Abstract
Multiple myeloma is characterized by the clonal expansion of malignant plasma cells (multiple myeloma cells [MMCs]), in the bone marrow. Osteolytic bone lesions are detected in 80% of patients because of increased osteoclastic bone resorption and reduced osteoblastic bone formation. MMCs are found closely associated with sites of increased bone resorption. Osteoclasts strongly support MMC survival in vitro. To further elucidate the mechanisms involved in osteoclast/MMC interaction, we have identified 552 genes overexpressed in osteoclasts compared with other bone marrow cell subpopulations. Osteoclasts express specifically genes coding for 4 CCR2-targeting chemokines and genes coding for MMC growth factors. An anti-CCR2 monoclonal antibody blocked osteoclast chemoattractant activity for MMC, and CCR2 chemokines are also MMC growth factors, promoting mitogen-activated protein kinase activation in MMC. An anti-insulin growth factor-1 receptor monoclonal antibody completely blocked the osteoclast-induced survival of MMC suppressing both osteoclast and MMC survival. Specific a proliferation-inducing ligand or IL-6 inhibitors partially blocked osteoclast-induced MMC survival. These data may explain why newly diagnosed patients whose MMC express high levels of CCR2 present numerous bone lesions. This study displays additional mechanisms involved in osteoclast/MMC interaction and suggests using CCR2 and/or insulin growth factor-1 targeting strategies to block this interaction and prevent drug resistance.
Introduction
Multiple myeloma (MM) is a plasma cell neoplasm characterized by the accumulation of malignant plasma cells primarily in the bone marrow (BM). A majority of patients with MM develop osteolytic bone disease characterized by bone pain, pathologic fractures, and hypercalcemia resulting from the disruption of the coupling of osteoclastic bone resorption and osteoblastic bone formation.1 An increase in bone turnover rate was reported to precede progression from monoclonal gammopathy of undetermined significance to overt MM.1 In patients with intramedullary MM, MM cells (MMCs) develop in close interaction with the BM microenvironment, mainly BM stromal cells,2 endothelial cells,3 and osteoclasts.4,5 In particular, MMCs promote osteoclast formation directly4,6 or indirectly7,8 and osteoclasts support MMC survival, producing a proliferation-inducing ligand (APRIL) or interleukin-6 (IL-6) particularly.9,10 Several factors are involved in myeloma bone disease,11 mainly the receptor activator of nuclear transcription factor-B (RANKL),8 macrophage inflammatory protein-1-α (MIP-1α/CCL3),12-14 tumor necrosis factor-α,15 interleukin-1α (IL-1α),15 and IL-6.15 A shift in RANKL versus osteoprotegerin expression in patients with MM favors osteoclast generation and activation,16 and blocking RANKL decreases tumor burden and bone destruction in patients with MM.4 MIP-1α/CCL3 is produced by MMCs, stromal cells, monocytes, and osteoclasts.13,17 Both osteoclasts and MMCs express CCR1, a receptor for MIP-1α/CCL3 receptor, which promotes osteoclast formation and activation, and inhibition of MIP-1α/CCL3 decreases markedly both tumor burden and bone destruction in a murine model of MM.14,18
Given the importance of the interaction of MMCs and osteoclasts to promote both MMC growth and osteoclast formation, this study aims to further characterize the cell communication mechanisms between these 2 cell types. We show here that osteoclasts specifically express a set of chemokines targeting CCR2 receptors that are overexpressed by MMCs compared with normal plasma cells. At the same time, newly diagnosed MM patients with a high CCR2 gene expression on MMCs exhibit a higher number of bone lesions than patients with a low CCR2 expression on MMCs. The chemoattractant activity of osteoclasts on MMCs is inhibited by an anti-CCR2 monoclonal antibody (mAb). These CCR2 chemokines are also myeloma growth factors (MGFs), promoting mitogen-activated protein kinase (MAPK) activation in MMCs. We also show that osteoclasts support MMCs by producing insulin growth factor-1 (IGF-1), APRIL, and IL-6. This study underscores the important role of osteoclasts in recruiting MMC and promoting their survival and emphasizes the interest of CCR2 targeting therapies.
