Abstract
The divalent metal ion transporter DMT1 is critical for nonheme iron import. We have previously shown that DMT1 is regulated in vitro by ubiquitination that is facilitated by the adaptor proteins Ndfip1 and Ndfip2. Here we report that in Ndfip1−/− mice fed a low- iron diet, DMT1 expression and activity in duodenal enterocytes are significant higher than in the wild-type animals. This correlates with an increase in serum iron levels and transferrin saturation. Liver and spleen iron stores were also increased in Ndfip1−/− mice fed a normal diet. Counterintuitive to the increase in iron uptake, Ndfip1−/− mice fed a low iron diet develop severe microcytic, hypochromic anemia. We demonstrate that this is due to a combination of iron deficiency and inflammatory disease in Ndfip1−/− mice, because Ndfip1−/−/Rag1−/− immunodeficient mice fed a low iron diet did not develop anemia and showed an iron overload phenotype. These data demonstrate that Ndfip1 is a critical mediator of DMT1 regulation in vivo, particularly under iron restricted conditions.
Introduction
Iron is essential for many biological processes. Iron homeostasis needs to be tightly controlled, because iron deficiency results in anemia, while iron overload leads to tissue damage and fibrosis.1 Iron uptake and metabolism are complex and highly regulated, involving a number of enzymes and transport proteins.2,3 The divalent metal ion transporter DMT1 plays a major role in both iron uptake into the body and iron release within cells.4 DMT1 is expressed at the apical membrane of duodenal enterocytes where it transports non-heme iron into the cell5 and in the endocytic compartment where it releases iron, internalized via the transferrin system, from the endosomes to the cytoplasm.6 Little is known about the posttranslational regulation of DMT1.
Ubiquitination plays an important role in the trafficking and degradation of proteins.7 The Nedd4 family of HECT ubiquitin ligases (E3s) is known to ubiquitinate a number of channels and transporters via a direct interaction between the WW domains of the E3s and PPxY (PY) motifs of the substrate. However, many Nedd4 family targets do not contain PY motifs, suggesting a need for adaptor or accessory proteins.8 We identified 2 proteins, Ndfip1 and Ndfip2,9-11 which regulate DMT1 in vitro via ubiquitination by the Nedd4 family members WWP2 and Nedd4-2.12,13 Both Ndfips bind to DMT1 and act as adapters for WWP2 and Nedd4-2, mediating the ubiquitination and subsequent degradation of DMT1.12,13
In this paper, we show that DMT1 regulation is impaired in Ndfip1−/− mice, especially when challenged with low iron availability. Ndfip1−/− mice fed a low-iron diet show increased duodenal DMT1 levels and DMT1 activity in isolated Ndfip1−/− enterocytes, leading to higher serum iron levels and greater transferrin saturation. The hepatic and splenic iron stores were significantly increased in Ndfip1−/− mice fed a normal iron diet. Together, this suggests an increase in iron uptake in Ndfip1−/− mice. Due to their known inflammatory phenotype,14 these mice develop anemia. By removing the inflammation and subsequent anemia using Ndfip1−/−/Rag1−/− mice, we further uncovered increased iron uptake in these mice, demonstrating a critical role for Ndfip1 in the regulation of DMT1 in vivo.
Methods
Animals
Ndfip1−/− mice have been described previously.14 Ndfip1−/−/Rag1−/− mice were generated by crossing Ndfip1+/− mice (backcrossed for 3 generations) with mouse strain B6.129S7-Rag1tm1Mom/J (Rag1−/− mice, Jackson Laboratory) to generate Ndfip1+/−/Rag1+/− mice. These mice were then backcrossed once with Rag1−/− mice to generate Ndfip1+/−/Rag1−/− mice, which were then intercrossed to generate Ndfip1−/−/Rag1−/− mice. On average, the genetic background of the Ndfip1−/− and the Ndfip1−/−/Rag1−/− mice used in this study would have approximately 95% and 97% contribution from C57Bl/6J, respectively. Animals were fed ad libitum on a standard (164 mg/kg iron), high iron (20 g/kg), or low iron (15 mg/kg; Specialty Feeds) rodent diet for 3 weeks. All studies were performed on 6-week-old mice. All animal studies were approved by the institutional animal ethics committee at the Center for Cancer Biology.
