Abstract
MicroRNAs are small RNA molecules that modulate protein expression by degrading mRNA or repressing translation. They have been shown to play important roles in hematopoiesis, including embryonic stem cell differentiation, erythropoiesis, granulocytopoiesis/monocytopoiesis, lymphopoiesis, and megakaryocytopoiesis. miR-150 and miR-155 play divergent roles in megakaryocytopoiesis, with the former promoting development of megakaryocytes at the expense of erythrocytes and the latter causing a reduction in megakaryocyte colony formation. Platelets also contain fully functional miRNA machinery, and certain miRNA levels in platelets have been found to coordinate with reactivity to specific agonists and to pathologic states. This review will cover the current state of knowledge of miRNAs in megakaryocytes and platelets and the exciting possibilities for future research.
Introduction
The linear string of nucleotides that forms a genome has a simplicity that is incongruous with the sometimes overwhelming complexity of organism anatomy and physiology. Some order is achieved by considering messenger RNA (mRNA), linking the proteins responsible for organism intricacy with specific genomic locations. But this “coding gene centric” viewpoint does not easily explain the large amount of noncoding DNA sequence, representing approximately 70% of the genome.1 Although protein coding genes may substantively contribute to the increased complexity of Caenorhabditis elegans compared with Escherichia coli (a 439% increase in gene number), differences in protein coding genes do not seem to sufficiently explain the greater complexity between Homo sapiens and C elegans (an increase of only 21%). On the other hand, unlike C elegans, humans have 21-fold more transcribed, noncoding sequence than transcribed, coding sequence, whereas C elegans has only 1.25-fold.2 This intriguing association between organism complexity and transcribed, noncoding sequence suggests a role for noncoding RNA, and the last decade has clearly established an important role for noncoding RNA in biology. Noncoding RNAs are classified as large (> 200 bp) or small (< 200 bp). Small regulatory RNAs include microRNAs (miRNAs), endogenous small interfering RNAs, piwi-interacting RNAs, small nucleolar RNAs, and others.3,4 The best studied noncoding RNA are miRNAs. miRNAs were discovered in C elegans in 19935,6 and have been shown to play important roles in developmental biology, cellular stress, circadian rhythm, and immunology, as well as numerous disease states, including Alzheimer disease, cancer, and heart failure. Abundant evidence demonstrates a critical role for miRNAs in normal human hematopoiesis, and dysregulated miRNA biology has been associated with and shown to cause numerous hematologic diseases (Table 1). This review will focus on the role of miRNAs in megakaryocytopoiesis and platelet biology.
miRNA biogenesis and function
miRNAs are 21 to 23 nucleotide regulatory RNAs expressed in multicellular organisms, from viruses to plants to humans.7 There are at least 400 human miRNAs, although some predictions estimate more than 1000.8,9 miRNAs regulate most (> 60%) mammalian protein coding genes primarily by repressing gene expression.10 Some miRNAs are expressed ubiquitously, but many are tissue and/or developmental stage–specific.11 Cell miRNA content is highly variable from 1 copy to 10 000 copies.12