Methods
XG-human myeloma cell lines (HMCLs) were obtained as described.19 SKMM, L363, OPM2, LP1, and RPMI8226 HMCLs were purchased from ATTC (LGC Promochem). MMCs were obtained in agreement with the French and German ethical laws. MMCs were purified from the BM of 206 patients with newly diagnosed MM (median age, 59 years) after written informed consent was given in accordance with the Declaration of Helsinki. The study has been approved by the ethic boards of Heidelberg University and Montpellier University Hospitals. These 206 patients were treated with high-dose therapy and autologous stem cell transplantation and were termed in the following Heidelberg-Montpellier series.20 We also used Affymetrix data of a cohort of 345 purified MMCs from previously untreated patients from the Arkansas Cancer Research Center. The patients were treated with total therapy 221 and termed in the following Arkansas Cancer Research Center-TT2 series. These data are publicly available via the online Gene Expression Omnibus (Gene Expression Profile of Multiple Myeloma, accession number GSE2658; http://www.ncbi.nlm.nih.gov/geo; accessed June 1, 2006). Normal BM plasma cells (BMPCs) and whole BM cells were obtained from healthy donors after informed consent was given. Whole BM cells were collected after lysis of red blood cells with NH4Cl. After Ficoll-density gradient centrifugation, plasma cells were purified using anti-CD138 MACS microbeads (Miltenyi Biotec). Bone marrow environment cells from 7 newly diagnosed patients were obtained after depletion of MMCs with anti-CD138 MACS microbeads (Miltenyi Biotec). For 5 newly diagnosed patients, BM T cells, monocytes, and polymorphonuclear neutrophils were purified. Bone marrow cells were labeled with a phycoerythrin (PE)-conjugated anti-CD3 mAb, allophycocyanin-conjugated anti-CD14 mAb, and a fluorescein isothiocyanate (FITC)-conjugated anti-CD15 mAb (all from BD Biosciences). CD3+, CD14+, and CD15+ cells were sorted with a FACSAria cell sorter (BD Biosciences). Memory B cells, polyclonal plasmablasts, and BM stromal cell lines were generated as described previously.9 The study was approved by the ethics boards of the Medical Faculty of the University of Heidelberg and the University of Montpellier.
Osteoclasts
Osteoclasts were generated as previously described.9 In brief, peripheral blood mononuclear cells were obtained from 7 patients with MM after informed consent. Cells were cultured at 2.5 × 106 cells/mL in α-minimum essential medium-10% fetal calf serum (FCS). After 12 hours of culture, nonadherent cells were eliminated and adherent cells were cultured in α-minimum essential medium-10% FCS, RANKL (50 ng/mL, PeproTech), macrophage colony-stimulating factor (25 ng/mL, PeproTech), and 10nM dexamethasone for 14 days. Before use, osteoclasts were phenotyped by reverse-transcribed polymerase chain reaction (TRAP and cathepsin K expression), by cytometry (integrin αvβ3 expression) and bone-resorbing activity (OsteoLyse assay kit, Cambrex).
Flow cytometry analysis
CCR1 and CCR2 expression on HMCLs was evaluated by incubating 5 × 105 cells with PE-conjugated anti-CCR1 or anti-CCR2 mAbs (BD Biosciences) in phosphate-buffered saline containing 30% human AB serum at 4°C for 30 minutes. For primary samples, cells were double stained with PE-conjugated anti-CCR1 or anti-CCR2 and FITC-conjugated anti-CD138 (Beckman Coulter) mAbs. Flow cytometry analysis was carried out on a FACScan flow cytometer (BD Biosciences).
In vitro cell migration assay
For Transwell migration assay, 24-well plates with transwell inserts (6.5-mm diameter, 5-μm pore size; Costar Corning Elscolab) and RPMI 1640 medium (Invitrogen) supplemented with 0.5% bovine serum albumin (Sigma-Aldrich) were used. The inserts were coated with 100 μL human fibronectin solution (Invitrogen) at a concentration of 10 μg/mL in distilled water and incubated for 1 hour at 37°C and 5% CO2. The solution was removed, and the inserts were dried for 2 hours at 37°C. The lower transwell chamber containing osteoclasts was filled with 600 μL of α-minimum essential medium-10% FCS. A total of 105 MMCs were added to the upper chamber. Cells were then allowed to migrate for 90 minutes at 37°C in a humid atmosphere (5% CO2). Finally, cells were collected from upper and lower wells, and the number of MMCs that transmigrated into lower wells was evaluated with a fluorescence-activated cell sorter flow cytometer (BD Biosciences) after incubation with an anti-CD138 mAb PE-conjugated at 4°C for 30 minutes. The frequencies of migrating cells (number of cells that migrated to the lower chamber divided by cell number in the upper and lower chamber) are indicated. For migration inhibition, neutralizing anti-CCR1 and CCR2 mAbs (R&D Systems) or a MAPK inhibitor (PD98059) (Cell Signaling) were used.
Growth assay for myeloma cells
HMCLs were IL-6- and serum-starved for 2 hours and cultured for 4 days in 96-well flat-bottom microtiter plates in serum-free culture medium without cytokine (control), with rIL-6 (2 ng/mL), or with graded CCL2, CCL7, CCL8, CCL13, or CCL23 concentrations. The growth of myeloma cells was evaluated by quantifying intracellular adenosine triphosphate with a Cell Titer Glo Luminescent Assay (Promega) with a Centro LB 960 luminometer (Berthold Technologies).
Enzyme-linked immunosorbent assay
The concentrations of B-cell-activating factor of the tumor necrosis factor family (BAFF), APRIL, IL-10, IL-6, and IGF-1 were assayed with enzyme-linked immunosorbent assays purchased from Bender MedSystems for BAFF and APRIL,22 from Diaclone for IL-6 and IL-10 and from R&D Systems for IGF-1 in accordance with the manufacturer's instructions.
Study of apoptosis
IL-6–dependent HMCLs were starved of IL-6 for 3 hours and cultured in 24-well flat-bottomed microtiter plates at 105 cells/well in RPMI 1640–10% FCS in the presence or not of osteoclasts (2.5 × 104 cells/well), with or without B-E8 anti–IL-6 mAb (10 μg/mL), TACI-Fc (20 μg/mL), anti–IL-10 mAb (10 μg/mL), anti-CCR2 mAb (10 μg/mL), or IGF-1 receptor mAb (4 μg/mL; Calbiochem). Recombinant IL-6 (2 ng/mL) was added in one culture group as positive control. After 4 days of culture, cells were washed twice in phosphate-buffered saline, and apoptosis was assayed with FITC-conjugated annexin V labeling (Boehringer). Fluorescence was analyzed on a FACScan flow cytometer (BD Biosciences).
cRNA and microarray hybridization
RNA extraction was performed using the RNeasy kit (QIAGEN), the SV-total RNA extraction kit (Promega), and Trizol (Invitrogen). Labeled cRNA was generated using the small sample-labeling protocol vII (Affymetrix), and hybridized to U133 2.0 plus arrays according to the manufacturer's instructions. Fluorescence intensities were quantified and analyzed using GECOS Version 1.4 software (Affymetrix).
All microarray data presented in this paper have been deposited in the ArrayExpress public database, under accession numbers E-MEXP-2360 for BMPCs23 and E-TABM-937 for B cells, polyclonal plasmablasts, MMCs, and environment population samples.
Real-time reverse-transcribed polymerase chain reaction
Total RNA was converted to cDNA using the Superscript II reverse transcriptase (Invitrogen). The assays-on-demand primers and probes and the TaqMan Universal Master Mix were used according to the manufacturer's instructions (Applied Biosystems). The measurement of gene expression was performed using the ABI Prism 7000 Sequence Detection System and analyzed using the ABI Prism 7000 SDS Version 1.0 software. For each set of primers, serial dilutions of a standard cDNA were amplified to create a standard curve, and values of unknown samples were estimated relative to this standard curve to assess the polymerase chain reaction efficiency. Threshold cycle (Ct) values were obtained for glyceraldehyde-3-phosphate dehydrogenase and the respective genes of interest during log phase of the cycle. Gene of interest levels were normalized to glyceraldehyde-3-phosphate dehydrogenase for each sample (δCt = Ct gene of interest − Ct glyceraldehyde-3-phosphate dehydrogenase) and compared with the values obtained for a known positive control using the following formula: 100/2δδCt where δδCt = δCt unknown − δCt positive control.
Western blot analysis
HMCLs were starved overnight in RPMI 1640–1% bovine serum albumin without IL-6 and then incubated with serum-free culture medium, recombinant IL-6 (30 ng/mL), recombinant CCL7, or CCL8 (2 μg/mL) for 10 and 30 minutes. Cells were then processed for Western blot analysis as detailed elsewhere.19 The primary antibodies (phospho-specific antibodies anti-ERK1/2, anti-STAT3, and anti-AKT; New England Biolabs) were diluted 1% bovine serum albumin Tris-buffered saline containing Tween 20 (1:1000 dilution). The primary antibodies were visualized with goat antirabbit (Sigma-Aldrich) or goat antimouse (Bio-Rad) peroxidase-conjugated antibodies using an enhanced chemiluminescence detection system. As a control for protein loading, we used anti-STAT3 (1:2000) (Transduction Laboratories), anti-ERK1/2 (1:2000) (Santa Cruz Biotechnology), and anti-AKT (New England Biolabs) antibodies.
Statistical analysis
Gene expression data were normalized with the MAS5 algorithm and analyzed with our bioinformatics platform (RAGE, http://rage.montp.inserm.fr24 ; and Amazonia, http://amazonia.montp.inserm.fr), the Significance Analysis of Microarrays software (as previously described),19 and the Maxstat package used in R software (http://cran.r-project.org). Statistical comparisons were done with Mann-Whitney, χ2, or unpaired or paired Student t tests.
Results
Identification of cell communication signals involved in MMC/osteoclast interactions
To identify osteoclast-associated factors that could promote MMC survival, we investigated genes overexpressed in osteoclasts compared with BM CD14 monocytes, BM CD15 polymorphonuclear cells, BM CD34 cells, BM stromal cells, or BM B cells using supervised Significance Analysis of Microarrays analysis (ratio ≥ 2, false discovery rate ≤ 1%, 1000 permutations). Genes overexpressed in osteoclasts compared with normal BM plasma cells or primary MMCs were also determined. Crossing these gene lists yielded 552 genes/expressed sequence tags significantly overexpressed in osteoclasts compared with the 7 BM cell populations (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These genes were significantly (P < .05) enriched in genes encoding for 2 major pathways: “cellular function and maintenance” and “cell-to cell signaling and interaction” as well as 10 other pathways (supplemental Figure 1).