Blood and bone marrow analyses
Blood was collected via cardiac puncture and analyzed for total serum iron and transferrin saturation using the Ferene method (Thermo Electron). Serum transferrin (Alpha Diagnostics), tumor necrosis factor-α (TNF-α), and interleukin-6 (IL-6; R&D Systems) were measured by enzyme-linked immunosorbent assay. Serum ferritin and complete blood count (including blood smears) were performed by the Department of Clinical Pathology, SA Pathology. Parameters measured in the complete blood count included hemoglobin (Hb), hematocrit (Hct), red blood cell count (RBC), white blood cell count (WBC), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), MCH concentration, and red cell distribution width.
Bone marrow was aspirated from femurs using a 23-gauge needle and 5-mL syringe. Bone marrow smears were stained with Perl Prussian Blue staining with 2% carbol fuchsin as the counterstain and May-Grünwald, Giemsa stain by the Department of Clinical Pathology, SA Pathology. Differential count was performed on 500 nucleated cells. Light images were captured at room temperature with an Olympus BX51 microscope, Olympus DP70 camera, and Olysia Bioreport software. The objectives used were 10× UPlanApo (NA 0.4), 20× UPlanApo (NA 0.7), and 40× UPlanApo (NA 0.85). Images were then compiled using Adobe Illustrator CS5 and Adobe Photoshop CS5 software. Statistical significance was determined using either 2-tailed unpaired t tests or 1-way analyses of variance with Tukey post-hoc tests (GraphPad Prism Version 4.03).
Cell isolation and fluorescence quenching assay
To isolate enterocytes, the proximal duodenum was removed and flushed through with solution A (1.5mM KCl, 96mM NaCl, 27mM sodium citrate, 8mM KH2PO4, 5.6mM Na2HPO4), followed by washing in solution A for 30 minutes at room temperature. The duodenum was minced in an enzyme cocktail (333 U/mL collagenase, 2.5 U/mL elastase, 10 μg/mL DNAse) in Krebs Ringer solution (120mM NaCl, 24mM NaHCO3, 5mM HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], 4.8mM KCl, 1.2mM MgSO4, 1.2mM KH2PO4, 20mM glucose, 1mM CaCl2) and incubated while shaking at 37°C for 30 minutes. Cells were filtered through a 40-μm filter and washed twice in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 1% nonessential amino acids, 1% β-mercaptoethanol, and 1% L-asparagine. Enterocytes were used immediately in a fluorescence quenching assay to measure DMT1 activity,12 and statistical significance determined using a 2-tailed unpaired t test (GraphPad Prism Version 4.03).
Immunohistochemistry and immunoblotting
Tissues were fixed in Histochoice (ProSciTech), cryopreserved in 30% sucrose, and embedded in Tissue-Tek Optimal Cutting Temperature compound. Ten-micrometer frozen sections were mounted on polysine slides, fixed with cold acetone for 10 minutes, then air dried and rehydrated with phosphate-buffered saline (PBS). Sections were then blocked with 5% skim milk in PBS for 2 hours and stained with either rabbit anti-DMT1 (specific for exon 1A)15 or rabbit anti-ferroportin (SLC40A1, Lifespan Biosciences) overnight at 4°C (1:200 in 5% skim milk in PBS), followed by rabbit AlexaFluor-488 (1:200; Molecular Probes) for 2 hours, and mounted in ProLong Gold antifade (Invitrogen). Confocal images were captured as previously described.12
For immunoblotting, tissues were snap frozen and protein extracted using lysis buffer (50mM Tris-HCl, pH 7.5, 150mM NaCl, 10% glycerol, 1% Triton X-100, 10mM EDTA [ethylenediaminetetraacetic acid]) containing a complete protease inhibitor mixture (Roche Applied Science). One hundred micrograms of protein were loaded per lane and immunoblotting performed as previously described.12 Blots were probed using rabbit anti-ferroportin or rabbit anti-ferritin light chain (Abcam), using goat anti–rabbit Cy5 (GE Healthcare Life Sciences) as the secondary antibody. Ferroportin was detected mostly as an apparent dimer of approximately 120 kDa.
