clones and the Jurkat lane to indicate repositioned lanes of Jurkat and LTR from the same gel.
The EF1α vector lacks LMO2 activation activity, whereas the Revgen vector activates LMO2 expression. (A) The provirus form of the Revgen and the EF1α vector was targeted to the first intron of the LMO2 gene locus in Jurkat T lymphocytes by Cre-mediated homologous recombination. The expression of LMO2 in the targeted Jurkat cell clones was analyzed by immunoblot. Clones 1 to 4 are 4 Jurkat clones targeted with the Revgen vector and clones 5 and 6 are 2 clones targeted with the EF1α vector. Human erythroid leukemic cell line K562 cells constitutively express high level of LMO2 and is used as positive control. Jurkat cell itself does not express detectable level of LMO2. LTR indicates Jurkat clone with a γ-retrovirus LTR-GFP cassette targeted into the first intron of the LMO2 gene in Jurkat cells; LTR(2F4), a subclone of the LTR clone that was culture for equivalent time as the Revgen and EF1α clones, showing the stability of LMO2 activation by the LTR enhancer; and ΔLTR, a Jurkat clone with the LTR-GFP cassette deleted by Cre-mediated recombination, showing that LMO2 expression is turned off. (B) Northern blot analysis of RNA preparations from the 4 Jurkat clones with targeted Revgen vector insertion. The full-length γc cDNA was used as probe. A single band of 1472 bp demonstrates that the γc transcript derived from the Revgen vector is indistinguishable from that of the endogenous γc allele of Jurkat cells. (C) Northern blot analysis of RNA preparations from the 2 Jurkat clones with targeted EF1α vector insertion. The full-length codon-optimized γc cDNA was used as probe. This probe cross-reacts with the endogenous γc RNA, which is expressed in Jurkat cells and is shown as a 1472-bp band. The RNA species transcribed from the EF1α vector is shown as another predicted main band of approximately 1.9 kb. A vertical line has been inserted between the EF1α clones and the Jurkat lane to indicate repositioned lanes of Jurkat and LTR from the same gel.
In contrast, the 2 EF1α clones showed no detectable LMO2 protein expression (Figure 5A). This was not the result of silencing of transcription from the EF1α vector as evidenced by significant levels of vector-driven γc transcripts in both clones on Northern blot analysis (Figure 5C). The size of the γc transcript from the EF1α vector was approximately 1.9 kb, whereas the lower band in the Northern blot represents the γc transcript from the endogenous allele of γc gene, which is approximately 1.5 kb. The lack of any other significant transcripts demonstrates lack of transcription driven by the SIN LTR and lack of significant transcriptional read-through.
The activation of LMO2 by the Revgen vector insertion could be the result of either an enhancer-mediated mechanism or transcriptional read-through, or both. To distinguish between these possibilities, we designed a PCR assay to detect read-through transcripts. The forward primer is localized in the first exon of the γc gene, and the reverse primer is localized in the exon 4 of the LMO2 gene. If transcriptional read-through had occurred, the first exon of the γc and the coding exons of the LMO2 would be the minimum components of the transcript, and the resultant RT-PCR product would be between 330 bp and 1873 bp, depending on how the splicing would occur. mRNA from the 2 of the highest LMO2 expressing clones (#3 and #4), as well as the control LTR clone were extracted and used for RT-PCR. In neither of the 2 Revgen clones was PCR product detected (supplemental Figure 5), demonstrating that transcriptional read-through was not likely responsible for the LMO2 activation.
The EF1α and Revgen vector lacks myeloid cell immortalization activity
We further tested the EF1α vector for safety using a myeloid cell immortalization assay in which primary bone marrow cells from mice are transduced, cultured for 2 weeks, and then replated at limiting dilution and scored for clonal proliferation.35 This assay assesses the overall proliferative consequences of vector integration and primarily reads vector integration within and activation of the Mds1-Evi1 proto-oncogene.35
In 2 independent experiments, immortalized clones were seen most frequently in the SFFV vector-transduced group, but also at lower numbers in the MSCV group (Table 2). In contrast, no immortalized clones were seen in any of the EF1α or Revgen vector groups (Table 2). Altogether, these results demonstrate that both lentiviral vectors lacked myeloid cell immortalization activity and demonstrate the lack of transactivation potential in a second independent assay system.
