Abstract
The Ets-related gene (ERG) is an Ets-transcription factor required for normal blood stem cell development. ERG expression is down-regulated during early T-lymphopoiesis but maintained in T-acute lymphoblastic leukemia (T-ALL), where it is recognized as an independent risk factor for adverse outcome. However, it is unclear whether ERG is directly involved in the pathogenesis of T-ALL and how its expression is regulated. Here we demonstrate that transgenic expression of ERG causes T-ALL in mice and that its knockdown reduces the proliferation of human MOLT4 T-ALL cells. We further demonstrate that ERG expression in primary human T-ALL cells is mediated by the binding of other T-cell oncogenes SCL/TAL1, LMO2, and LYL1 in concert with ERG, FLI1, and GATA3 to the ERG +85 enhancer. This enhancer is not active in normal T cells but in transgenic mice targets expression to fetal liver c-kit+ cells, adult bone marrow stem/progenitors and early CD4−CD8− double-negative thymic progenitors. Taken together, these data illustrate that ERG promotes T-ALL and that failure to extinguish activity of stem cell enhancers associated with regulatory transcription factors such as ERG can contribute to the development of leukemia.
Introduction
Dynamic patterns of chromatin accessibility, transcription factor (TF) binding, and DNA methylation all contribute to the gene expression signature of a cell. These changes are plastic and potentially reversible,1 with perturbation of the underlying molecular machinery thought to be a major contributor to the development of malignancies. Acute leukemia development occurs within a particular cellular context where genetic and epigenetic changes in stem cells, progenitor cells, or stem and progenitor cells facilitate the generation and perpetuation of a leukemic clone.2
The Ets-related gene (ERG) is an ETS-family TF required for normal hematopoiesis. It is also a potent oncogene associated with both solid organ and hematologic malignancies.3–8 Although the precise role of ERG during normal hematopoiesis has yet to be fully elucidated, it has been shown that mice with 1 functional allele have reduced circulating HSCs, whereas the loss of both results in embryonic lethality because of impaired proliferation of HSCs that appear to be normally specified in the embryo.3,8 The presence of an extra copy of Erg, on the other hand, has been implicated in the myeloproliferative phenotype of the mouse model of Down syndrome.9 Chromosomal translocations that result in the expression of oncogenic ERG fusion proteins have been identified in leukemia, in Ewing sarcoma, and in 40%-80% of prostate carcinomas.4–6 The Ewing sarcoma gene (EWS)–ERG fusion protein can not only transform mesenchymal progenitors into sarcomas, but also transform committed lymphocytes into T-cell leukemias in vivo.10 Notably, ERG overexpression in T-ALL was shown to be an independent risk factor for an adverse outcome,11 whereas low expression is associated with a highly favorable outcome. Indeed, high ERG expression correlates with disease relapse and unfavorable characteristics in T-ALL (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Comparable observations have also been made in relation to cytogenetically normal acute myeloid leukemia and ERG expression. However, the transforming effects of ERG have only recently been demonstrated in lymphoid cells12 and the precise mechanism of ERG overexpression is not known. As ERG overexpression does not appear to be caused by an increase in DNA content, it may be a consequence of epigenetic, transcriptional, or posttranscriptional or posttranslational regulatory mechanisms driving increased ERG activity in this patient cohort.
The upstream regulators of ERG have not been systematically analyzed in hematopoietic cells. In a survey of Scl targets that used high-output sequencing of chromatin immunoprecipitation (ChIP-seq) material from HPC-7 cells (a stem cell factor–dependent, multipotential, mouse hematopoietic cell line), a region 85 kb downstream of the mouse promoter, mErg +85, showed strong Scl binding.13 We have previously shown that this region targets fetal liver blood cells in F0 mouse transgenics.13 We now report that transgenic mice expressing ERG under control of the vav promoter die from T-ALL and that ERG plays a nonredundant role that promotes the proliferation of T-ALL cells. We also show that the ERG +85 enhancer targets adult HSCs and thymic T-cell progenitors. This enhancer is active in primary human T-ALL cells and is regulated by the T-ALL oncogenes, SCL, LYL1, and LMO2.
Methods
Generation of transgenic animals
The vav-ERG3 transgenic lines were generated by subcloning a 1.4-kb fragment of human ERG3 complementary DNA7 into the HS21/45-vav vector replacing hCD4.14,15 This construct was microinjected into pronuclei of fertilized C57BL6 F1 oocytes and several founders were identified by PCR. Transgenic ERG expression was verified by RT-PCR and Western blot (data not shown). The data presented here are based on analysis of 3 different founders with no difference in the phenotypes identified. Multiple SV/Erg+85/venus lines were generated by amplifying enhancer sequences from mouse genomic DNA and generating founders by pronuclear injection of fertilized ova with the enhancer reporter fragment. The data shown in this manuscript are from L8373.
Cell lines, primary cells, and xenografts
Molt4, REH and 416B cells were maintained as outlined by German Collection of Microorganisms and Cell Cultures (German abbreviation DSMZ). OP9-DL1 cells were maintained as described.16 CD34+/CD38− cells from cord blood were isolated as described.16 To obtain pro-T and pre-T cells, CD34+/CD38− cells were cocultured with OP9-DL1 cells as described.16 Pro-T (CD34+CD7+CD1a−) and pre-T (CD34+CD7+CD1a+) cells were isolated by staining with fluorescence conjugated antibodies and FACS sorting at day 8 and day 14, respectively. The process by which continuous xenografts from childhood ALL biopsies have been established in immunodeficient NOD/SCID mice has been previously described in detail.17
Histology
Thymus tissues were fixed in 10% neutral buffered formalin, transferred to 70% ethanol the next day and paraffin-embedded. H&E staining was carried out using a conventional technique. MOLT4 cells were cytocentrifuged, fixed, and stained using a Wright stain. Images of mouse thymus were acquired using a Leica 40×/0.65 air objective and those of MOLT4 cells using a Leica 100×/1.25 oil objective and a Leica DFC 420C digital camera attached to a Leica DM2500 microscope. Images were acquired using the Leica Application Suite and processed using Adobe Photoshop (Adobe Systems).
FACS
Single cell suspensions prepared from the thymus and bone marrow were washed in PBS with 0.05M EDTA and 0.1% BSA and then stained with fluorochrome-conjugated monoclonal antibodies. For analysis of murine hematopoietic precursors, bone marrow was isolated from 6- to 8-week-old mice and bone marrow cells were depleted with Miltenyi lineage depletion columns (Miltenyi Biotec). Cells were sorted, analyzed, or both using either a MoFlo cell sorter (Dako) or FACSVantage (BD Biosciences). See supplemental Methods for details.
ChIP
ChIP assays were performed as described.18 Enrichment was measured by real-time PCR using SYBR green (chromatin immunoprecipitation-PCR; Invitrogen) or by Agilent custom HD-ChIP arrays (ChIP-chip; ArrayExpress/A-MEXP-1853). See supplemental Methods for details.
ERG expression
RNA isolation (QIAGEN) and complementary DNA synthesis was performed using standard protocols. Relative ERG mRNA levels were determined by quantitative SYBR Green (Stratagene) qPCR or semiquantitative RT-PCR, and normalized against a control gene. ERG protein levels were determined by Western blot using an anti-ERG antibody (Santa Cruz Biotechnology, sc-354X) and anti-TOPO 1 control (BD Pharmingen, 556597).
Stable transfection assays
The ERG +85 fragment was PCR amplified from human genomic DNA and subcloned into the pGL2-promoter vector (Promega). The plasmid was electroporated into MOLT4, REH, and 416B cells with pGKneo and tested in stable transfection assays as described.18 Mutations were introduced by PCR to replace the element indicated with a XhoI restriction site, and confirmed by sequencing.
ERG knockdown
Stable knockdown of ERG in MOLT4 cells was performed as previously described.19 The 19-bp ERG target sequence is ATGCGCATCTCTTTCTTTG and as control, a nontargeting sequence TCCAACACGACACTCACTA was used.19 Briefly, 293T cells were transiently transfected with pRetroSuper-Control or pRetroSuper-ERG retroviral plasmids in combination with expression vectors coding for the Gag, Pol, and Env proteins to produce virus and the viral supernatant was used to infect MOLT4 cells. The cells were doubly infected within a 24-hour interval and allowed to recover for a further 48 hours before selection. Transduced cells were selected for 3 weeks with puromycin. Cell lines with stable knockdown of ERG were assessed for altered growth rate, cell-cycle characteristics, and morphology.
T-cell receptorβ2 rearrangements and Notch1 mutations
Genomic DNA was purified from the thymus and tail of wild-type and ERG transgenic mice and PCR amplified using primers to examine Dβ2-Jβ rearrangements as described.20 Germline PCR product was obtained from tail DNA. The heterodimerization (HD) and PEST domains of NOTCH1 were amplified by PCR, sequenced, and compared with wild-type Notch1 sequence. See supplemental Methods for details.
Statistical analysis
Data was analyzed using GraphPad Prism 5 software and was represented as mean ± SD.
Results
ERG induces T-acute lymphoblastic leukemia in mice
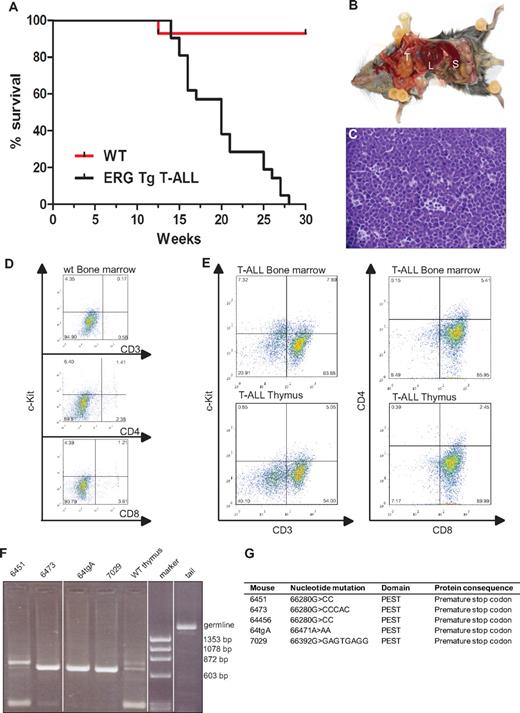
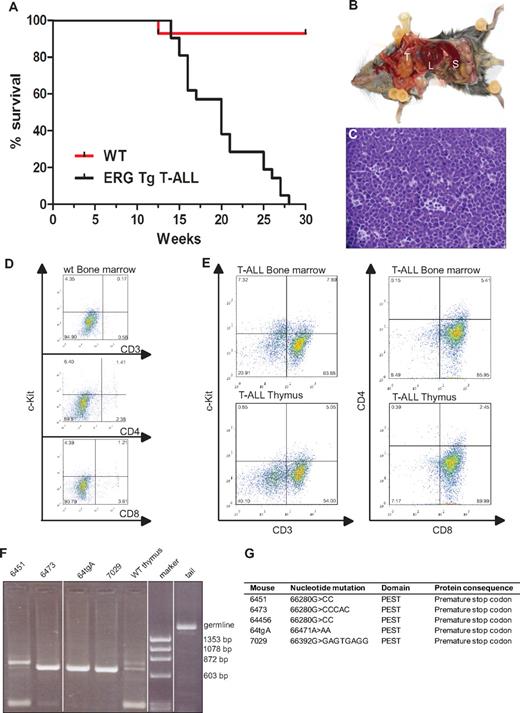
Mice transplanted with syngeneic hematopoietic progenitors overexpressing ERG develop megakaryoblastic leukemia and die within 100 days.19 To assess whether the spectrum of ERG-induced leukemia may be wider following transgenic expression of ERG in the hematopoietic system throughout ontology, we generated transgenic mice expressing the human ERG3 hematopoietic isoform7 under the control of the vav promoter using the HS21/45-vav vector.14,15 All animals expressing ERG developed acute leukemia. Approximately 30% of leukemias were characterized as T-ALL. All animals that developed T-ALL died by the age of 7 months with a median survival age of 20 weeks (Figure 1A). Macroscopic examination of diseased mice revealed thymic and splenic enlargement (Figure 1B). Histologic examination of the thymus showed dense infiltration with lymphoblasts and almost complete disorganization of the normal tissue architecture (Figure 1C). Compared with wild-type littermates (Figure 1D), flow cytometric analysis of the thymus and bone marrow of T-ALL transgenic mice (n = 5) showed that the immunophenotype of the T-ALL blasts was cKit−CD3+CD8+CD4− (Figure 1E). To assess clonality, we extracted DNA from thymuses of wild-type and T-ALL mice and PCR amplified a region within T-cell receptor β2.20 Three mice had a monoclonal blast population whereas 1 (6451) was oligoclonal (Figure 1F).
ERG induces T-ALL in mice. (A) Life expectancy of ERG transgenic mice. Kaplan-Meier survival curves of ERG transgenic mice with T-ALL and wild-type littermate controls (WT). (B) An ERG transgenic mouse with T-ALL dissected to show its internal organs. There is enlargement of the thymus (T), spleen (S), and liver (L). (C) Histology of the thymus of an ERG transgenic mouse with T-ALL. An H&E-stained thymus showing dense leukemic cell infiltration. (D) Flow cytometric analysis of bone marrow from wild-type mice. (E) Flow cytometric analysis of bone marrow and thymus from transgenic ERG mice. The cell surface markers were measured relative to isotype control. The percentage of positive cells is indicated in the corner of each quadrant. (F) T-cell receptorβ rearrangements. PCR products showing Dβ2-Jβ rearrangements in thymus from 4 ERG transgenic mice compared with wild-type thymus and tail (germline). (G) Notch1 mutations. Insertion or deletion mutations in the Notch1 PEST domain in 5 ERG transgenic mice.
ERG induces T-ALL in mice. (A) Life expectancy of ERG transgenic mice. Kaplan-Meier survival curves of ERG transgenic mice with T-ALL and wild-type littermate controls (WT). (B) An ERG transgenic mouse with T-ALL dissected to show its internal organs. There is enlargement of the thymus (T), spleen (S), and liver (L). (C) Histology of the thymus of an ERG transgenic mouse with T-ALL. An H&E-stained thymus showing dense leukemic cell infiltration. (D) Flow cytometric analysis of bone marrow from wild-type mice. (E) Flow cytometric analysis of bone marrow and thymus from transgenic ERG mice. The cell surface markers were measured relative to isotype control. The percentage of positive cells is indicated in the corner of each quadrant. (F) T-cell receptorβ rearrangements. PCR products showing Dβ2-Jβ rearrangements in thymus from 4 ERG transgenic mice compared with wild-type thymus and tail (germline). (G) Notch1 mutations. Insertion or deletion mutations in the Notch1 PEST domain in 5 ERG transgenic mice.
The Notch signaling pathway is important for cell fate decisions in many different cell types, including the T-cell lineage,21 and mutations in NOTCH1 with aberrantly high intracellular activity are a common feature in T-ALL.22 One class of mutations are in the heterodimerization domain (HD), which makes the receptor more prone to proteolytic cleavage and less dependent on ligand-mediated ICD release. The other class of mutations occurs in the PEST domain of NOTCH1, generating truncated versions of the NOTCH1 ICD, which increases the stability of the normally short-lived ICD. Mutations in the heterodimerization and PEST domains of Notch1 have been found in both human and mouse models of T-ALL.22,23 We therefore examined Notch1 mutations in ERG transgenic mice showing signs of T-ALL. DNA was extracted from the thymus and spleen of 5 transgenic animals and sequence analysis of the HD and PEST domains of Notch1 were performed. All 5 mice showed insertions or deletions in the PEST domain, but no mutations were found in the HD domain (Figure 1G). An identical mutation was found in 2 of the mice (6451 and 64456). This specific mutation has been previously reported,24 suggesting it is a hot spot. All mutations caused a frame shift and resulted in a premature stop codon. Taken together, these data show that constitutive overexpression of ERG in hematopoietic cells, drives the development of T-ALL, and cooperates with Notch1 mutations in leukemogenesis. These results are concordant with recent data showing that mice transplanted with bone marrow cells overexpressing ERG also develop T-ALL in association with Notch1 mutations.12
ERG plays a nonredundant role in proliferating human T-ALL cells
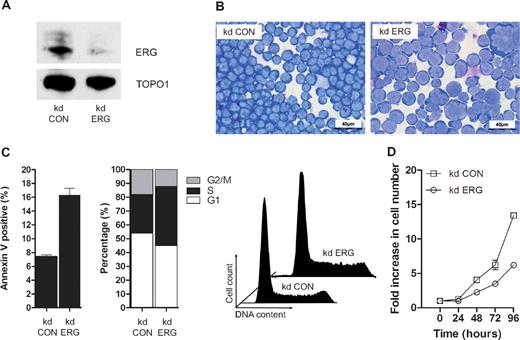
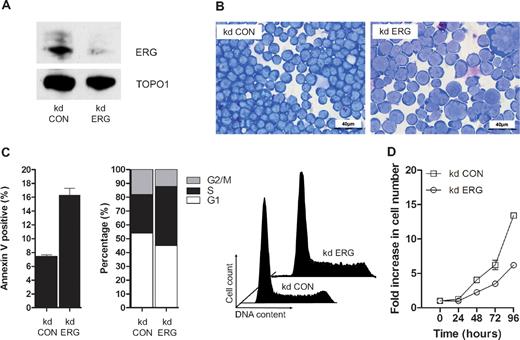
ETS domain genes encode a super-family of TFs organized into 11 subfamilies based on domain structures, which are conserved from worm and fly to mammals.25 Twelve of the 31 ETS genes including erg are expressed in blood/endothelial precursors in zebrafish embryos,26 but despite similarities in structure and expression pattern, Erg plays a nonredundant role in blood/endothelial development in both fish and mice.3,26 ERG is also required for the proliferation and maintenance of acute megakaryocytic leukemic cells.7,19,27 To establish whether ERG plays a nonredundant role in T-ALL cells, we performed knockdown experiments in MOLT-4 cells using previously characterized anti-ERG and control shRNA vectors.19 Knockdown of ERG was evident both by RT-PCR (data not shown) and immunoblot analysis of MOLT4 cells following stable transduction with a retrovirus expressing shRNA that targets ERG (Figure 2A). There was no reduction in FLI1 protein, with which ERG shares approximately 80% amino acid homology (data not shown). ERG knockdown led to a significant change in cell morphology width; increased cell size and nuclear/cytoplasmic ratio, appearance of multinucleate cells and cells with abnormal nuclei and multiple nucleoli; and increased cytoplasmic vacuolation and blebbing (compare kd ERG with kd Con in Figure 2B). These changes were accompanied by increased apoptosis and impaired cell-cycle progression with prolongation of the S-phase (Figure 2C) and a significant reduction in growth potential (Figure 2D). Taken together with data showing impaired proliferation of ERG-deficient Jurkat T-ALL cells,12 these results show that endogenous ERG expression in T-ALL cells plays a nonredundant role in supporting leukemic cell proliferation.
ERG plays a nonredundant role in proliferating T-ALL cells. (A) ERG knockdown (kd) in MOLT4 cells. A Western blot of nuclear lysates from MOLT4 clones transduced with retroviral vectors encoding either control shRNA (kd CON) or shRNA targeting ERG (kd ERG) vectors showing reduced ERG protein in the latter relative to DNA topoisomerase 1 control. (B) Morphology; cytospins of MOLT4 cells transduced with either the control vector (kd CON) or knockdown vector (kd ERG) stained with Wright stain. (C) Cell-cycle kinetics and viability: kd ERG MOLT4 cells show increased apoptosis and impaired progression through the cell cycle. These data are representative of duplicate experiments each recording 105 events. (D) Comparative growth curves: MOLT4 cells stably transduced with an ERG knockdown vector show a significant reduction in growth rate compared with those transduced with a control vector. The data show replicates of 3 separate experiments each using a starting number of 105 control and ERG knockdown cells.
ERG plays a nonredundant role in proliferating T-ALL cells. (A) ERG knockdown (kd) in MOLT4 cells. A Western blot of nuclear lysates from MOLT4 clones transduced with retroviral vectors encoding either control shRNA (kd CON) or shRNA targeting ERG (kd ERG) vectors showing reduced ERG protein in the latter relative to DNA topoisomerase 1 control. (B) Morphology; cytospins of MOLT4 cells transduced with either the control vector (kd CON) or knockdown vector (kd ERG) stained with Wright stain. (C) Cell-cycle kinetics and viability: kd ERG MOLT4 cells show increased apoptosis and impaired progression through the cell cycle. These data are representative of duplicate experiments each recording 105 events. (D) Comparative growth curves: MOLT4 cells stably transduced with an ERG knockdown vector show a significant reduction in growth rate compared with those transduced with a control vector. The data show replicates of 3 separate experiments each using a starting number of 105 control and ERG knockdown cells.
The ERG stem cell enhancer is active in human T-cell progenitors and in human T-ALL
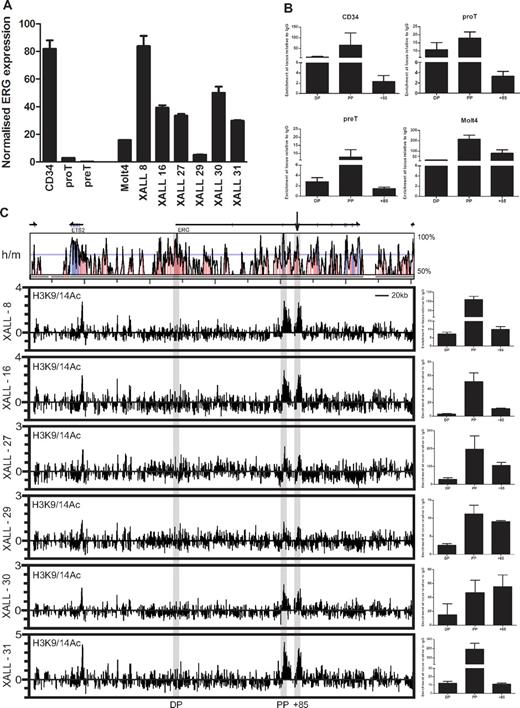
The transcriptional control of ERG expression during normal T-lymphopoiesis and in T-ALL is poorly understood. To gain insights into the regulation of ERG in these cells, we used both an in vitro T-cell differentiation system and patient T-ALL cells derived from xenografted NOD/SCID mice. Human T-lineage differentiation can be induced from umbilical cord blood (CB)–derived CD34+/38− HSCs cocultured with OP9-DL1 cells and serves as a useful tool to purify populations of T-cell progenitors.16 The transition from CD34+CD7+CD1a− (Pro-Ts) to CD34+CD7+CD1a+ (Pre-Ts) by early thymocytes is associated with T-cell commitment (reviewed in Blom and Spits28 ). We used this culture system to harvest pro-T- and pre-T-cell populations to assess ERG expression and chromatin accessibility at the ERG locus.
ERG expression in pro-T cells was considerably lower than in undifferentiated cord blood CD34+ cells but higher than in pre-T cells in which expression was barely detectable by RT-PCR (Figure 3A). ERG expression in the MOLT4 T-ALL cell line and in primary T-ALL cells harvested from NOD/SCID xenografts (5 pro-T-ALLs and one pre-T-ALL), was considerably higher than in normal pro- and pre-T cells and comparable with the expression levels in CD34+ cells (Figure 3A). To assess chromatin accessibility at the ERG locus, we performed ChIP assays for acetylated lysine residues 9 and 14 on histone H3 (H3K9/14Ac). XALL-29 which shows the lowest level of acetylation at the proximal promoter and enhancer also shows the lowest level of ERG mRNA and protein expression (Figure 3A and supplemental Figure 2). The human ERG gene has at least 2 recognized promoters separated by approximately 165 kb. The distal promoter (DP) has previously been reported to be inactive in T-ALL with promoter CpG methylation.29 Consistent with this and other data, active chromatin marks are present on the proximal promoter (PP) and depleted at the DP in CD34+ stem/progenitors, pro- and pre-T cells, and MOLT4 leukemic cells (Figure 3B). Significantly, chromatin accessibility at the ERG enhancer diminishes during the pro-T to pre-T transition but is significantly elevated in MOLT4 T-ALL cells. Consistent with the lack of ERG expression in mature T cells, there is no AcH3 enrichment at the ERG promoters and enhancer in CD4 and CD8 single-positive T cells (supplemental Figure 3).
The ERG stem cell enhancer is active in human T-cell progenitors and in T-ALL. (A) Relative ERG expression in normal and leukemic cells. RNA was isolated from MACS- or FACS-sorted cord blood CD34+ cells, cultured T-cell progenitors and T-ALL xenografts, and ERG expression quantified by SYBR Green qPCR and expressed relative to GAPDH or β actin. ERG is expressed in CD34+ stem cells, but is rapidly down-regulated during T-cell differentiation. However, ERG is highly expressed in MOLT4 and xenograft T-ALL cells. (B) Chromatin accessibility at the ERG promoters and +85 enhancer in normal and leukemic cells. Chromatin accessibility measured by histone H3K9/K14 acetylation (AcH3) in CD34+ stem/progenitors, developing T cells, and MOLT4 T-ALL cells. AcH3 enrichment at the ERG proximal promoter (PP) and +85 element in the various cell types corresponds with ERG expression. (C) Chromatin accessibility across the ERG locus in NOD/SCID T-ALL xenografts. The top panel shows a VISTA plot of human/mouse sequence conservation > 50% across the ERG locus and flanking genes. The ERG promoters and enhancer are highlighted by gray bars. The +85 enhancer is also marked by an arrow. The bottom graphs show chromatin accessibility as determined by AcH3 ChIP-chip and ChIP-PCR in T-ALL xenografts. ChIP-chip plots show AcH3 enrichment along the ERG locus and flanking genes; clusters of AcH3 enrichment are seen at the ERG PP and +85 element in 6/6 T-ALL xenografts. Graphs at right show these peaks of enrichment confirmed by ChIP-PCR.
The ERG stem cell enhancer is active in human T-cell progenitors and in T-ALL. (A) Relative ERG expression in normal and leukemic cells. RNA was isolated from MACS- or FACS-sorted cord blood CD34+ cells, cultured T-cell progenitors and T-ALL xenografts, and ERG expression quantified by SYBR Green qPCR and expressed relative to GAPDH or β actin. ERG is expressed in CD34+ stem cells, but is rapidly down-regulated during T-cell differentiation. However, ERG is highly expressed in MOLT4 and xenograft T-ALL cells. (B) Chromatin accessibility at the ERG promoters and +85 enhancer in normal and leukemic cells. Chromatin accessibility measured by histone H3K9/K14 acetylation (AcH3) in CD34+ stem/progenitors, developing T cells, and MOLT4 T-ALL cells. AcH3 enrichment at the ERG proximal promoter (PP) and +85 element in the various cell types corresponds with ERG expression. (C) Chromatin accessibility across the ERG locus in NOD/SCID T-ALL xenografts. The top panel shows a VISTA plot of human/mouse sequence conservation > 50% across the ERG locus and flanking genes. The ERG promoters and enhancer are highlighted by gray bars. The +85 enhancer is also marked by an arrow. The bottom graphs show chromatin accessibility as determined by AcH3 ChIP-chip and ChIP-PCR in T-ALL xenografts. ChIP-chip plots show AcH3 enrichment along the ERG locus and flanking genes; clusters of AcH3 enrichment are seen at the ERG PP and +85 element in 6/6 T-ALL xenografts. Graphs at right show these peaks of enrichment confirmed by ChIP-PCR.
To survey the chromatin accessibility profiles across the ERG loci in primary T-ALL cells, we performed ChIP-chip using H3K9/K14Ac ChIP material from a panel of NOD/SCID T-ALL xenografts hybridized to high-density Agilent custom arrays with oligomers tiled across the ERG locus to its flanking genes (Figure 3C). AcH3 enrichments are prominent at the ERG PP and +85 enhancer (marked with an arrow above the Vista sequence conservation plots) in all 6 T-ALL xenografts assayed by ChIP-chip. These enrichments were confirmed by quantitative PCR analysis of ChIP material. The promoter of ETS2, a related member of the Ets TF family that is immediately upstream of the ERG locus and known to be expressed in hematopoietic tissues, also has an active chromatin mark. Taken together, these results show that genomic sequences in the human ERG locus corresponding to the mouse Erg +85 enhancer are in an active configuration in CD34+ stem/progenitors, pro-T, and T-ALL cells.
The mErg +85 enhancer targets fetal and adult hematopoietic immature progenitors and shows selective activity in lineage committed cells
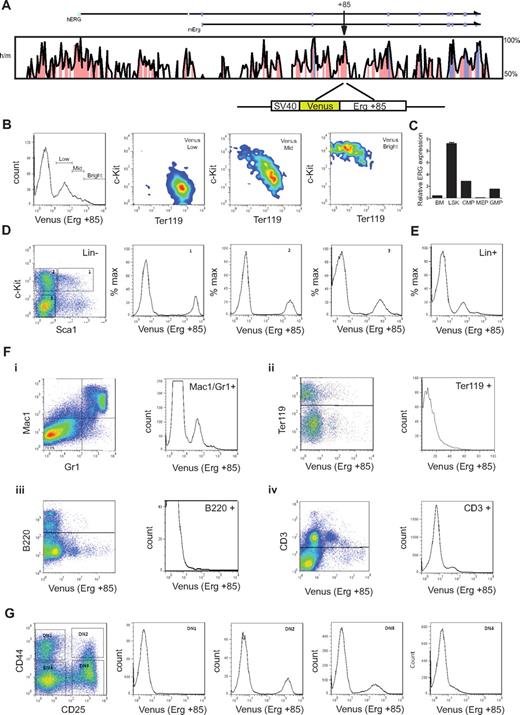
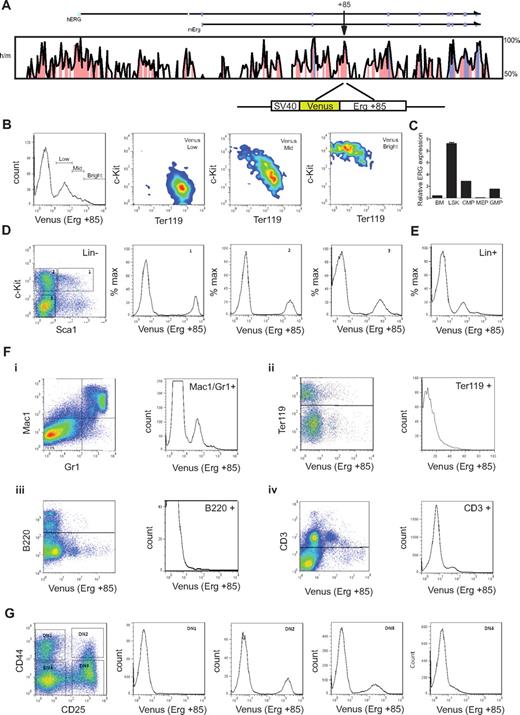
T-cell commitment in the human thymus parallels that in the mouse (reviewed in Blom and Spits28 ). To understand the activity of the Erg +85 enhancer during embryonic and adult hematopoiesis, we generated transgenic mouse lines expressing the Venus fluorescent reporter under control of the SV40 heterologous promoter and mErg +85 enhancer (SV/Venus/+85; see schematic in Figure 4A). Mice homozygous for a germline mutation in Erg (ErgMld2/Mld2) that disrupts its transcriptional activity die in utero at E11.5-E13.5 with defects in definitive hematopoiesis.3 ErgMld2/+ mice survive to adulthood with cell intrinsic defects in HSC number and function. We have previously shown that the mErg +85 enhancer is active in fetal liver cells in transient transgenic assays.13 In E11.5 fetal livers, the activity of the enhancer was most prominent in c-Kit+ progenitors and diminishes progressively with erythroid commitment (Figure 4B). In adult bone marrow, Erg expression monitored by quantitative PCR of sorted cell populations is higher in lineage negative c-Kit+ Sca1+ (LSK) cells, which represent long- and short-term repopulating HSCs (LT- and ST-HSCs), than whole bone marrow (bone marrow), common myeloid progenitors (cytidine 5′-monophosphate), granulocyte-monocyte progenitors (GMP), or megakaryocyte-erythroid progenitors (MEP) (Figure 4C). The +85 enhancer is equally active in LSK cells and in c-Kit+ Sca1− multipotent hematopoietic progenitors but its activity diminishes with lineage commitment (Figure 4D-E). The enhancer also targets the myeloid and T-cell lineages but has little or no activity in the erythroid (Ter119+) or B-cell lineages (B220+; Figure 4F). This pattern of enhancer activity was interesting given the known association between both AML and T-ALL with high ERG expression.11,30
The mErg +85 enhancer targets fetal and adult hematopoietic stem/progenitors and shows selective activity in lineage committed cells. (A) Cross species sequence conservation at the ERG locus. The top panel shows a VISTA plot of human/mouse sequence conservation > 50% across the ERG locus. Noncoding sequences with > 75% conservation are shaded in pink, exons in magenta, and the untranslated regions in cyan; arrow indicates location of the +85 element. Bottom schematic shows the Venus reporter construct driven by the SV40 promoter and mouse Erg +85 element. (B) Erg +85 activity in E11.5 fetal liver (FL) cells. Fetal livers were dissociated and single cell suspensions stained with fluorescent-labeled antibodies and FACS analyzed to assess Venus reporter expression in fetal liver cells. Enhancer activity is highest in c-Kit+Ter119− FL stem/progenitors and decreases in c-Kit−Ter119+ erythroid cells. (C) Erg expression in bone marrow (BM) cells. Bone marrow cells were harvested from approximately 3-month-old C57BL/6J mice, lineage depleted, stained with fluorescent-labeled antibodies, and FACS sorted into various stem/progenitor fractions for RNA isolation. Erg expression was quantified by SYBR Green qPCR and is expressed relative to GAPDH. Erg expression is higher in bone marrow Lin−Sca1+c-Kit+ (LSK) stem progenitor cells than in whole bone marrow. (D) Erg +85 activity in lineage negative adult bone marrow cells. Bone marrow from +85 enhancer transgenic mice was harvested and sorted into lineage-negative and -positive fractions using magnetic beads. The lineage-negative fraction was FACS analyzed following incubation with fluorescent-labeled antibodies to establish Venus reporter expression in gated cell fractions. Erg +85 is most active in c-Kit+ stem/progenitors. (E) Erg +85 activity in lineage positive adult bone marrow cells. Unsorted Lin+ cells show mid-low reporter activity (F) Erg +85 activity in specific subpopulations of lineage committed bone marrow cells. (i) Erg +85 is active in a subset of Mac1+/Gr1+ macrophage/granulocyte precursors. (ii) Erg +85 has low activity in a small fraction of Ter119 erythrocyte precursors. (iii) Erg +85 is inactive in the B-cell lineage. (iv) Erg +85 has low activity in a subset of CD3+ T cells. (G) Erg +85 activity during early T-cell development. The thymus was dissected from 6-week-old +85 enhancer transgenic mice and single-cell suspensions stained for FACS analysis. Venus reporter activity is absent during the DN1 stage, then rapidly increases in DN2 cells before reducing in DN3 and DN4 cells.
The mErg +85 enhancer targets fetal and adult hematopoietic stem/progenitors and shows selective activity in lineage committed cells. (A) Cross species sequence conservation at the ERG locus. The top panel shows a VISTA plot of human/mouse sequence conservation > 50% across the ERG locus. Noncoding sequences with > 75% conservation are shaded in pink, exons in magenta, and the untranslated regions in cyan; arrow indicates location of the +85 element. Bottom schematic shows the Venus reporter construct driven by the SV40 promoter and mouse Erg +85 element. (B) Erg +85 activity in E11.5 fetal liver (FL) cells. Fetal livers were dissociated and single cell suspensions stained with fluorescent-labeled antibodies and FACS analyzed to assess Venus reporter expression in fetal liver cells. Enhancer activity is highest in c-Kit+Ter119− FL stem/progenitors and decreases in c-Kit−Ter119+ erythroid cells. (C) Erg expression in bone marrow (BM) cells. Bone marrow cells were harvested from approximately 3-month-old C57BL/6J mice, lineage depleted, stained with fluorescent-labeled antibodies, and FACS sorted into various stem/progenitor fractions for RNA isolation. Erg expression was quantified by SYBR Green qPCR and is expressed relative to GAPDH. Erg expression is higher in bone marrow Lin−Sca1+c-Kit+ (LSK) stem progenitor cells than in whole bone marrow. (D) Erg +85 activity in lineage negative adult bone marrow cells. Bone marrow from +85 enhancer transgenic mice was harvested and sorted into lineage-negative and -positive fractions using magnetic beads. The lineage-negative fraction was FACS analyzed following incubation with fluorescent-labeled antibodies to establish Venus reporter expression in gated cell fractions. Erg +85 is most active in c-Kit+ stem/progenitors. (E) Erg +85 activity in lineage positive adult bone marrow cells. Unsorted Lin+ cells show mid-low reporter activity (F) Erg +85 activity in specific subpopulations of lineage committed bone marrow cells. (i) Erg +85 is active in a subset of Mac1+/Gr1+ macrophage/granulocyte precursors. (ii) Erg +85 has low activity in a small fraction of Ter119 erythrocyte precursors. (iii) Erg +85 is inactive in the B-cell lineage. (iv) Erg +85 has low activity in a subset of CD3+ T cells. (G) Erg +85 activity during early T-cell development. The thymus was dissected from 6-week-old +85 enhancer transgenic mice and single-cell suspensions stained for FACS analysis. Venus reporter activity is absent during the DN1 stage, then rapidly increases in DN2 cells before reducing in DN3 and DN4 cells.
T cells develop within the thymus from bone marrow–derived hematopoietic progenitors and follow a series of specification events marked by the loss of alternative lineage choices and the coordinated expression of CD4 and CD8 (reviewed in Rothenberg and Yui31 ). As T-cell progenitors develop, double-negative cells (CD4−CD8−) can be subdivided into developmental stages referred to as DN1 to DN4 based on CD25 and CD44 expression (reviewed in Rothenberg and Yui31 ). T-cell commitment occurs between the DN2 and DN3 stages in mouse early T cells and is accompanied by increased dependence on Notch-Delta signaling for viability (reviewed in Rothenberg and Yui31 ). This transition is characterized by the down-regulation of HSC transcriptional regulators and proto-oncogenes such as Scl/Tal1, Lyl1, Lmo2, and Gf1b; persistence of TFs such as Gata3, Tcf7, Ets1, Runx1, and Lef1 that are known to be important for T-cell development32 ; and the up-regulation of Bcl11b, a zinc finger TF recently shown to be a major determinant of T-lineage commitment.33 The precise role of Erg in T-cell biology is unknown but significantly, Erg is expressed in CD25+ (DN2/3) but not in early thymic progenitors (ETP) or CD44−/ CD25− (DN4) thymocytes.34 The activity of the Erg enhancer mirrors this pattern of expression (Figure 4G). Taken together, these data show that the Erg +85 HSC enhancer has stage specific activity that is restricted to immature thymic T-lymphocyte precursors.
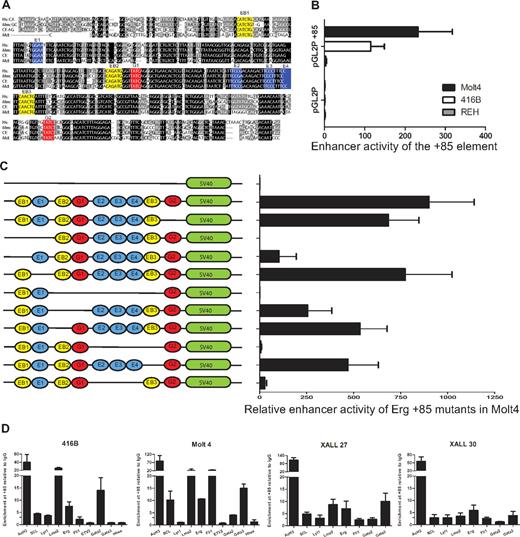
The ERG +85 stem cell enhancer is active in T-ALL cells and relies on ETS/GATA/E-Box binding motifs
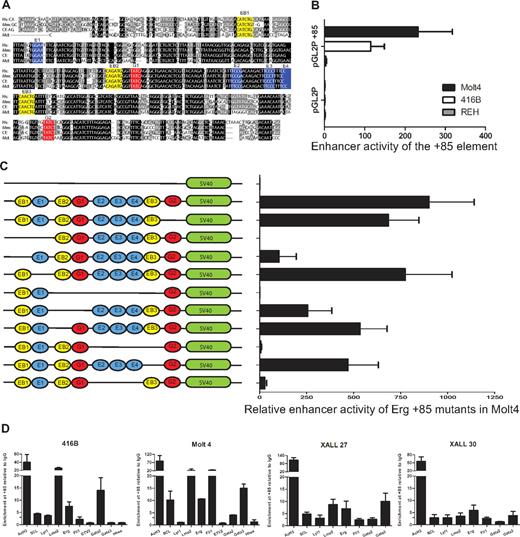
The ERG+85 region has blocks of sequences that are highly conserved from human to opossum including ETS (blue), GATA (red), and E-Box (yellow) consensus motifs (Figure 5A). To test enhancer activity of this region in T-ALL cells, a 496-bp fragment of PCR amplified human genomic sequence was subcloned into the pGL2P reporter vector and tested in stable transfection assays in mouse 416B blood progenitors, human MOLT4 T-ALL cells, and human REH B-ALL cells (Figure 5B). In contrast to the pGL2promoter vector alone, the pGL2P/ERG+85 construct showed approximately 100-fold to 200-fold greater activity in MOLT4 T-ALL cells and in 416B progenitors. There was little or no enhancement in REH B-ALL cells.
The ERG +85 stem cell enhancer is active in T-ALL cells, relies on E-Box and Ets binding motifs, and is bound by oncogenic transcription factors in vivo. (A) Nucleotide sequence alignment of the Erg +85 enhancer region. Alignment of human (Hs), mouse (Mm), dog (cf), and opossum (Md) sequences extracted from the UCSC genome browser with conserved Ets (blue), Gata (red), E-Box (yellow) motifs colored for clarity. Black boxes indicate 100% cross-species sequence conservation, and gray boxes show less conserved sequences. (B) Enhancer activity of the hERG +85 region. Results of stable transfection assays in MOLT4 T-ALL, 416B hematopoietic precursors, and REH B-ALL cells showing luciferase activity of the +85 enhancer/pGL2P construct relative to pGL2P alone. The +85 fragment was active in MOLT4 and 416B, but not REH cells. (C) Mutational analysis of +85 enhancer activity in MOLT4 cells. Schematic on the left shows TF binding motifs of WT and mutant enhancers as detailed in panel A. (D) ChIP-PCR analysis of TF binding. The enhancer has active chromatin marks (AcH3) in 416B blood progenitors as well as in T-ALL cells. There is strong LMO2 binding in all cell types, whereas SCL and LYL1 enrichments are more pronounced in T-ALL cells. The Ets factors, FLI1 and ERG, are also enriched in T-ALL cells. Consistent with their expression profiles, Gata2 is enriched at the +85 enhancer in 416B progenitors, whereas GATA3 is enriched in T-ALL cells.
The ERG +85 stem cell enhancer is active in T-ALL cells, relies on E-Box and Ets binding motifs, and is bound by oncogenic transcription factors in vivo. (A) Nucleotide sequence alignment of the Erg +85 enhancer region. Alignment of human (Hs), mouse (Mm), dog (cf), and opossum (Md) sequences extracted from the UCSC genome browser with conserved Ets (blue), Gata (red), E-Box (yellow) motifs colored for clarity. Black boxes indicate 100% cross-species sequence conservation, and gray boxes show less conserved sequences. (B) Enhancer activity of the hERG +85 region. Results of stable transfection assays in MOLT4 T-ALL, 416B hematopoietic precursors, and REH B-ALL cells showing luciferase activity of the +85 enhancer/pGL2P construct relative to pGL2P alone. The +85 fragment was active in MOLT4 and 416B, but not REH cells. (C) Mutational analysis of +85 enhancer activity in MOLT4 cells. Schematic on the left shows TF binding motifs of WT and mutant enhancers as detailed in panel A. (D) ChIP-PCR analysis of TF binding. The enhancer has active chromatin marks (AcH3) in 416B blood progenitors as well as in T-ALL cells. There is strong LMO2 binding in all cell types, whereas SCL and LYL1 enrichments are more pronounced in T-ALL cells. The Ets factors, FLI1 and ERG, are also enriched in T-ALL cells. Consistent with their expression profiles, Gata2 is enriched at the +85 enhancer in 416B progenitors, whereas GATA3 is enriched in T-ALL cells.
To test the contribution of the conserved ETS/GATA and E-Box motifs to enhancer activity, a series of mutation constructs were generated and tested in stable transfection assays in MOLT4 cells (Figure 5C). The GATA binding motif at the 3′ end of the enhancer (G2) does not contribute significantly to the activity of the enhancer. In contrast, the activity of the enhancer was abolished with the deletion of the 5′ conserved E-Box and Ets (EB1-E1) motifs or a conserved block of sequences, which include a cluster of GATA, 3 conserved ETS, and 2 flanking E-Box motifs (EB2-G1-E2/3/4-EB3). Within the EB2-G1-E2/3/4-EB3 block, loss of the second E-Box motif (EB2) has a moderate effect whereas loss of both EB2 and the first Gata motif (G1) reduces activity to less than 50%, implying that both these motifs contribute to the activity of the enhancer. The trio of Ets binding sites (E2/3/4) and the third E-Box motif (EB3) collectively and independently contribute to the activity of the enhancer, with the trio of Ets binding sites showing particular importance. Taken together, these data show that the activity of the ERG +85 enhancer in T-ALL cells is dependent on specific highly conserved Ets, Gata, and E-box motifs.
The proto-oncogenes SCL, LYL1, and LMO2 as well as ETS and GATA factors bind the ERG +85 enhancer in T-ALL cells
Our understanding of the molecular basis of T-ALL is derived largely from analysis of recurrent chromosomal translocations and intrachromosomal rearrangements that juxtapose gene regulatory elements that drive expression of T-cell receptor genes with a small set of TFs that are important during normal hematopoiesis, and include SCL/TAL1, LYL1, and LMO2.35 Although the majority of T-ALLs aberrantly express one or more of these TFs, only a minority have identifiable chromosomal abnormalities.36 SCL and its paralogue, LYL1, are bHLH proteins that bind E-box motifs, whereas LMO2 is a transcriptional cofactor containing 2 LIM domain zinc fingers that does not bind DNA directly but in a complex that may include either of the former and E2A, LDB1, and GATA factors.37
The transactivating factors that bind the ERG +85 enhancer and drive ERG expression in T-ALL cells are not known. Given the activity of the ERG +85 enhancer during normal hematopoiesis and in T-ALL cells, as well as the contribution of the E-Box motifs to this activity, we assayed binding of SCL, LYL1, and LMO2 in 416B blood progenitors and T-leukemic cells by RT-PCR ChIP assays (Figure 5D). We also assessed binding of FLI1 and ERG based on our previous observations in T-ALL cells,38 and the enrichment of GATA factors, based on their known involvement in the composition of the SCL/LMO2 transcriptional complex.37 A variable but definite in vivo enrichment of each of these TFs were identified at the ERG stem cell enhancer relative to IgG and a control region. Taken together, these data show that known T-ALL proto-oncogenes, SCL, LYL1, and LMO2 together with FLI1, ERG, and GATA3, bind an accessible cis-regulatory element that drives ERG expression in leukemic cells.
Discussion
ERG is an Ets TF that is required for normal blood stem cell development.3 It is overexpressed in T-ALL cells38 and is recognized as an independent risk factor for an adverse outcome.11 We report that transgenic mice overexpressing ERG in hematopoietic cells die with T-ALL and that ERG promotes the proliferation of human T-ALL cells. We show using locus-wide chromatin accessibility assays in primary human T-ALL xenografts, in vitro T-cell differentiation assays and transgenic reporter mice that ERG overexpression in T-ALL is probably caused by aberrant activation of the +85 enhancer. We show that this enhancer is transiently activated during normal T-cell ontogeny and in leukemic cells is bound by transcription factors that are typically activated in T-ALL.
The biologic role of aberrant ERG expression in oncogenesis has been the focus of much investigation in prostate cancer. ERG has been shown to disrupt lineage-specific differentiation of the prostate and to facilitate a stem cell–like program by perturbing polycomb-mediated H3K27 trimethylation.39 Together, these events are thought to promote cell proliferation leading to oncogenic transformation with other cooperating events. Indeed, the overexpression of ERG in mouse prostate tissue promotes marked acceleration and progression of high-grade prostatic intraepithelial neoplasia to prostatic adenocarcinoma in conjunction with deficiency of the tumor suppressor PTEN.40 In the present study, we show that overexpression of ERG in hematopoietic cells results in T-ALL, and consistent with other mouse models, occurs in the context of activating mutations of NOTCH1. The ongoing role for ERG in proliferating T-ALL cells is evident from the slowing of cell growth, cell-cycle arrest, and increased apoptosis following ERG depletion. Taken as a whole, aberrant ERG expression in blood progenitors is likely to promote the expansion and impede differentiation of a population of cells that is then ripe to acquire additional mutations that lead to full -blown leukemia.
ERG is not a gene that is known to be expressed during the specification of prostatic tissue, and overexpression in prostate cancer occurs in the context of a specific gene rearrangement. On the other hand, ERG is expressed transiently during T-cell ontogeny and is maintained in T-ALL. The evidence that a stem cell enhancer, which is transiently active during normal T-cell ontogeny, is active in T-ALL provides a mechanism by which ERG can be expressed in the absence of genomic translocation or amplification. Our data show that the Erg +85 enhancer is active in a population of T-cell progenitors, which are known to also express high levels of Scl, Lyl1, and Lmo2.32 Binding of these TFs to the enhancer regulates Erg expression in these cells. However, the activity of the enhancer and the “stem cell” TFs such as Scl, Lyl1, and Lmo2 that bind the enhancer are normally shut down during T-cell differentiation. The mechanisms by which ERG expression may persist in the absence of genomic translocation or amplification of the locus and facilitate the development of T-ALL are not known. One mechanism could be the mutational activation of an upstream regulator such as SCL, LYL1, or LMO2, which could bind and inappropriately maintain the enhancer in an active configuration. Chromosomal rearrangements, sub-microscopic DNA copy number alterations, or sequence mutations resulting in the overexpression of each of these transcription factors in T-ALL have of course been well documented.41 Another possibility is that clones of high ERG-expressing T-cell progenitors are generated stochastically during T-ALL development and act as cells primed for subsequent mutational events and poorer prognosis. ERG positively regulates its own expression via the +85 enhancer and as a component of the FLI1-ERG-LMO2-HHEX transcriptional sub-circuit could help facilitate the LMO2-HHEX axis, which has been shown to confer greater self-renewal capacity to committed thymic progenitors.42
Overexpression is not the only mechanism by which aberrant TF activity can be maintained in a cell. Another ETS transcription factor, ETV1, has recently been reported to mediate the development of gastrointestinal stromal tumors (GISTs) in the context of abnormal KIT-MEK signaling in interstitial cells of Cajal (ICCs), the presumed cell of origin for GIST.43 This again differs from prostate cancer and Ewing sarcoma in that aberrant activity of ETS expression and activation of an oncogenic transcription program occurs in the absence of genomic translocation or amplification of the ETS locus but rather in the context of prolonged activation of a TF that is normally expressed in the cell of origin. The MAP kinases are activated in response to a variety of mitogens/growth factors and stress/cytokines and are known to phosphorylate and activate the ETS family of transcription factors as a group.44 Whether ERG activity in T-ALL is prolonged by a similar mechanism is not known.
The Erg +85 enhancer in HPC7 stem cells is bound by a number of hematopoietic TFs including Scl, Lyl1, Lmo2, Gata2, Fli1, Erg, and Runx1, which constitute a heptad of TFs that show overlapping DNA binding45 (supplemental Figure 4). Interestingly, a number of these factors, or in the case of GATA2, a related family member GATA3, bind the enhancer in T-ALL. The stem cell signatures that appear to be reactivated in T-ALL42 could in part be related to the activity of these HSC transcriptional sub-circuits. Early T-cell precursor acute lymphoblastic leukemia (ETP-ALL) is a subtype of very-high-risk leukemia with a distinct immunophenotype and stem cell–like features.46 It is thought to arise from a cell equivalent to a mouse early thymocyte precursor (ETP), a population of cells that are recent immigrants to the thymus from the bone marrow that retain multilineage differentiation potential suggestive of recent derivation from bone marrow HSCs.47–49 The xenografts analyzed in this study were either pro-T-ALL (X8, 27, 29, 30, and 31) or pre-T ALL (X16) and overexpressed ERG. ETP-ALLs express significantly more ERG than typical ALLs.46 The mechanism of high ERG expression in ETP-ALLs is unexplained. One possibility is that these leukemias arise from a thymocyte with multilineage potential. Alternatively these leukemias could in fact arise from a later-stage thymocyte in which these HSC transcriptional subcircuits have been activated. The dismal prognosis associated with ETP-ALLs could therefore reflect not so much their cell of origin but the effectiveness with which these networks are activated and the stem cell genes they express. Taken together, our data show that failure to extinguish the activity of a stem cell enhancer of a potent regulatory TF contributes to the development of leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The HS21/45-vav vector was provided by Dr P. Aplan with the approval of Dr J. Adams; the venus reporter plasmid was from Dr A. Miyawaki. The authors thank Dr J. Schibi (Sheba Medical Centre, Israel) for the pathologic assessment of the ERG transgenic mice.
This work was supported by the National Health and Medical Research Council of Australia, the Australian Research Council, the Leukemia Foundation, the Viertel Foundation, the Ramaciotti Foundation, the Cancer Institute of New South Wales, Cancer Council New South Wales, the Israel Science Foundation, Waxman Foundation, Children with Leukemia United Kingdom, Leukemia Research Foundation Chicago, the European Hematology Association, and the Medical Research Council and Leukemia & Lymphoma Research United Kingdom.
Y.B. is a European Hematology Association Fellow. This study is partial fulfillment of the requirements for completing an MSc degree (Y.K.) from the Sackler Medical School, Tel Aviv University.
Authorship
Contribution: J.A.I.T., Y.B., S.F., K.K., Y.K., V.C., G.J., D.S., S.H.O., and S.J.K. performed experiments; J.A.I.T., Y.B., S.F., K.K., Y.K., V.C., J.W.W., and J.E.P. analyzed data; J.A.I.T., Y.B., B.G., and S.I. contributed to writing the paper; Y.G. provided research assistance; R.L. provided experimental material for analysis; K.L.M., B.G., and S.I. designed experiments; and J.E.P. designed the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John E. Pimanda, Lowy Cancer Research Centre, University of New South Wales, Sydney, Australia 2052; Shai Izraeli, Sheba Medical Centre, Tel-Hashomer, and Tel Aviv University, Israel 52621; and Berthold Gottens, Cambridge Institute of Medical Research, University of Cambridge, Cambridge CB2 OXY, United Kingdom; e-mail: jpimanda@unsw.edu.au, Shai.Izraeli@sheba.health.gov.il, or bg200@cam.ac.uk.
References
Author notes
J.A.I.T., Y.B., and S.F. contributed equally to this study.