Real-time qPCR
Total RNA was extracted using Trizol reagent (Invitrogen) and used for cDNA synthesis using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). cDNA was analyzed by real-time quantitative polymerase chain reaction (qPCR) in a Rotor-Gene 6000 system (QIAGEN) using RT2 Real-TimeTM SYBR Green PCR master mix (SABiosciences, QIAGEN) with primers shown in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Data were analyzed using Rotor-Gene 6000 Series Software 1.7 and normalized to β-actin.
Tissue iron measurement using inductively coupled plasma mass spectroscopy
Small pieces of tissue (liver or spleen) were dried down for 24 hours at 85°C after which they were weighed and then digested in 25% nitric acid in a glass tube at 65°C for 16 hours. The samples were then diluted in 1% nitric acid and analyzed by inductively coupled plasma mass spectroscopy by SA Water. Values were then normalized to dry weight.
Results
Previously, we have reported that Ndfip1 regulates DMT1 by acting as an adaptor between DMT1 and the ubiquitin ligase WWP2, thereby mediating its ubiquitination and subsequent degradation.12 Ndfip1−/− mice fed a standard rodent diet had increased DMT1 levels and activity in the liver, with a concomitant increase in liver iron levels.12 To further dissect the physiological role of Ndfip1 in iron homeostasis in vivo, we subjected Ndfip1−/− mice to a low-, normal, or high-iron diet. Immunostaining of the duodenum showed an increase in DMT1 levels in Ndfip1−/− mice fed a low- iron diet compared with Ndfip1+/+ mice (Figure 1A-B). DMT1 activity in enterocytes isolated from mice fed a low-iron diet was also dramatically increased in Ndfip1−/− mice compared with Ndfip1+/+ mice (Figure 1C). As a result, serum iron levels and transferrin saturation were significantly increased in Ndfip1−/− compared with Ndfip1+/+ mice (Figure 2A-B). Total iron binding capacity, serum transferrin, and serum ferritin were not significantly different (supplemental Table 2).
Ndfip1 is important for regulating DMT1 under iron-deficient conditions in vivo. DMT1 expression in the duodenum of (A) Ndfip1+/+ mice and (B) Ndfip1−/− mice fed a low iron diet for 3 weeks. DMT1 levels are increased in Ndfip1−/− mice compared with Ndfip1+/+ mice. Scale bar, 50 μm. (C) DMT1 relative transport activity in enterocytes isolated from Ndfip1+/+ and Ndfip1−/− mice fed a low iron diet. DMT1 activity is increased in Ndfip1−/− mice compared with Ndfip1+/+ mice. Data represented as mean ± SD,*P < .05, n = 3-7.
Ndfip1 is important for regulating DMT1 under iron-deficient conditions in vivo. DMT1 expression in the duodenum of (A) Ndfip1+/+ mice and (B) Ndfip1−/− mice fed a low iron diet for 3 weeks. DMT1 levels are increased in Ndfip1−/− mice compared with Ndfip1+/+ mice. Scale bar, 50 μm. (C) DMT1 relative transport activity in enterocytes isolated from Ndfip1+/+ and Ndfip1−/− mice fed a low iron diet. DMT1 activity is increased in Ndfip1−/− mice compared with Ndfip1+/+ mice. Data represented as mean ± SD,*P < .05, n = 3-7.