Lack of myeloid cell immortalization activity by the EF1α vector
| . | Positive wells on 96-well plate . | Copy number . | |
|---|---|---|---|
| 100 cells/well . | 500 cells/well . | ||
| Experiment 1 | N = 1 | n = 1 | |
| Mock | 0 | 0 | — |
| EF1α | 0 | 0 | 1.57 |
| Revgen | 0 | 0 | 0.31 |
| SFFV-GFP | 6 | 16 | 2.32 |
| MSCV-hγc | 0 | 5 | 1.46 |
| Experiment 2 | N = 3 | n = 3 | |
| Mock | 0, 0, 0 | 0, 0, 0 | — |
| EF1α | 0, 0, 0 | 0, 0, 0 | 1.6 |
| Revgen | 0, 0, 0 | 0, 0, 0 | 1.12 |
| SFFV-GFP | 3, 3, 6 | 14, 21, 13 | 1.45 |
| . | Positive wells on 96-well plate . | Copy number . | |
|---|---|---|---|
| 100 cells/well . | 500 cells/well . | ||
| Experiment 1 | N = 1 | n = 1 | |
| Mock | 0 | 0 | — |
| EF1α | 0 | 0 | 1.57 |
| Revgen | 0 | 0 | 0.31 |
| SFFV-GFP | 6 | 16 | 2.32 |
| MSCV-hγc | 0 | 5 | 1.46 |
| Experiment 2 | N = 3 | n = 3 | |
| Mock | 0, 0, 0 | 0, 0, 0 | — |
| EF1α | 0, 0, 0 | 0, 0, 0 | 1.6 |
| Revgen | 0, 0, 0 | 0, 0, 0 | 1.12 |
| SFFV-GFP | 3, 3, 6 | 14, 21, 13 | 1.45 |
— indicates not applicable.
Long-term follow-up of transplanted mice
The transplanted primary recipient mice in the Revgen vector group and in the EF1α group were followed for 5 to 12 months and 5 to 8 months, respectively (supplemental Tables 1A, 2A). One mouse in the γ-retroviral control group (MSCV-hγc-IRES-GFP) developed a vector+ (GFP+) tumor. In the only other leukemia case, 1 mouse reconstituted with Revgen vector-transduced cells developed CD4+CD8+ T-cell leukemia 18 weeks after transplantation. The mouse had enlarged spleen, an abnormal and high proportion of CD4+/CD8+ T cells in the spleen, bone marrow, and the peripheral blood. Molecular analysis of the Revgen vector leukemia cells identified a single vector insertion in the intron 10 of Nars2 gene, in the sense orientation of the vector backbone, but antisense orientation relative to the direction of the γc gene. This insertion is 120 kb downstream of the transcriptional start site of the Nars2 gene but only 27 kb upstream of the start site of the adjacent Gab2 gene, which is an adaptor protein involved in hematopoietic cell signaling.42 Quantitative PCR measurement of the Nars2 and Gab2 transcripts in sorted CD4+CD8+ leukemic cells showed no up-regulation of these 2 genes relative to their level seen in CD4+CD8+ thymocytes from a nontransplanted healthy C57BL6 mouse (data not shown), indicating that development of this tumor may be vector integration independent, as has been seen in other studies.43 Three mice transplanted with the Revgen vector were killed at 5 months and found to have no abnormalities at autopsy. Bone marrow cells from these mice were transplanted into 9 secondary recipients, which were then followed for another 7 to 8 months without seeing any hematologic malignancies (supplemental Table 1A-B).
None of the mice in the primary EF1α group has developed any hematologic malignancies. Fifteen of the primary recipient mice with the EF1α vector were killed at various time points up to 8 months after transplantation and had no evidence of abnormalities at autopsy. Bone marrow cells from each of these EF1α mice were used to transplant 2 or 3 secondary recipients, and these secondary transplantation experiments are still in progress (supplemental Table 2A-B).
Discussion
Although the pioneering clinical trials for SCID-X1 gene therapy provided clear proof of efficacy for gene therapy, the 5 unexpected cases of leukemia have demonstrated that insertional mutagenesis is a serious adverse event that must be addressed. The detailed molecular data collected from these tumors illustrate that LMO2 activation was the driving event in most of these cases.17,18 Furthermore, it is also clear that the enhancer elements in the viral LTR were responsible for proto-oncogene activation in most, if not all, of these cases. These observations have prompted us and others to develop vectors that use nonviral transcriptional elements to express the vector-encoded γc gene and that lack the ability to transactivate cellular proto-oncogenes, particularly LMO2. An important question remains as to what are the most appropriate expression and safety elements in these newly designed SCID-X 1 vectors.
In this study, we evaluated 2 very different types of self-inactivating lentiviral vectors for SCID-X1 gene therapy. Our data show that both the Revgen and the EF1α vectors were effective in restoring the T, B differentiation in SCID-X1 mouse models, and in T-cell differentiation assays using human SCID-X1 CD34+ cells. Both vectors partially restored NK-cell numbers in the mouse transplantation model, and the EF1α vector was shown to restore NK-cell differentiation in human SCID-X1 CD34+ cells. Based on our preclinical efficacy data, both vectors appear satisfactory in their ability to correct the SCID-X1 phenotype.
However, in our safety experiments, only the EF1α vector lacked the capacity for LMO2 activation in Jurkat T cells. Further evidence for the relative safety of this vector comes from our data showing no myeloid cell immortalization activity. The Revgen vector led to easily detectable levels of LMO2 transactivation in Jurkat T cells, but it was noted that the transactivation potential of Revgen appeared quantitatively less than that seen with the MFG γ-retroviral LTR. We conclude that the EF1α vector may be less probable to cause transformation than the Revgen vector.