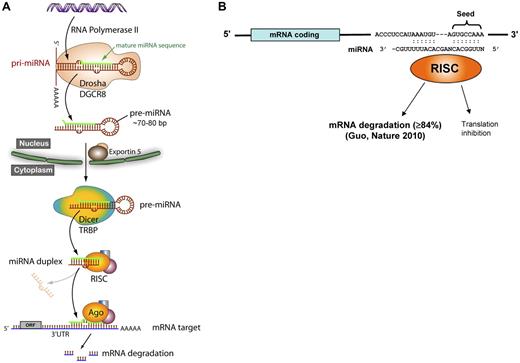
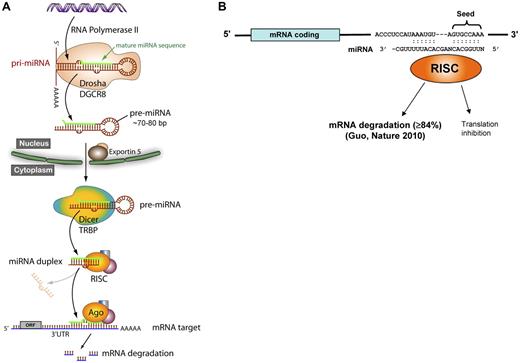
Approximately 30% of miRNA genes are located in intergenic regions, and approximately 70% are located within introns or exons of protein coding genes. Figure 1A illustrates the canonical understanding of intergenic miRNA biogenesis, although exceptions have been described to some of these steps. miRNAs located within introns are transcribed as part of the surrounding coding gene's mRNA, whereas miRNAs located intergenically use an independent transcriptional apparatus. RNA polymerase II is responsible for most miRNA transcription, although RNA polymerase III has been identified transcribing miRNAs located within repetitive elements.13 A several-kilobase primary transcript (pri-miRNA) is then capped and polyadenylated, and forms a hairpin structure.14,15 The pri-miRNA is cleaved into an approximately 60- to 70-bp pre-miRNA by Drosha, an RNase type III endonuclease, which is complexed with DiGeorge syndrome critical region 8 (DGCR8). The pre-miRNA is transported out of the nucleus via exportin 5. In the cytoplasm, the 3′ overhang of the pre-miRNA is recognized by the Dicer-TAR RNA-binding protein (TRBP) complex. Dicer is another RNase type III endonuclease, which generates the miRNA duplex for many miRNAs. The strands separate and the mature miRNA associates with a macromolecular complex called the RNA-induced silencing complex (RISC), which guides the miRNA to its mRNA target. The process of miRNA generation is regulated at both transcriptional and posttranscriptional levels, occasionally composed of positive feed-forward or negative feedback circuitry, in which the miRNA targets a transcriptional activator or repressor of itself. The defining RISC protein is Argonaute 2 (Ago2), which catalyzes mRNA cleavage. Notably, Ago2-dependent, dicer-independent miRNA processing pathways have been identified.16,17
miRNA biogenesis and function. (A) The canonical miRNA biosynthesis pathway. Drosha indicates an RNase type III endonuclease; DGCR8, DiGeorge syndrome critical region 8; Dicer, another RNase type III endonuclease; TRBP, TAR RNA-binding protein; RISC, RNA-induced silencing complex; Ago, Argonaute 2; and ORF, open reading frame. (B) The seed region of the miRNA (bp 2-8) binds to complementary sites on the 3′-UTR of target mRNAs where it predominantly causes a decrease in mRNA levels but may also inhibit translation.
miRNA biogenesis and function. (A) The canonical miRNA biosynthesis pathway. Drosha indicates an RNase type III endonuclease; DGCR8, DiGeorge syndrome critical region 8; Dicer, another RNase type III endonuclease; TRBP, TAR RNA-binding protein; RISC, RNA-induced silencing complex; Ago, Argonaute 2; and ORF, open reading frame. (B) The seed region of the miRNA (bp 2-8) binds to complementary sites on the 3′-UTR of target mRNAs where it predominantly causes a decrease in mRNA levels but may also inhibit translation.
A fundamental aspect of miRNA function relates to mRNA targeting: most miRNAs are predicted to target multiple mRNAs, and most mRNAs have predicted targets for many miRNAs. Different algorithms are publicly available that predict miRNA binding sites, including Miranda, TargetScan, PicTar, and miRBase. The miRNA associates with mRNA by complementary Watson-Crick base pairing (Figure 1B), with sequences usually, but not always,18 in the 3′-untranslated region (UTR) of mRNA. The seed region of nucleotides 2 to 8 at the 5′ end of the miRNA has perfect complementarity, and this sequence defines families of miRNAs. The miRNA sequence 3′ to the seed sequence has variable degrees of complementarity with the mRNA. The traditional view of miRNA-mediated translation repression posits that lower complementarity induces translation inhibition. But recent work by Guo et al indicates that miRNA knock-down of protein expression is primarily via mRNA degradation, with translation inhibition representing only a minor mechanism.19 miRNAs have been aptly referred to as “rheostats” because their regulatory impact is generally to fine-tune but not abolish protein expression.20,21 The significance of proper miRNA synthesis and function is underscored by diseases caused by genetic defects at virtually all steps in miRNA biogenesis and targeting, including miRNA gene deletions/duplications, single nucleotide polymorphisms in the target mRNA 3′-UTR, miRNA “decoys,” and genetic variants in the proteins mediating miRNA biogenesis.22-24 Importantly, small differences (as little as a 20% change) in miRNA levels have been shown to cause autoimmune disease and predispose to malignancy.25
miRNAs and megakaryocytopoiesis
Traditionally, mechanistic studies on hematopoietic cell self-renewal and differentiation have focused primarily on transcription factors.26 Over the past 6 to 7 years, numerous laboratories have established the essential role of Dicer127 and miRNAs in hematopoiesis,28-30 including embryonic stem cell differentiation,18 erythropoiesis,31-36 granulocytopoiesis/monocytopoiesis,37-40 and lymphopoiesis.28,41-44 Since the first report by Garzon et al in 2006,45 nearly 20 studies have addressed various aspects of miRNAs in megakaryocytopoiesis using megakaryocytes generated from in vitro cultured CD34+ hematopoietic stem cells (HSCs) or transformed cell lines with megakaryocytic properties (Table 2). Both unbiased miRNA profiling and candidate miRNA studies have established associations between specific miRNAs and the developmental stage of megakaryocyte progenitors. Functionality has been tested by assessing the effects of candidate miRNAs on in vitro and in vivo proliferation and differentiation after overexpression or knock-down. Mechanistic assessment of these miRNAs has included target protein knock-down and reporter gene assays using constructs with the putative target 3′-UTR. Several of the best-studied miRNAs will be discussed for their role in megakaryocytopoiesis.
miR-155 inhibits megakaryocytopoiesis
Georgantas et al performed both mRNA and miRNA expression profiling on CD34+ HSCs from healthy subjects29 and used a bioinformatic approach to predict candidate miRNAs that target mRNAs encoding transcription factors associated with hematopoietic differentiation. miR-155 was predicted to repress expression of 9 different CD34+ mRNAs that regulate myelopoiesis. miR-155 expression was dramatically reduced when CD34+ HSCs were differentiated along the megakaryocyte lineage.29,46 Forced overexpression of miR-155 inhibited K562 (a chronic myelogenous leukemia cell line) differentiation and reduced CD34+ HSC-derived myeloid and erythroid colony formation in vitro.29 Transplantation of HSCs overexpressing miR-155 into irradiated mice caused reduced numbers of megakaryocytes in recipient bone marrow.40 Although the effect of miR-155 on platelet count has not been studied, these studies provide strong support that miR-155 inhibits megakaryocytopoiesis.
miR-150 enhances megakaryocytopoiesis
By profiling primary cells from human umbilical cord blood, Lu et al discovered miR-150 levels increased as megakaryocyte-erythrocyte progenitors (MEPs) differentiated toward the megakaryocyte lineage, but not the erythroid lineage.47 Overexpression of miR-150 enhanced both in vitro and in vivo megakaryocyte differentiation at the expense of erythroid differentiation, suggesting a critical switching function at the level of the MEP. miR-150 also knocked down expression of MYB via its 3′-UTR, consistent with data showing that low c-Myb levels promote megakaryocytopoiesis.48 These findings, coupled with work from the Kaushansky laboratory showing thrombopoietin up-regulates miR-150,49 underscore a critical role for miR-150 in promoting megakaryocytopoiesis.
miR-146a modulates megakaryocytopoiesis
miR-146a levels have been reported to dramatically change on megakaryocytic differentiation of HSCs, but there is conflicting evidence as to the direction of change.50-53 Opalinska et al52 reported that, in murine and human hematopoietic stem cells induced to differentiate into megakaryocytes, the expression level of miR-146a increased. However, forced expression of miR-146a had no effect on megakaryocyte colony forming units (CFU-MK), marker expression, or platelet activation.52 In contrast, Labbaye et al50 found that miR-146a decreased in human cord blood stem cells induced to differentiate into megakaryocytes and that forced expression caused a reduction in the number of polyploid cells. Conversely, inhibition of miR-146a by an antagomir caused an increase in the number of polyploid cells.50 Two recent reports by Starczynowski et al53 did not clarify the matter. They report that the level of miR-146a is lower in megakaryocyte/erythroid precursors relative to hematopoietic stem cells, similar to Labbaye et al,50 but that forced expression had no effect on platelet number, similar to Opalinska et al52 However, when down-regulated by decoy targets or anti-miRNA locked nucleic acids, CFU-MK and platelet number increase and the ploidy of the megakaryocytes decreases.51 These apparently conflicting findings may stem from species differences or differing experimental culture conditions (because miR-146a levels vary according to lineage). It is also possible that different experimental conditions result in functionally different levels of miR-146a. This is because Ago proteins preferentially associate with transcripts that contain targets for the highly expressed miRNAs, and mRNAs with a greater number of target sites in their 3′-UTR are subject to stricter control.54,55 Therefore, differing levels of miRNA expression can result in different mRNA levels and experimental results. Lastly, because single nucleotide polymorphisms in the 3′-UTR binding site as well as the surrounding sequence8 can significantly alter miRNA effects,23 it may be necessary to consider the precise mRNA target sequences in these different studies. Recent work by our group77 and by Landry et al56 profiling platelets both found miR-146a to be in the top quartile of expressed miRNAs (Figure 2). An interesting possibility is that the effect of miR-146a is not cell autologous. Opalinska et al found than the expression levels of tumor necrosis factor-α, interferon-β, and interleukin-1β were all repressed in macrophages overexpressing miR-146a.52 In addition, Starczynowski et al53 reported that miR-146a directly targets TRAF6, and down-regulating it results in increased TRAF6 and interleukin-6 levels. Interleukin-6 has been reported to have an effect on megakaryocyte development, and it is possible that differences in the reports are because of complications resulting from changes in cytokine signaling pathways.57
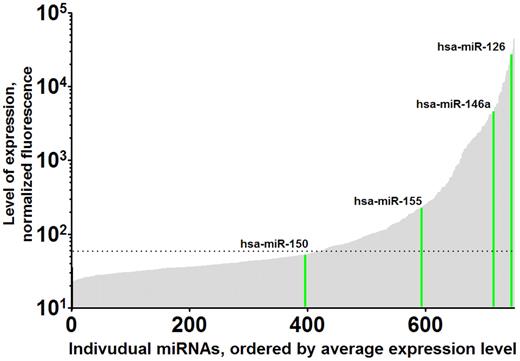
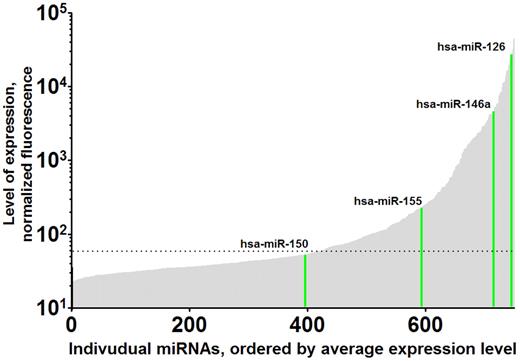
Variation in levels of platelet miRNAs. Levels of individual human platelet miRNAs are shown, arbitrarily ordered from lowest to highest expression (unpublished data). As examples, the expression levels of miRNA-150, miRNA-155, miRNA-126, and miRNA-146a are highlighted in green. A total of 750 human miRNAs were profiled from leukocyte-depleted platelet RNAs from 19 human volunteers using the miRCURY LNA Array, Version 11.0 (Exiqon). Expression levels ranged over 4 orders on magnitude. The horizontal dashed line indicates background level.
Variation in levels of platelet miRNAs. Levels of individual human platelet miRNAs are shown, arbitrarily ordered from lowest to highest expression (unpublished data). As examples, the expression levels of miRNA-150, miRNA-155, miRNA-126, and miRNA-146a are highlighted in green. A total of 750 human miRNAs were profiled from leukocyte-depleted platelet RNAs from 19 human volunteers using the miRCURY LNA Array, Version 11.0 (Exiqon). Expression levels ranged over 4 orders on magnitude. The horizontal dashed line indicates background level.
miR-34a and megakaryocytopoiesis
Increased levels of miR-34a have consistently been observed in K562 cells stimulated to differentiate with phorbol myristate acetate.58,59 When overexpressed in K562 cells, miR-34a inhibited K562 cell proliferation and promoted differentiation. Importantly, miR-34a increased megakaryocyte colony formation from CD34+ HSCs.59 Thus, miR-34a appears to enhance megakaryocytopoiesis.
Other miRNAs and megakaryocytopoiesis
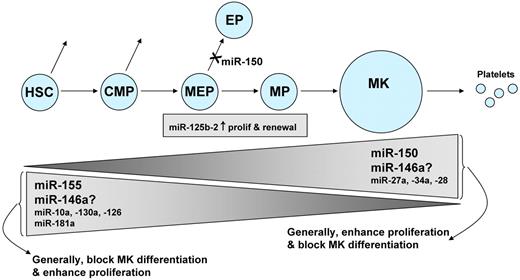
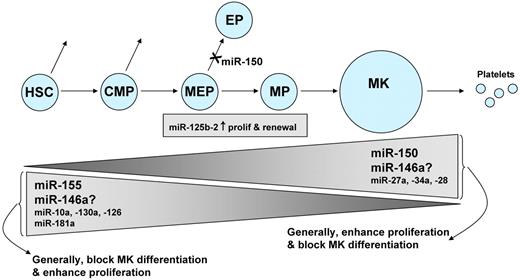
Several other miRNAs may regulate megakaryocytopoiesis, although these miRNAs have been less well studied. Several investigators have observed that more miRNAs were down-regulated than up-regulated during megakaryocytopoiesis, but biphasic patterns of expression have also been observed.31,45,52 Several studies have linked reciprocal associations between miRNA levels and target mRNA levels, a particularly interesting finding when the mRNA encodes a transcription factor known to be involved in hematopoiesis. Examples include miR-130a and MAFB, miR-10a and HOXA1, and miR-27a and Runx1.45,60 Forced expression of the oncomiR miR-125b-2 increased proliferation and self-renewal of MEP and megakaryocyte progenitors,61 and miR-28 appears to exert a negative effect on megakaryocyte differentiation.62 Lastly, both decreasing miR-181 and increasing miR-27a levels have been associated with megakaryocytic differentiation.60,63 In the case of megakaryocytic cell lines, such associations should be viewed cautiously because the correlation between miRNA expression in primary megakaryocytes and cell lines may not always be strong.64 Figure 3 summarizes the studies of miRNAs in megakaryocytopoiesis.
miRNAs and megakaryocytopoiesis. This diagram summarizes the miRNAs known to participate in megakaryocytopoiesis. Megakaryocytes (MK) originate from self-renewing HSC, which differentiate progressively into common myeloid precursors (CMP), MEPs, and megakaryocyte precursors (MP). The height of the triangles indicates the level of the miRNA, with some miRNAs decreasing during megakaryocytopoiesis, and others increasing. There is conflicting evidence regarding the role of miR-146a. Levels of miR-125b-2 do not change during progression from MEP to MP.
miRNAs and megakaryocytopoiesis. This diagram summarizes the miRNAs known to participate in megakaryocytopoiesis. Megakaryocytes (MK) originate from self-renewing HSC, which differentiate progressively into common myeloid precursors (CMP), MEPs, and megakaryocyte precursors (MP). The height of the triangles indicates the level of the miRNA, with some miRNAs decreasing during megakaryocytopoiesis, and others increasing. There is conflicting evidence regarding the role of miR-146a. Levels of miR-125b-2 do not change during progression from MEP to MP.
Platelet miRNAs
Platelets contain miRNAs,31,56,65-68 and we and others have used genome-wide profiling to demonstrate normal human platelets express high levels of miRNA (Figure 2).56 Microarray profile levels are readily validated using a novel stem-loop reverse-transcribed polymerase chain reaction that allows quantification of specific miRNAs.12 Emerging evidence over the past year suggests that platelet miRNAs are biologically and clinically relevant as (1) potential regulators of platelet protein translation and expression, (2) markers of mature megakaryocyte miRNA levels, (3) biomarkers for hematologic disease and platelet reactivity, and (4) a tool for understanding basic mechanisms of megakaryocyte/platelet gene expression.
It is well known that platelets have mRNA and mRNA splicing machinery, and translate mRNA into proteins relevant to hemostasis and inflammation.69-71 Notably, platelets stored in blood banks increase synthesis of integrin-β3.72 Work from the Provost laboratory has demonstrated that human platelets also contain miRNA processing machinery, including Dicer, TAR RNA-binding protein 2, and Ago2, and that platelets are able to process pre-miRNA into mature miRNA.56 Our in silico analysis indicates that each platelet miRNA targets an average of 307 distinct mRNAs (range, 12-1417; unpublished data), consistent with prior predictions.73,77 Thus, platelet miRNAs have ample opportunity to regulate platelet function, although direct evidence has yet to be reported.
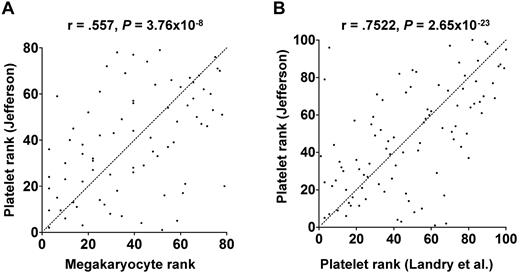
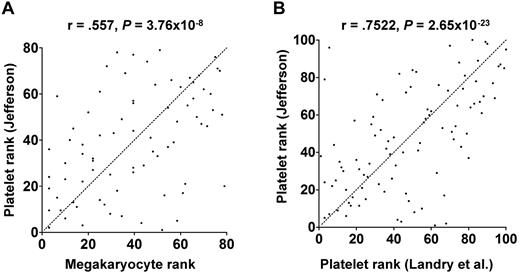
As described in “miRNAs and megakaryocytopoiesis,” functional miRNA levels change during megakaryocyte differentiation of cultured CD34+ HSCs. It is not difficult to imagine similar changes occurring in vivo, but this has not been formally studied. In vitro culture conditions lack numerous in vivo factors that could alter miRNA levels, including other marrow niche cells, innervation, plasma components, spatial constraints, and environmental factors. Hussein et al attempted to directly assess in vivo megakaryocyte miRNA levels using laser microdissection to isolate mature megakaryocytes from patient bone marrow biopsies.74 The extent to which platelet and mature megakaryocyte miRNA profiles correlate is unknown. As an initial attempt to address this issue, we compared platelet miRNA profiles from 19 healthy subjects with the megakaryocyte miRNA profiles from the 2 patients reported by Hussein et al.74 As shown in Figure 4A, there was a significant correlation between the platelet and megakaryocyte miRNAs. We suspect that the true correlation is even stronger because different profiling platforms and RNA preparations were used and only 2 megakaryocyte samples were assessed. Furthermore, the megakaryocyte samples were from patients, whereas the platelets were from healthy donors. As expected, when we compared our dataset with the platelet miRNAs reported by Landry et al,56 we found a greater degree of correlation (Figure 4B).
Correlation between human platelet and megakaryocyte miRNAs. miRNA expression levels were determined in platelets from 19 healthy donors by microarray (our unpublished data) and in patient megakaryocytes by quantitative reverse-transcribed polymerase chain reaction.74 The miRNAs that were queried in both studies were rank ordered and plotted. Statistical significance was calculated by Pearson rank correlation. Dotted line represents 1:1 ratio of rank. (A) Platelet miRNAs correlated with megakaryocyte miRNAs. (B) Platelet miRNAs from the author's laboratory correlated with platelet miRNAs from the Provost laboratory.56
Correlation between human platelet and megakaryocyte miRNAs. miRNA expression levels were determined in platelets from 19 healthy donors by microarray (our unpublished data) and in patient megakaryocytes by quantitative reverse-transcribed polymerase chain reaction.74 The miRNAs that were queried in both studies were rank ordered and plotted. Statistical significance was calculated by Pearson rank correlation. Dotted line represents 1:1 ratio of rank. (A) Platelet miRNAs correlated with megakaryocyte miRNAs. (B) Platelet miRNAs from the author's laboratory correlated with platelet miRNAs from the Provost laboratory.56
miRNAs are very stable and, compared with mRNAs, have superior performance characteristics as biomarkers for disease activity.47,75,76 The first human platelet miRNA profiling study was performed in 2008 by Bruchova et al as part of a study testing for differentially expressed miRNAs in patients with polycythemia vera.65 These investigators found that miR-26b was significantly higher in polycythemia vera platelets than in platelets from healthy donors. Landry et al described how miR-223 could target the mRNA of the adenosine 5′-diphosphate receptor, P2Y12, and that P2Y12 mRNA was found in Ago2 immunoprecipitates in both megakaryocytes and platelets.56 Our group has investigated associations between miRNAs and platelet reactivity, both to better understand megakaryocyte/platelet gene expression and to identify potential biomarkers for thrombotic risk. Because a single nucleotide polymorphism in the VAMP8 3′-UTR has been associated with coronary artery disease and because VAMP8 mRNA is differentially expressed between platelets of differing reactivity,67 we considered whether a miRNA might affect VAMP8 expression. Indeed, miR-96 was shown to knock down VAMP8 mRNA and protein; and in a small number of subjects, miR-96 was differentially expressed between platelets of differing reactivity in a manner that was consistent with its effect on VAMP8 expression.67 Recent data from our laboratory demonstrate the use of differentially expressed mRNA-miRNA pairs for identifying functional miRNAs, as assessed by the ability of the miRNA to target and knock down the mRNA was confirmed in cell culture.77
In conclusion, miRNAs have an established role in hematopoiesis and megakaryocytopoiesis, and platelet miRNAs have potential as tools for understanding basic mechanisms of megakaryocyte/platelet gene expression. Genome-wide association studies have identified orphan loci not in or near protein-coding genes, and at least one such variant proved to be caused by a mutation in a miRNA gene.78,79 It is probable that additional megakaryocyte/platelet disease-producing genetic variants in miRNA biogenesis will be identified. Very exciting opportunities exist for future translational research involving miRNAs in megakaryocytopoiesis and platelet biology. Environmental stresses and aging can affect miRNA levels,80,81 and it will be important to test whether these factors influence miRNA-mediated megakaryocyte/platelet physiology. Gene therapy approaches are using tissue-specific or developmental stage-specific miRNA expression to avoid off-target effects.82 For example, Gentner et al have incorporated miRNA target sequences into an expression vector that results in suppressed ectopic gene expression in HSCs but enhanced expression in mature hematopoietic cells.83 miRNAs can be released via shed microvesicles,84 raising the possibility of platelet-mediated delivery of miRNAs to targeted vascular sites. Platelets have been engineered to ectopically deliver recombinant factor VIII and reduce bleeding85 ; perhaps P-selectin bearing platelet microvesicles could target miRNAs to PSGL-1 expressing leukocytes at sites of inflammation. Other potential directions include manipulating platelet miRNAs to modify platelet life span or the platelet storage defect in banked platelets.
Authorship
Contribution: L.C.E. and P.F.B. analyzed and interpreted data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul F. Bray, Thomas Jefferson University, Cardeza Foundation for Hematologic Research and the Department of Medicine, Jefferson Medical College, 1015 Walnut St, Curtis Bldg, Rm 324, Philadelphia, PA 19107; e-mail: paul.bray@jefferson.edu.
References
National Institutes of Health