Ndfip1−/− mice show increased iron uptake under low iron conditions and increased iron storage under normal iron conditions. (A) Serum iron levels and (B) transferrin saturation (Tf sat) in Ndfip1+/+ and Ndfip1−/− mice fed a low- or normal iron diet. Both serum iron and transferrin saturation are increased in Ndfip1−/− mice on the low-iron diet compared with Ndfip1+/+ mice. Iron stores in (C) liver and (D) spleen. Data represent mean ± SD, *P < .05, n = 3-7. Bone marrow iron levels in (E) Ndfip1+/+ mice and (F) Ndfip1−/− mice on the low-iron diet and in (G) Ndfip1+/+ mice and (H) Ndfip1−/− mice on the normal diet. Scale bar, 50 μm. Iron stores are significantly increased in Ndfip1−/− mice on the normal diet compared with Ndfip1+/+ mice.
Ndfip1−/− mice show increased iron uptake under low iron conditions and increased iron storage under normal iron conditions. (A) Serum iron levels and (B) transferrin saturation (Tf sat) in Ndfip1+/+ and Ndfip1−/− mice fed a low- or normal iron diet. Both serum iron and transferrin saturation are increased in Ndfip1−/− mice on the low-iron diet compared with Ndfip1+/+ mice. Iron stores in (C) liver and (D) spleen. Data represent mean ± SD, *P < .05, n = 3-7. Bone marrow iron levels in (E) Ndfip1+/+ mice and (F) Ndfip1−/− mice on the low-iron diet and in (G) Ndfip1+/+ mice and (H) Ndfip1−/− mice on the normal diet. Scale bar, 50 μm. Iron stores are significantly increased in Ndfip1−/− mice on the normal diet compared with Ndfip1+/+ mice.
Because serum iron levels can be highly variable in mice, we measured tissue iron stores as a more accurate indication of iron balance. In mice fed a normal diet, hepatic and splenic iron stores were significantly higher in Ndfip1−/− mice compared with Ndfip1+/+ mice (Figure 2C-D). Stainable iron could also be found in the bone marrow (Figure 2G-H). The increase in liver and splenic iron in Ndfip1−/− mice on the normal iron diet with no significant increase in serum iron and transferrin saturation (Figure 2) may be attributable to a maximum transferrin saturation maintained by hepcidin and other homeostatic regulators. As expected, the hepatic, splenic, and bone marrow iron stores were lower in Ndfip1−/− mice and Ndfip1+/+ mice fed low-iron diet compared with mice fed with normal iron diet. There was no significant difference in hepatic and bone marrow iron stores in Ndfip1−/− mice and Ndfip1+/+ mice fed low iron diet (Figure 2C-F). However, spleen iron stores were reduced in Ndfip1−/− mice compared with Ndfip1+/+ mice. This may be due to increased iron demand in Ndfip1−/− mice due to severe anemia and also the difference in splenic architecture between the 2 genotypes.14
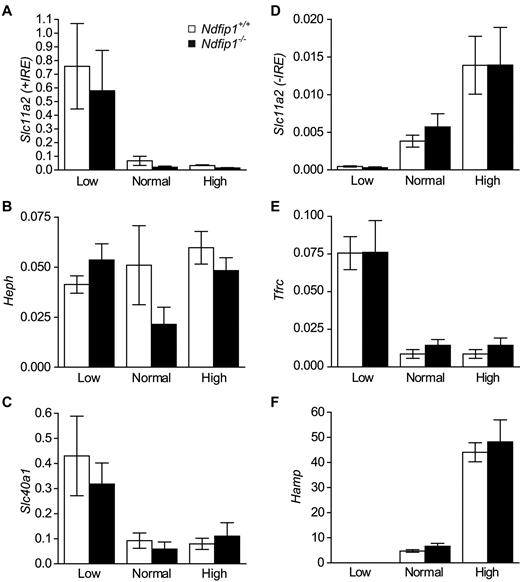
The body maintains normal iron balance by fine tuning the expression of a number of genes in response to changes in dietary iron levels. We therefore examined the expression of genes involved in iron uptake in the duodenum and iron storage in the liver of mice fed a low-, normal, or high-iron diet using qPCR. At the transcript level, both Ndfip1−/− and Ndfip1+/+ mice respond as expected16,17 to varying levels of iron in the diet (Figure 3). Given that there is no difference between the genotypes, these data support a role for Ndfip1 in the posttranscriptional regulation of DMT1.
mRNA levels of genes involved in regulating iron homeostasis in Ndfip1+/+ and Ndfip1−/− mice fed either a low-, normal, or high-iron diet. (A) Slc11a2 (+IRE isoform), (B) Heph, and (C) Slc40a1 in the duodenum. These genes are involved in iron uptake from the diet. (D) Slc11a2 (-IRE isoform), (E) Tfrc1, and (F) Hamp in the liver. These genes are involved in iron storage. Slc11a2, DMT1; Heph, hephaestin; Slc40a1, ferroportin 1; Tfrc, transferrin receptor 1; Hamp, Hepcidin. All genes respond to the different diets as expected. Data represent mean ± SE, *P < .05, n = 7-9.
mRNA levels of genes involved in regulating iron homeostasis in Ndfip1+/+ and Ndfip1−/− mice fed either a low-, normal, or high-iron diet. (A) Slc11a2 (+IRE isoform), (B) Heph, and (C) Slc40a1 in the duodenum. These genes are involved in iron uptake from the diet. (D) Slc11a2 (-IRE isoform), (E) Tfrc1, and (F) Hamp in the liver. These genes are involved in iron storage. Slc11a2, DMT1; Heph, hephaestin; Slc40a1, ferroportin 1; Tfrc, transferrin receptor 1; Hamp, Hepcidin. All genes respond to the different diets as expected. Data represent mean ± SE, *P < .05, n = 7-9.
To investigate further the effects of loss of Ndfip1, a complete blood count was performed. Surprisingly, Ndfip1−/− mice fed a low iron diet developed severe microcytic, hypochromic anemia as indicated by significantly reduced Hb and Hct (Figure 4A-B). This was also clearly evident in the blood smears (Figure 4D-E). The MCV, MCH, and MCH concentration were significantly lower in mice fed the low-iron diet compared with mice fed a normal iron diet. Although there was no significant difference in MCV and MCH between the genotypes, the RBC count and Hb were significantly lower in Ndfip1−/− mice compared with Ndfip1+/+ mice fed the low-iron diet (Figure 4A,C). This may suggest suppressed effective erythropoiesis in Ndfip1−/− mice fed the low-iron diet (Figure 4A-C and supplemental Table 3). This is further supported by examination of the bone marrow, which demonstrated reduced erythropoiesis in Ndfip1−/− mice compared with Ndfip1+/+ mice fed low-iron, as indicated by a reduction in the percentage of erythroid series cells and an increase in the myeloid to erythroid ratio (Figure 4F-G). Although lymphoid and myeloid series cells were increased in Ndfip1−/− mice compared with Ndfip1+/+ mice fed low iron, the difference was not statistically significant (Figure 4F).
Ndfip1−/− mice fed a low-iron diet develop hypochromic, microcytic anemia. (A) Hemoglobin (Hb), (B) hematocrit (Hct), and (C) RBC count are decreased in Ndfip1−/− mice compared with Ndfip1+/+ mice. Blood smears from (D) Ndfip1+/+ mice and (E) Ndfip1−/− mice show RBCs from Ndfip1−/− mice are microcytic and hypochromic compared with their wild-type counterparts. Scale bar, 50 μm. (F) Erythroid, lymphoid, and myeloid series as a percentage of total cells in the bone marrow smears. (G) The myeloid to erythroid ratio in bone marrow. The percentage of erythroid series cells is decreased in Ndfip1−/− mice, and the myeloid to erythroid ratio is increased, indicating impaired erythropoiesis. Data represent mean ± SD, *P < .05, n = 4-7.
Ndfip1−/− mice fed a low-iron diet develop hypochromic, microcytic anemia. (A) Hemoglobin (Hb), (B) hematocrit (Hct), and (C) RBC count are decreased in Ndfip1−/− mice compared with Ndfip1+/+ mice. Blood smears from (D) Ndfip1+/+ mice and (E) Ndfip1−/− mice show RBCs from Ndfip1−/− mice are microcytic and hypochromic compared with their wild-type counterparts. Scale bar, 50 μm. (F) Erythroid, lymphoid, and myeloid series as a percentage of total cells in the bone marrow smears. (G) The myeloid to erythroid ratio in bone marrow. The percentage of erythroid series cells is decreased in Ndfip1−/− mice, and the myeloid to erythroid ratio is increased, indicating impaired erythropoiesis. Data represent mean ± SD, *P < .05, n = 4-7.
Ndfip1−/− mice develop inflammatory disease due to hyperactivated T cells.14 Ndfip1−/− mice on the low-iron diet showed significant increases in IL-6 and TNF-α compared with Ndfip1+/+ mice (with no significant differences seen in mice on the normal diet; Figure 5A-B). WBC count was increased in Ndfip1−/− mice compared with Ndfip1+/+ mice on both the low-iron and normal diets; Figure 5C), indicative of anemia of chronic disease.18 This did not seem to be attributable to any single cell type, although neutrophils (myeloid series) and lymphocytes (lymphoid series) made up the majority of the cells (supplemental Table 3 and Figure 4). To further investigate the contribution of each disease process (iron deficiency and inflammation) toward the severe anemia of Ndfip1−/− mice fed the low-iron diet, we measured ferritin and ferroportin levels in the liver and ferroportin levels in duodenum (supplemental Figure 1). As expected, the hepatic ferritin expression was higher in all mice fed the normal iron diet compared with mice fed the low-iron diet; however, there was no difference between Ndfip1−/− mice and Ndfip1+/+ mice fed with the same diet. The hepatic ferroportin expression was increased in mice fed the low-iron diet (supplemental Figure 1A) compared with mice fed the normal iron diet (supplemental Figure 1B), but there was no difference between genotypes on the same diet. The duodenal ferroportin expression was not different between Ndfip1−/− mice and Ndfip1+/+ mice fed with low-iron diet (supplemental Figure 1C-D). Although Ndfip1−/− mice fed the low-iron diet have higher transferrin saturation and serum iron compared with Ndfip1+/+ mice fed the low-iron diet, both groups of animals were still iron deficient. Iron deficiency and anemia down-regulate hepcidin (Hamp) expression (Figure 3F), which in turn increases ferroportin stability (Hamp being a negative regulator of ferroportin).19 Thus, the lack of difference in Hamp expression in Ndfip1+/+ and Ndfip1−/− mice fed the low-iron diet would explain why ferroportin expression (both RNA and protein) was not significantly different between the 2 genotypes.
The anemia of Ndfip1−/− mice under iron-deficient conditions is partly contributable to inflammation. (A) IL-6 and (B) TNF-α are increased in Ndfip1−/− mice compared with Ndfip1+/+ mice fed a low-iron diet but are not significantly different on the normal diet. (C) WBC count is significantly increased in Ndfip1−/− mice compared with Ndfip1+/+ mice fed both the low-iron and normal diet. These factors are indicative of anemia of inflammation. nd, not detected. Data represent mean ± SD, *P < .05, n = 3-7.
The anemia of Ndfip1−/− mice under iron-deficient conditions is partly contributable to inflammation. (A) IL-6 and (B) TNF-α are increased in Ndfip1−/− mice compared with Ndfip1+/+ mice fed a low-iron diet but are not significantly different on the normal diet. (C) WBC count is significantly increased in Ndfip1−/− mice compared with Ndfip1+/+ mice fed both the low-iron and normal diet. These factors are indicative of anemia of inflammation. nd, not detected. Data represent mean ± SD, *P < .05, n = 3-7.
The inflammatory markers (IL-6, TNF-α, and WBC) were significantly higher in Ndfip1−/− mice fed the low-iron diet compared with Ndfip1−/− mice fed the normal iron diet (Figure 5). Because Ndfip1−/− mice eventually develop inflammatory disease regardless of diet, these data suggest that iron deficiency may accentuate the inflammatory phenotype. Normally, iron deficiency down-regulates while inflammation up-regulates Hamp expression.19 We observed no significant difference in Hamp levels in the livers of Ndfip1−/− and Ndfip1+/+ mice fed the low-iron diet (Figure 3F), and, consistent with previous work,20 this suggests that iron deficiency may be a more powerful regulator of Hamp compared with inflammation.
To overcome the confounding effects of inflammation, we crossed Ndfip1 knockout mice into a Rag1−/− immunodeficient background. Interestingly, compared with Ndfip1−/−/Rag1+/+ and Ndfip1+/+/Rag1−/− mice, Ndfip1−/−/Rag1−/− mice fed a low-iron diet showed significantly higher Hb, Hct, and RBC (Figure 6A-C). These parameters in Ndfip1−/−/Rag1−/− mice were comparable with Ndfip1+/+/Rag1+/+ animals (Figure 6A-C). These results suggest that the anemia in Ndfip1−/− mice on the low-iron diet is, at least in part, due to inflammation. As predicted, Ndfip1−/−/Rag1−/− mice exhibited increased DMT1 levels in the duodenum compared with single knockout mice (Figure 6F-H), as well as elevated serum iron levels (although not significant, Figure 6D) and a significant increase in transferrin saturation (Figure 6E).
Removing inflammation reveals further evidence for the importance of Ndfip1 in the regulation of DMT1 in vivo. (A) Hb, (B) Hct, and (C) RBC count are significantly increased in Ndfip1−/−/Rag1−/− mice compared with single knockout Ndfip1+/+/Rag1−/− mice and Ndfip1−/−/Rag1+/+ and are restored to Ndfip1+/+/Rag1+/+ levels. (D) Serum iron and (E) transferrin saturation in (Ndfip1+/+/Rag1+/+) as for other genotypes Ndfip1−/−/Rag1+/+, Ndfip1+/+/Rag1−/−, and Ndfip1−/−/Rag1−/− mice. Both serum iron and transferrin saturation are elevated in the double knockout mice. DMT1 expression in the duodenum of (F) Ndfip1+/+/Rag1−/− mice, (G) Ndfip1−/−/Rag1+/+ mice, and (H) Ndfip1−/−/Rag1−/− mice. DMT1 levels are increased in mice lacking Ndfip1 (Ndfip1−/−/Rag1+/+ and Ndfip1−/−/Rag1−/− mice) compared with Ndfip1+/+/Rag1−/− controls. All mice were fed a low-iron diet for 3 weeks, data represent mean ± SD, *P < .05, n = 3-4.
Removing inflammation reveals further evidence for the importance of Ndfip1 in the regulation of DMT1 in vivo. (A) Hb, (B) Hct, and (C) RBC count are significantly increased in Ndfip1−/−/Rag1−/− mice compared with single knockout Ndfip1+/+/Rag1−/− mice and Ndfip1−/−/Rag1+/+ and are restored to Ndfip1+/+/Rag1+/+ levels. (D) Serum iron and (E) transferrin saturation in (Ndfip1+/+/Rag1+/+) as for other genotypes Ndfip1−/−/Rag1+/+, Ndfip1+/+/Rag1−/−, and Ndfip1−/−/Rag1−/− mice. Both serum iron and transferrin saturation are elevated in the double knockout mice. DMT1 expression in the duodenum of (F) Ndfip1+/+/Rag1−/− mice, (G) Ndfip1−/−/Rag1+/+ mice, and (H) Ndfip1−/−/Rag1−/− mice. DMT1 levels are increased in mice lacking Ndfip1 (Ndfip1−/−/Rag1+/+ and Ndfip1−/−/Rag1−/− mice) compared with Ndfip1+/+/Rag1−/− controls. All mice were fed a low-iron diet for 3 weeks, data represent mean ± SD, *P < .05, n = 3-4.
Discussion
In previous studies, we had demonstrated that Ndfip1 and Ndfip2 proteins act as adaptors to regulate the ubiquitination of DMT1 by Nedd4 family members, such as WWP2 and Nedd4-2.12,13 Those observations provided a novel mechanism of the regulation of iron transport via ubiquitination and possible degradation of DMT1. In the current study, we have extended previous observations to show that Ndfip1 is a key regulator of DMT1 expression in vivo. As such Ndfip1−/− mice fed a normal diet have increased iron stores in the liver, spleen, and bone marrow and higher serum iron concentration. These observations are consistent with iron overload due to increased DMT1 levels and activity, as would be predicted in Ndfip1−/− mice.