Transcript analysis of the Jurkat/Revgen clones with a sensitive RT-PCR showed no transcriptional read-through from the Revgen vector into the endogenous LMO2 allele. Even if any transcriptional read-through had occurred, the ATG start codon of the γc transgene, which is localized at 5 bp in the first vector exon, would be expected to preclude translation from the downstream start codon of LMO2. Instead, our favored but unproven explanation for Revgen-induced LMO2 activation would be the presence of a lymphoid-specific enhancer within the genomic γc fragment of Revgen. Informatic analysis of the γc genomic fragment shows highly conserved sequences in introns 3 and 7 with an increased density of putative transcription factor-binding sites (data not shown), consistent with potential enhancer elements within these regions. If an enhancer within the γc genomic allele indeed exists, it would suggest that the 400-bp cHS4 insulator fragment present in the SIN LTRs may be insufficient for blocking interactions with the LMO2 promoter(s). The cHS4 insulator is relatively tissue specific, but there are new synthetic insulator fragments that may be more effective in lymphoid versus myeloid cells.36 We are currently testing whether such elements can be used to eliminate LMO2 expression and to increase the potential safety of the Revgen vector.
Our EF1α vector shares the same promoter tested in a self-inactivating γ-retroviral vector for SCID-X1.44 This vector was also shown to effectively restore lymphocyte development in an SCID-X1 model and lack myeloid cell immortalization activity.23,44 Because the vector used in the SIN γ-retrovirus study did not have an insulator fragment present, it seems most probable that the lack of transactivation seen with our EF1α lentiviral vector is best explained by a lack of enhancer activity in the EF1α promoter rather than an insulator effect provided by the 400-bp cHS4 element. Although it is unclear whether insulator fragments are needed when using enhancer-free cellular promoters, such as EF1α, the 400-bp element has been shown to protect against transgene variegation and chromatin effects. Therefore, it is possible that inclusion of this insulator sequence in our vector may enhance expression of the vector-encoded γc gene and increase the probability of expression at different integration sites. The high levels of transcription of the γc cDNA from the EF1α vector in the otherwise silenced LMO2 locus of Jurkat cells is consistent with this possibility.
Mouse models have been used to assess safety, but they are limited by several disadvantages. SCID-X1 mouse models lack sensitivity for transformation as demonstrated by the fact that several independent groups reported the correction of the SCID-X1 phenotype with γ-retroviral vectors, but no tumors were reported.45-47 In our positive control MSCV-hγc vector group, which is very similar to the MFG-γc vector that was used in the previous SCID-X1 clinical trials, only 1 case of leukemia occurred in 21 evaluable primary recipients, further supporting the lack of sensitivity and relevance of mouse model for vector safety evaluation. Tumor-prone mice are much more sensitive to vector-induced transformation and have been successfully used to define the relative risk associated with different vector types.20,40,48 However, the disadvantage of these models is a high background rate of tumor formation resulting from the loss of tumor-suppressor gene function, making it difficult to determine whether any given transformation event is definitely associated with a vector-insertion event. Another problem relating to the specificity of tumor formation is that irradiation of recipient mice can itself cause tumors, in part because of mobilization of endogenous mouse retroviruses.49 Therefore, we think that, although mouse models can be useful, they have a number of problematic features that need to be addressed for vector-mediated oncogenesis studies to be informative.43
In conclusion, we have defined an SIN lentiviral vector based on the EF1α promoter that is effective in correcting the SCID-X1 phenotype in both mouse and human cells. This vector lacks the ability to transactivate LMO2 resulting from enhancer-mediated effects and appears to be relatively safer than LTR-containing γ-retroviral vectors in other assays as well. Although these preclinical studies demonstrate the relative safety of this vector, the definitive test of safety and efficacy will require testing in appropriately designed clinical trials.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Satoru Kumaki and Adrian Thrasher for providing the ED7R cells, Christopher Baum for providing the SFFV-GFP.pre vector and the EF1α core promoter element, Mary Ellen Conley for providing EBV transformed B cells, and Laura Janke and Jerold Rehg for pathologic review of tissue sections.
This work was supported by the National Heart, Lung, and Blood Institute (grant P01 HL 53749), Cancer Center (support grant P30 CA 21765), the Assisi Foundation of Memphis, and the American Lebanese Syrian Associated Charities.
National Institutes of Health
Authorship
Contribution: S.Z., S.S.D., J.T.G., M.C.-C., H.L.M., and B.P.S. designed the study; S.Z., D.M., S.S.D., J.H., T.L., Z.M., S.H.-B.A., J.T.G., and M.R.G. performed the study; and S.Z. and B.P.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sheng Zhou, Division of Experimental Hematology, Department of Hematology, St Jude Children's Research Hospital, 262 Danny Thomas Pl, Memphis, TN 38105; e-mail: sheng.zhou@stjude.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal