Abstract
Adult T-cell leukemia/lymphoma (ATLL) is the neoplasm caused by human T-cell leukemia virus type 1 (HTLV-1). We performed oligo-array comparative genomic hybridization (CGH) against paired samples comprising peripheral blood (PB) and lymph node (LN) samples from 13 patients with acute ATLL. We found that the genome profiles of the PB frequently differed from those of the LN samples. The results showed that 9 of 13 cases investigated had a log2 ratio imbalance among chromosomes, and that chromosome imbalances were more frequent in LN samples. Detailed analysis revealed that the imbalances were likely caused by the presence of multiple subclones in the LN samples. Five of 13 cases showed homozygous loss regions in PB samples, which were not found in the LN samples, indicating that tumors in the PB were derived from LN subclones in most cases. Southern blot analysis of TCRγ showed that these multiple subclones originated from a common clone. We concluded that in many ATLL cases, multiple subclones in the LNs originate from a common clone, and that a selected subclone among the LN subclones appears in the PB.
Introduction
Adult T-cell leukemia/lymphoma (ATLL) is the neoplasm caused by human T-cell leukemia virus type 1 (HTLV-1). The disease is associated with poor prognosis due to drug resistance, the occurrence of opportunistic infections, a large tumor burden with multi-organ failure, and hypercalcemia. Shimoyama et al1 classified ATLL into 4 subtypes: smoldering, chronic, lymphoma, and acute. It is also known that HTLV-1 infection alone does not facilitate the progress of infected CD4+ T cells to fully malignant ATLL cells. Therefore, the search for genes involved in ATLL development and for the specific genes involved in each ATLL type has been actively pursued, albeit with limited success. ATLL-specific chromosomal abnormalities have yet to be found; however, a frequent abnormality found in ATLL is 14q11, which has also been found in other types of T-cell malignancies.2,3 HTLV-1 provirus integration sites have also been extensively sought, and the sites identified were found to be randomly located. Investigations relying on G-band and fluorescence in situ hybridization analyses have not been fruitful in providing a detailed delineation of the genomic aberrations involved.4 The use of high-resolution, array-based comparative genomic hybridization (CGH) for comprehensive chromosome analysis should prove useful in the search for genomic aberrations. We showed previously that acute and lymphoma ATLL types possess distinct genomic profiles, as determined by bacterial artificial chromosome array CGH.5 It should be noted, however, that when lymphoma-type ATLL progresses to manifest more than 2% flower cells in the peripheral blood (PB), it is then classified as the acute type. We set out to analyze the genomic aberrations of acute-type ATLL with paired PB and lymph node (LN) samples in more detail by oligo-array CGH.
An important factor in the diagnosis of ATLL is the identification of monoclonal integration of HTLV-1. It has been reported that the same HTLV-1–infected clone was detected over several years in a chronic-type ATLL patient.6,7 These types of HTLV-1–infected CD4+ T lymphocytes are believed to accumulate various changes during an extensive latency period of over 50 years.8 Alterations in genomic copy number represent one example of the type of accumulated genomic changes that can occur. In the present study, we performed high-resolution oligo-array CGH (Agilent Technologies) using a 44 000-probe set against paired samples obtained from the PB and LNs of 13 patients with acute-type ATLL.
Methods
ATLL patients and cell lines
We conducted a survey of genomic profiles by examination of PB and LN samples taken from 13 patients with acute-type ATLL. Paired samples were collected from each patient within 14 days of diagnosis. The PB and LN samples, together with clinical data, were obtained from 13 patients under a protocol approved by the institutional review board of the Aichi Cancer Center. Informed consent was provided according to the Declaration of Helsinki. Patients were diagnosed from those hospitalized between 1988 and 2010 at Imamura-Bunin Hospital and Nagasaki University School of Medicine. The diagnosis of ATLL was based on clinical features, hematologic characteristics, immunophenotype, and the presence of serum antibodies to ATLL-associated antigens. The median age of the patients was 57 years (range, 32-74 years). Detailed patient information is provided in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Four cell lines, SP-49,9 HANK1,10 ATN-1,11 and Jurkat,12 were also analyzed. SP-49 is a mantle cell lymphoma cell line, HANK1 is a natural killer/T-cell lymphoma line, ATN-1 is an ATLL cell line, and Jurkat is a T-cell lymphoblast–like cell line.
Peripheral blood samples were obtained from the blood of 8 healthy male donors. PBMCs were isolated by Ficoll-Paque PLUS centrifugation (GE Healthcare).
DNA extraction
CD4+ cells in PB samples were purified using a magnetic-activated cell-sorting protocol (Miltenyi Biotec). High–molecular-weight DNA was extracted from CD4+ cells, frozen LNs, and from the SP-49, HANK1, ATN-1, and Jurkat cell lines using standard proteinase K treatment and phenol-chloroform extraction.13 Normal DNA was obtained from PBMC samples of 8 healthy male donors.
Oligo-array CGH
Characterization of the genomic aberrations was performed using Agilent 44K Whole Human Genome CGH arrays (Agilent Technologies) containing 44 000 probes. Procedures for DNA digestion, labeling, hybridization, scanning, and data analyses were performed according to the manufacturer's protocol (Agilent Technologies).
CGH data analysis
CGH data were extracted from scanned images using Feature Extraction software (version 10.3; Agilent Technologies). Raw data were transferred to the Genomic Workbench v5.0 software (Agilent Technologies) for further analysis. We defined gains and losses over a continuous 15-probe dataset as a linear log2 ratio average of ≥ 0.05 or ≤ −0.05, respectively, and microdeletion for a range of 3-15 probes as a linear log2 ratio average of ≤ −0.4. A detailed explanation of the log2 ratio is available in the supplemental data. The array CGH data have been deposited in ArrayExpress under the accession number E-MEXP-3042.
Southern blot analysis of HTLV-1 integration and TCRγ rearrangement
Integration of the HTLV-1 provirus genome and TCRγ rearrangement were assayed as described previously.5,14 In brief, DNA samples (5 μg) of LNs were digested with restriction enzymes (PstI) and electrophoresed through 0.7% agarose gels. The DNA was then transferred onto a Hybond N+ membrane (Amersham Pharmacia Biotech) and hybridized to randomly primed 32P-labeled DNA probes specific for the HTLV-1 and TCRγ genes. Blots were then washed at the appropriate stringency and visualized by autoradiography. The HTLV-1 probe comprised a 1.0-kb fragment of the pX region, which was PCR amplified using the primers 5′-ccacttcccagggtttggacag-3′ and 5′-tctgcctctttttcgttaaaaagtagagaaatggg-3′, and the TCRγ probe comprised a 0.6-kb fragment of Jγ2.1.14
Results
Oligo-array CGH analysis against paired samples obtained from the PB and LNs
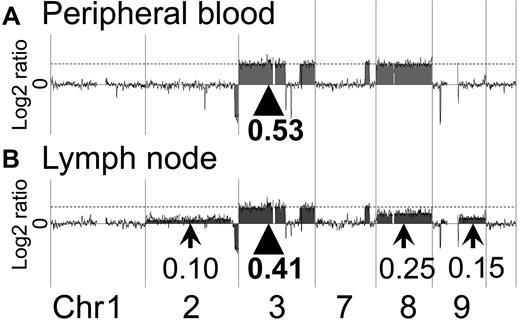
In all of the 13 acute-type ATLL cases, genomic aberrations were detected by oligo-array CGH. Representative profiles of the paired samples obtained from the PB and LNs in cases 1 and 2 are shown in Figure 1A and B and Figure 2A and B, respectively.
Representative profiles of the PB and LN samples of case 1. Array CGH results for case 1 are shown. (A) In the PB sample of case 1, regions of gain were detected. The log2 ratio of chromosome 3 was 0.53 (arrowhead). The log2 ratios of chromosomes 7 and 8 were the same as for chromosome 3 (dotted line). (B) In the LN sample of case 1, a log2 ratio imbalance was found. Log2 ratios among chromosomes 2, 3, 7, 8, and 9 differed. The log2 ratios of chromosome 3 and 7 were 0.41 (arrowhead and dotted line). Arrows show different log2 ratios: chromosome 2 = 0.10, chromosome 8 = 0.25, and chromosome 9 = 0.15.
Representative profiles of the PB and LN samples of case 1. Array CGH results for case 1 are shown. (A) In the PB sample of case 1, regions of gain were detected. The log2 ratio of chromosome 3 was 0.53 (arrowhead). The log2 ratios of chromosomes 7 and 8 were the same as for chromosome 3 (dotted line). (B) In the LN sample of case 1, a log2 ratio imbalance was found. Log2 ratios among chromosomes 2, 3, 7, 8, and 9 differed. The log2 ratios of chromosome 3 and 7 were 0.41 (arrowhead and dotted line). Arrows show different log2 ratios: chromosome 2 = 0.10, chromosome 8 = 0.25, and chromosome 9 = 0.15.
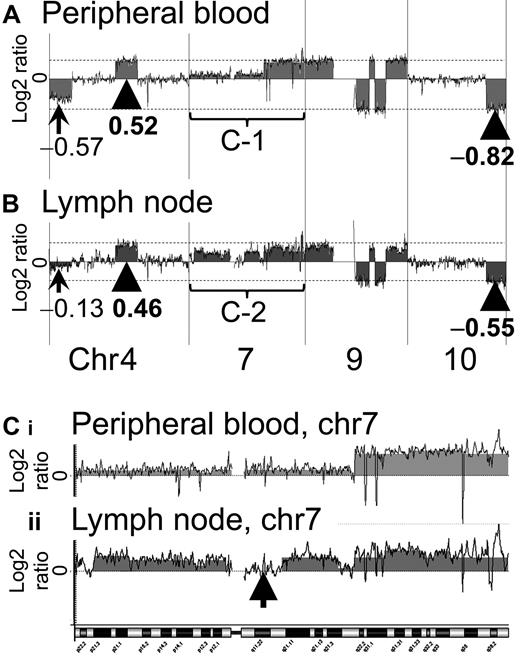
Representative profiles of the PB and LN samples of case 2. The results for case 2 were more complex than those for case 1. In both the PB and the LN samples of case 2, a log2 ratio imbalance was found. (A) In the PB sample, the arrowhead and dotted line indicate the majority of log2 ratios of gain and loss regions. Log2 ratios of the majority of loss regions were −0.82. The log2 ratio of chromosome 4 was −0.57. (B) In the LN sample, the arrowhead and dotted line indicate the majority of log2 ratios of gain and loss regions. Log2 ratios of the majority of loss regions were −0.55. The log2 ratio of chromosome 4 was −0.13. Chromosome 7 regions of PB and LN samples are magnified as Ci and Cii, respectively. (C) Chromosome 7 of the case 2 PB sample shows complex aberrations (i). This result also indicates a log2 ratio imbalance. Chromosome 7 of the case 2 LN sample shows more complex aberrations (ii). An arrow indicates a region (7q11.21-11q.23) without genomic aberration. Ci and Cii suggest that the genomic profiles of the PB and LN samples differ.
Representative profiles of the PB and LN samples of case 2. The results for case 2 were more complex than those for case 1. In both the PB and the LN samples of case 2, a log2 ratio imbalance was found. (A) In the PB sample, the arrowhead and dotted line indicate the majority of log2 ratios of gain and loss regions. Log2 ratios of the majority of loss regions were −0.82. The log2 ratio of chromosome 4 was −0.57. (B) In the LN sample, the arrowhead and dotted line indicate the majority of log2 ratios of gain and loss regions. Log2 ratios of the majority of loss regions were −0.55. The log2 ratio of chromosome 4 was −0.13. Chromosome 7 regions of PB and LN samples are magnified as Ci and Cii, respectively. (C) Chromosome 7 of the case 2 PB sample shows complex aberrations (i). This result also indicates a log2 ratio imbalance. Chromosome 7 of the case 2 LN sample shows more complex aberrations (ii). An arrow indicates a region (7q11.21-11q.23) without genomic aberration. Ci and Cii suggest that the genomic profiles of the PB and LN samples differ.
In the PB sample of case 1, genomic aberration regions showed a constant log2 ratio. Regions of gain were detected on chromosomes 3, 7, and 8. The log2 ratios corresponding to these regions were 0.53, suggesting that there was no imbalance (Figure 1A arrowhead). On the other hand, imbalance of the log2 ratio among chromosomes was found for the LN sample of case 1. Genomic aberrations of the case 1 LN sample were similar to that of the PB sample. However, the log2 ratios among chromosomes 2, 3, 7, 8, and 9 differed as follows. Regions of gain were detected on chromosomes 2, 3, 7, 8, and 9, as shown by the log2 ratios: chromosome 2 = 0.10, chromosomes 3 and 7 = 0.41, chromosome 8 = 0.25, and chromosome 9 = 0.15 (Figure 1B arrowhead and arrows). The log2 ratio of chromosome 8 was lower than that of chromosomes 3 and 7. Gains of chromosomes 2 and 9 were detected in the LN sample, but not in the PB sample. These results indicated that a log2 ratio imbalance occurred in the LN sample.
Case 2 had a log2 ratio imbalance in both the PB and LN samples (Figure 2A). The genomic aberrations of the case 2 PB sample differed from those of the LN sample, as was also found with case 1. In the case 2 PB sample, regions of loss were detected on chromosomes 4, 9, and 10, as shown by the log2 ratios: chromosome 4 = −0.57 (Figure 2A arrow) and chromosomes 9 and 10 = −0.82 (Figure 2A arrowhead and dotted line). In the case 2 LN sample, regions of loss were also detected on chromosomes 4, 9, and 10, as shown by the log2 ratios: chromosome 4 = −0.13 (Figure 2B arrow) and, chromosomes 9 and 10 = −0.55 (Figure 2B arrowhead and dotted line). These data indicated that both samples had a log2 ratio imbalance. Complex genome aberrations were found for chromosome 7 in the paired samples of case 2. Consecutive gain regions were found in the whole of chromosome 7 of the PB sample (Figure 2Ci, and a region (7q11.21-11q.23) without genomic aberrations was found in chromosome 7 of the LN sample (Figure 2Cii arrow).
A log2 ratio imbalance among chromosomes was present in many other samples of acute-type ATLL, as summarized in Table 1.
Confirmation of log2 ratio imbalance among chromosomes
A log2 ratio imbalance among chromosomes was found in many ATLL clinical samples. We expected that a log2 ratio imbalance would indicate the presence of clones with different genomic aberrations. Therefore, we prepared 2 cell lines, SP-49 and HANK1, which possess different genomic aberrations. The genomic DNA of SP-49 was mixed with that of HANK1. We then conducted oligo-array CGH using the mixed-genomic DNA samples at various ratios.
Array CGH analysis of the SP-49 genome showed some genomic aberration regions, which were consistent with the G-band result that had been reported.9 Log2 ratios of all 1-copy gain regions were 0.55, and log2 ratios of all 1-copy loss regions were −0.80. Imbalance of the log2 ratio among the chromosomes was not found. The same was true for HANK1, in which genomic aberration regions were consistent with the G-band result that had been reported and an imbalance of the log2 ratio among the chromosomes was not found.10
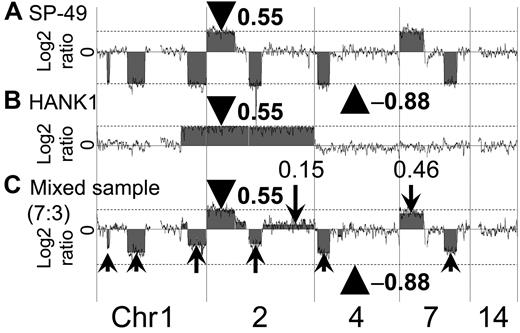
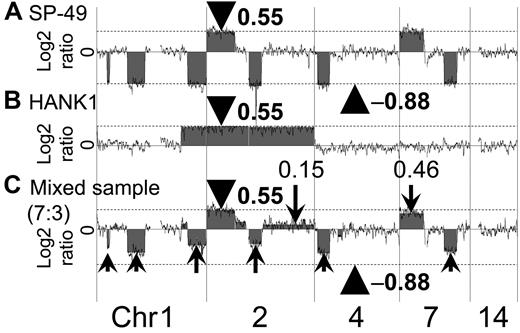
A representative array CGH result using a mixed-DNA sample at a ratio of 7:3 (SP-49:HANK1) is shown in Figure 3. The results showed an imbalance of the log2 ratio among chromosomes. It was possible to reproduce the log2 ratio imbalance. For example, the log2 ratios of chromosomes 2p14-pter, 2q14.3-qter, and 7p were 0.55, 0.15, and 0.46, respectively. These log2 ratios clearly differed. Furthermore, additional regions with different log2 ratios were found.
Confirmation of log2 ratio imbalance among chromosomes. The manner in which the log2 ratio imbalance occurred was confirmed. (A) SP-49 showed no imbalance. Log2 ratios of gain regions were 0.55 (arrowhead and dotted line). Log2 ratios of loss regions were −0.88 (arrowhead and dotted line). (B) HANK1 showed no imbalance. Log2 ratios of gain regions were 0.55 (arrowhead and dotted line). (C) Mixed-genomic DNA at a ratio of 7:3 reproduced the log2 ratio imbalance. The log2 ratio of chromosome 2p14-pter of the mixed DNA sample was 0.55 (arrowhead). Chromosome 2p had a copy region identical to both SP-49 and HANK1. Arrows indicate the log2 ratio imbalance.
Confirmation of log2 ratio imbalance among chromosomes. The manner in which the log2 ratio imbalance occurred was confirmed. (A) SP-49 showed no imbalance. Log2 ratios of gain regions were 0.55 (arrowhead and dotted line). Log2 ratios of loss regions were −0.88 (arrowhead and dotted line). (B) HANK1 showed no imbalance. Log2 ratios of gain regions were 0.55 (arrowhead and dotted line). (C) Mixed-genomic DNA at a ratio of 7:3 reproduced the log2 ratio imbalance. The log2 ratio of chromosome 2p14-pter of the mixed DNA sample was 0.55 (arrowhead). Chromosome 2p had a copy region identical to both SP-49 and HANK1. Arrows indicate the log2 ratio imbalance.
These results indicated that some of the clones present in the sample that had different genome profiles caused a log2 ratio imbalance in the array CGH result. The log2 ratio did not differ in chromosome 2p, which had a copy region identical to both SP-49 and HANK1.
Log2 ratios reflect the ratio of tumor
The genome profiles of mixed-DNA samples comprising various ratios were superimposed (Figure 4). The ratios of SP-49 to HANK1 were 7:3, 6:4, and 3:7. These results clearly revealed that the log2 ratio reflected the ratio of the tumor. When tumors included in a sample had identical genomic aberration regions, the log2 ratio never changed in these regions.
Log2 ratio reflects the ratio of tumor. The genome profiles of mixed-DNA samples comprising various ratios were superimposed. Gain was detected in chromosome 7 of all mixed samples, as shown by the log2 ratio: SP-49 = 0.55; 100%, 7:3 = 0.46; 70%, 6:4 = 0.32; 60%, 3:7 = 0.20; 30%. Loss was also detected in chromosome 7 of all mixed samples as shown by the log2 ratio: Sp-49 = −0.88; 100%, 7:3 = −0.62; 70%, 6:4 = −0.39; 60%, 3:7 = −0.14; 30%. Chromosome 2p had a copy region identical to both SP-49 and HANK1. The log2 ratios never changed in these regions.
Log2 ratio reflects the ratio of tumor. The genome profiles of mixed-DNA samples comprising various ratios were superimposed. Gain was detected in chromosome 7 of all mixed samples, as shown by the log2 ratio: SP-49 = 0.55; 100%, 7:3 = 0.46; 70%, 6:4 = 0.32; 60%, 3:7 = 0.20; 30%. Loss was also detected in chromosome 7 of all mixed samples as shown by the log2 ratio: Sp-49 = −0.88; 100%, 7:3 = −0.62; 70%, 6:4 = −0.39; 60%, 3:7 = −0.14; 30%. Chromosome 2p had a copy region identical to both SP-49 and HANK1. The log2 ratios never changed in these regions.
Southern blot analysis of HTLV-1 integration and TCRγ rearrangement
HTLV-1 integration
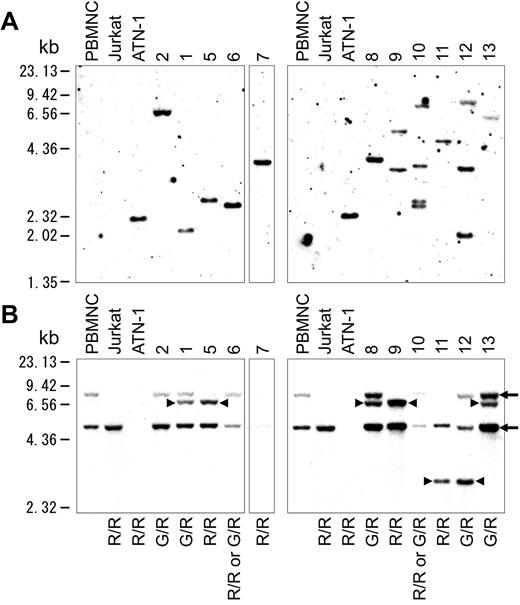
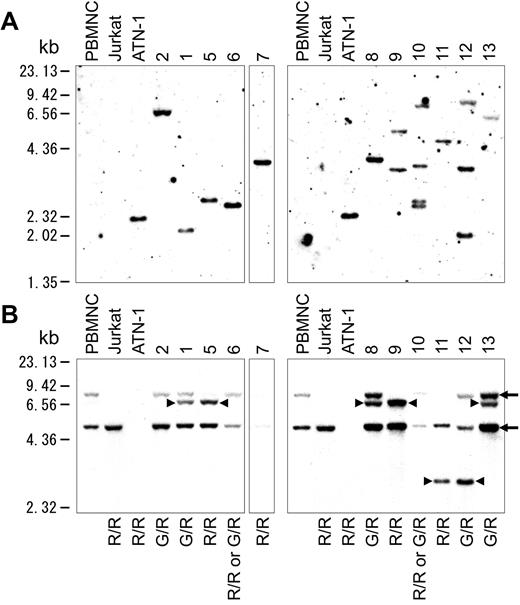
HTLV-1 integration was examined using Southern blot analysis, and the results showed HTLV-1 integration in all of the 11 cases examined. Eight of the 11 cases examined comprised a mono-integration band, whereas the others showed multi-integration bands. (Figure 5A)
Southern blot analysis. (A) Southern blot analysis of HTLV-1 integration in 11 of 13 cases. (B) Southern blot analysis of TCRγ rearrangement. Arrows indicate the 8.00- and 4.9-kb germline bands. Arrowheads indicate the 6.8- and 2.9-kb rearrangement bands. G indicates a germinal allele; R, rearrangement allele; G/R, rearrangement of one allele; R/R, rearrangement of both alleles.
Southern blot analysis. (A) Southern blot analysis of HTLV-1 integration in 11 of 13 cases. (B) Southern blot analysis of TCRγ rearrangement. Arrows indicate the 8.00- and 4.9-kb germline bands. Arrowheads indicate the 6.8- and 2.9-kb rearrangement bands. G indicates a germinal allele; R, rearrangement allele; G/R, rearrangement of one allele; R/R, rearrangement of both alleles.
TCRγ rearrangement
Southern blot analysis of TCR Jγ rearrangement was also conducted and evaluated as described previously by Moreau et al.14 The results indicated that all samples were monoclonal (Figure 5B). Five of the 11 cases examined had a 6.8-kb rearrangement band, and 2 had a 2.9-kb rearrangement band. The others showed loss of germinal bands. In case 2, one allele of TCRγ was rearranged, because the germinal band of 8.0 kb was weaker than that of 4.9 kb. Case 7 lost all germinal bands, such as ATN-1, which is an ATLL cell line. This result indicated that both alleles of TCRγ were rearranged at Jγ2.3, because no deletion was found in case 7 by array CGH. Given that 3 or more TCR rearrangement bands were not found, no cases showed definite multi-clonality in tumor cells. These results indicated that the acute-type ATLL examined represented a monoclonal tumor comprising TCR rearrangements and with some possessing multiple integrations of HTLV-1.
Appearance of LN subclones before PB subclones
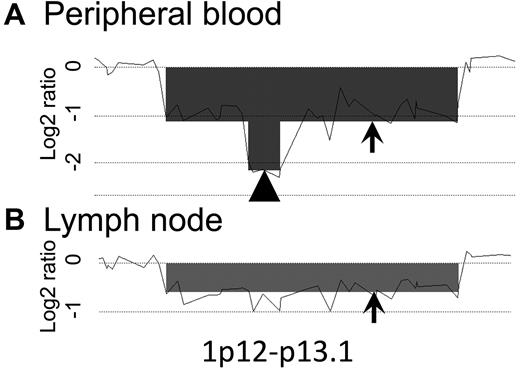
Array CGH analysis revealed that PB samples from 5 of 13 cases had homozygous loss regions that were not found in the corresponding LN samples of each case. In case 2, 1p12-1p13.1 of the PB sample was seen to represent homozygous loss, unlike the case with the LN sample (Figure 6). However, log2 ratios of same region in the LN sample seemed to be slightly lower than those of neighboring regions. This raised the possibility that a minor subclone was present. The PB samples from cases 1, 4, 8, and 10 also had homozygous loss regions that were not clearly found in the corresponding LN samples (Table 2).
Homozygous loss region analysis. A representative homozygous loss region of case 2 is shown (1p12-p13.1). The total scale of the figure is approximately 2 Mb. The arrowhead indicates a homozygous loss region; arrows indicate heterozygous loss regions. Homozygous loss was found only in the PB sample. The log2 ratio of this region in the LN sample was slightly lower than that in the neighboring regions, suggesting the possibility that a minor subclone may exist in the LNs.
Homozygous loss region analysis. A representative homozygous loss region of case 2 is shown (1p12-p13.1). The total scale of the figure is approximately 2 Mb. The arrowhead indicates a homozygous loss region; arrows indicate heterozygous loss regions. Homozygous loss was found only in the PB sample. The log2 ratio of this region in the LN sample was slightly lower than that in the neighboring regions, suggesting the possibility that a minor subclone may exist in the LNs.
No cases had a homozygous loss region in the LN samples when the PB samples had a heterozygous loss in the same regions. Array CGH and Southern blotting results indicated that multiple subclones had developed from one clone. Therefore, when 2 clones were found in a patient, the clone with homozygous loss must have developed from the clone with heterozygous loss. The homozygous loss analysis revealed that in about 40% of ATLL patients, subclones that had appeared in the PB were derived from LN subclones.
Selected subclone of LNs in the PB
Tumor cells in the PB samples of some cases (eg, cases 1 and 9) appeared to have been selected from multiple subclones. In these cases, a log2 ratio imbalance was not found in the PB sample but was found in the LN sample. This indicated that PB samples were monoclonal and that the LN samples contained multiple subclones. Both samples from each case had common aberrations, and the LN samples had aberrations that were not found in the PB samples. These results may indicate that the LNs contain multiple subclones with different genomic aberrations, and that one of these subclones then appears in the PB (Figure 7).
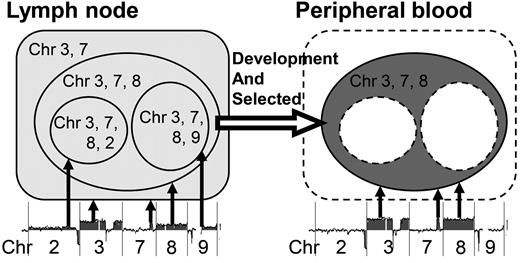
Selected subclone from the LN in the PB. Shown is a schematic representation of a selected subclone from the LN sample in the PB of case 1. In the LN sample of case 1, at least 4 subclones exist: a subclone with chromosome 3 and 7 aberrations; a subclone with additional chromosome 8 aberrations; a subclone with chromosome 3, 7, 8, and 2 aberrations; and a subclone with chromosome 3, 7, 8, and 9 aberrations. Among these subclones, a subclone with chromosome 3, 7, and 8 aberrations appeared in the PB sample.
Selected subclone from the LN in the PB. Shown is a schematic representation of a selected subclone from the LN sample in the PB of case 1. In the LN sample of case 1, at least 4 subclones exist: a subclone with chromosome 3 and 7 aberrations; a subclone with additional chromosome 8 aberrations; a subclone with chromosome 3, 7, 8, and 2 aberrations; and a subclone with chromosome 3, 7, 8, and 9 aberrations. Among these subclones, a subclone with chromosome 3, 7, and 8 aberrations appeared in the PB sample.
Discussion
The imbalance and differing genomic profiles of PB and LN samples indicate that acute-type ATLL comprises multiple subclones
In this study, we revealed the presence of a log2 ratio imbalance among chromosomes of LN samples in many patients with acute-type ATLL. Most of the genomic profiles were found to differ from those of the PB samples. Although monoclonal proliferation of acute-type ATLL is referred to in the World Health Organization classification,15 these data clearly show that acute-type ATLL contains multiple subclones that originate as a result of clonal evolution in ATLL patients.
Shinawi et al16 reported a case of pediatric AML in which 2 clones with different chromosome aberrations showed a log2 ratio imbalance as detected by array CGH. We were able to reproduce a log2 ratio imbalance among chromosomes by mixing different ratios of DNA prepared from 2 different cell lines. The log2 ratio reflected the ratio of tumor clones. Based on these data, we analyzed the acute-type ATLL data and identified that a log2 ratio imbalance indicated the presence of multiple subclones in a sample. Minority clones with low log2 ratios could be found in this experiment by taking advantage of the high sensitivity associated with the use of array CGH. As a result, the presence of multiple subclones was unambiguously determined.
Cases showing different genomic profiles between PB and LN samples reached as high as 69%. We reported previously that paired samples obtained from different sites had different chromosomal aberrations in some cases.17 We also reported that sequential samples at chronic and crisis or acute onset and relapse in each case showed different chromosome aberrations or integrations as determined by chromosomal CGH or Southern blot analysis.17 Similar clonal change has been reported previously in some cases of B-cell lymphoma.18 Although analysis of sequential samples is important when examining the stability of multiple subclones, it is difficult to acquire sequential samples from acute-type ATLL patients because these patients require immediate chemotherapy. However, chronic-type ATLL can be treated with “watchful waiting,” so the clonal stability of ATLL may be explored in these patients.
Our data indicate that acute-type ATLL comprises multiple subclones with differing genomic aberrations. Several morphologic variants of ATLL have been described,15 and the presence of a mixture of cells of different sizes has been reported. However, the histological type does not correspond to the clinical subtype.19 Therefore, it is reasonable to postulate that the histological type does not always reflect the clinical features because the tumor subclones may differ at various sites.
HTLV-1 integration and TCRγ rearrangement determined by Southern blotting
We focused on the cell origin of the multiple subclones in each patient. Southern blot analysis revealed a monoclonal band of HTLV-1 integration or monoclonal rearrangement of TCRγ in all samples examined. These data indicated that the ATLL clones in each case had a common tumor cell origin. ATLL research and treatment utilize the Shimoyama classification. Acute-type ATLL represents one subtype in the classification, and is considered to be a monoclonal tumor. Our data are also consistent with this classification. However, it is possible that multiple subclones in the LNs possess a diversity that may account for the variable clinical manifestations and drug resistance that can occur during the treatment of ATLL.
Selection of leukemic clone and diversity in LNs
Array CGH suggested that the subclones in the PB and LNs differed even though they are derived from an identical monoclonal tumor cell, as determined by in Southern blot analysis. Given that the clones are derived from one clone, theoretically the clone with heterozygous loss is never derived from a cell with homozygous loss. Homozygous loss regions were only present in the PB samples examined at a frequency of 38% (5 of 13 cases examined). None of the 5 samples showed homozygous loss regions found in the LN samples, indicating that in these cases, subclones present in the LNs were not derived from those in the PB. These results suggested that the selected subclones appeared in the PB after subclones developed in the LNs. However, it remains to be determined how these clones in the PB become stable during the course of disease. It is also important to determine whether the tumor cells in the PB can proliferate at the level of tumor cells in the LNs.
In conclusion, the results of the present study showed that there are multiple subclones in acute-type ATLL, all of which possess a common TCR rearrangement and the genomic profiles of which often differ between the PB and LNs. Cases were identified in which a selected subclone from multiple subclones in the LN samples was also identified in the PB samples. ATLL was clinically classified into 4 subtypes by Shimoyama. However, the specific genes that characterize acute-type ATLL have not been identified. Our results reveal that acute-type ATLL is a genetically heterogeneous neoplasm and that clonal evolution of ATLL takes place in the LNs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Koichi Ohshima for diagnoses; Dr Shinobu Tsuzuki, Dr Kennosuke Karube, and Dr Keiichiro Honma for discussions and encouragement throughout this study; and Kyoko Hirano for outstanding technical assistance.
This work was supported in part by grants-in-aid from the Ministry of Health, Labor and Welfare of Japan; from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and from the Japan Society for the Promotion of Science.
Authorship
Contribution: A. Umino performed the experiments, analyzed data, and wrote the paper; M.N. analyzed data and performed experiments; A. Utsunomiya provided advice, discussed clinical data, and treated patients; K.T. provided advice, discussed clinical data, treated patients, and wrote the paper; N.T. analyzed and discussed clinical data; N.K. discussed clinical data; and M.S. supervised the research, discussed clinical data, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masao Seto, MD, PhD, Division of Molecular Medicine, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chikusa-ku, Nagoya, Aichi 464-8681, Japan; e-mail: mseto@aichi-cc.jp.