The control of iron transport and homeostasis are highly dependent on dietary iron.2,3 As reported in this study, Ndfip1 plays a critical role in iron homeostasis, especially under low-iron dietary conditions. We found that Ndfip1−/− mice fed a low-iron diet had significantly more DMT1 expression and transport activity in duodenal enterocytes than in the Ndfip1+/+ mice under the same conditions. More importantly and contrary to our expectations, Ndfip1−/− mice fed a low-iron diet developed more severe microcytic, hypochromic anemia than their wild-type Ndfip1+/+ counterparts. Although Ndfip1−/− mice had higher serum iron and transferrin saturation than Ndfip1+/+ mice, when fed a low-iron diet, the iron stores were significantly lower than in the mice of either genotype fed the normal iron diet. This suggests that Ndfip1−/− mice fed a low-iron diet are still iron deficient, and increased DMT1 activity in these mice is unable to compensate completely for reduced iron supplement. An important contributing factor for the severe anemia is inflammatory diseases in Ndfip1−/− mice.14 Although the iron feeding experiments were carried out before the onset of severe inflammatory disease, Ndfip1−/− mice fed low-iron diet showed more inflammation compared with Ndfip1−/− mice fed normal iron diet. These data suggest that iron deficiency may accentuate the inflammatory phenotype in Ndfip1−/− mice. The removal of inflammation by crossing Ndfip1−/− mice into a Rag1−/− background resulted in preventing the onset of anemia, confirming that inflammation is important for the anemic phenotype.
Although the effect of inflammation on iron homeostasis is well established, there is limited literature assessing the effect of iron deficiency on the inflammatory process. In vitro studies demonstrate that iron chelation triggers inflammatory response in human intestinal epithelial cells, alveolar macrophages, and human acute monocytic leukemia cell line, THP1.21,22 Another study showed that iron deficiency enhances inflammation through p38 MAPK–NF-κB–extracellular matrix metalloproteinase inducer and matrix metalloproteinase-9 pathways.23 The data presented here suggest that a combination of iron deficiency and inflammation results in a more severe anemia in Ndfip1−/− mice. This is a unique murine model linking iron deficiency, inflammation, and anemia. The inflammation leads to suppression of erythropoiesis and ineffective use of iron. On the other hand, iron deficiency seems to exacerbate the inflammatory phenotype of Ndfip1−/− mice. The combination of iron deficiency and inflammation is a common clinical problem. Most of the patients suffering with systemic illness such as systemic lupus erythematosus, inflammatory bowel disease, and malignancies frequently develop iron deficiency either due to low iron intake and/or blood loss.
In conclusion, our study provides strong evidence that Ndfip1 is critical for DMT1 regulation and iron homeostasis in vivo. We predict that deregulated levels of Ndfip1 expression or modulation of its function by mutations or polymorphisms may have pathological consequences in humans.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the staff of SA Pathology animal house for help with animal care and maintenance.
This work was supported by a project grant (508085) from the National Health and Medical Research Council of Australia.
Authorship
Contribution: N.J.F., L.E.D., H.E.D., and S.K. designed experiments; N.J.F., Y.A.L., L.E.D., H.E.D., and K.H. performed the experimental work; N.J.F., Y.A.L., L.E.D., H.E.D., D.H., and S.K. analyzed data and wrote the paper; D.H. analyzed all hematological data; L.Z. and M.D.G. generated antibodies and provided technical advice; and B.Y. generated genetically modified mice.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Professor Sharad Kumar, Department of Hematology, Centre for Cancer Biology, SA Pathology, PO Box 14, Rundle Mall, Adelaide, Australia; e-mail: sharad.kumar@health.sa.gov.au.
References
Author notes
N.J.F. and Y.A.L. contributed equally to this work