Osteoclasts express genes coding for CCR2 chemokines specifically, and high CCR2 gene expression in myeloma cells is associated with increased bone lesions
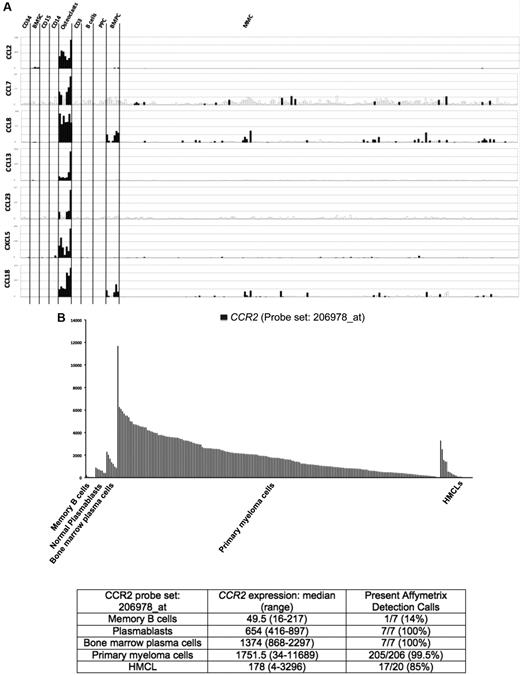
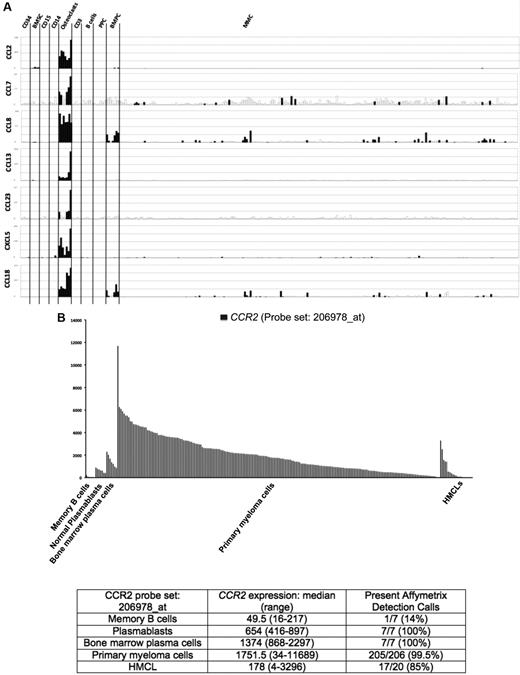
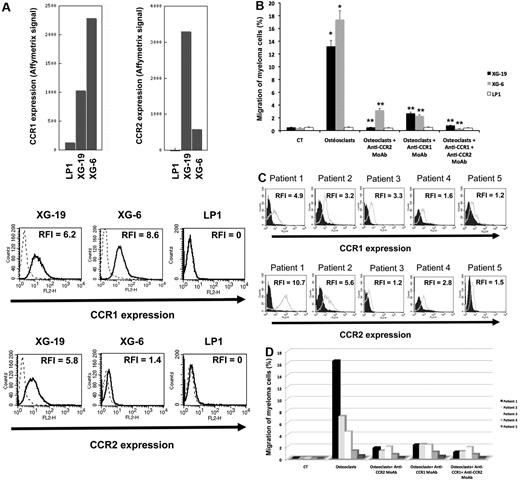
Seven of the 552 osteoclast genes encoded for chemokines (Figure 1A; supplemental Table 1). Four of these chemokines (CCL2, CCL7, CCL8, and CCL13) target the CCR2 receptor expressed by myeloma cells.25,26 One gene encodes for CCL23 chemokine, which is a weak activator of CCR1,27 a chemokine receptor also expressed by myeloma cells.13,14,28 The 2 other chemokine genes encode for CXCL5 whose receptor CXCR2 is not expressed by MMC (http://amazonia.transcriptome.eu) and CCL10 whose receptor is unknown. Besides these osteoclast-specific genes, osteoclasts expressed additional chemokine genes also expressed by other components of the BM environment, in particular the MIP1-α/CCL3 gene expressed by monocytes, BM stromal cells, and MMCs (supplemental Figure 2) as previously described.29 Given the very specific expression of genes coding for CCR2 chemokines by osteoclasts and that myeloma cells stimulate bone resorption,30 we questioned whether CCR2 gene expression of MMCs from newly diagnosed patients with MM could be linked with increased bone lesions. The CCR2 gene expression on normal and malignant plasma cells is shown in Figure 1B. The reliability of the Affymetrix probe set (206978_at) to assay for CCR2 gene expression was validated characterizing protein expression by flow cytometry in HMCLs. A strong correlation between CCR2 gene and protein expression was found (r = .90, P < .001, supplemental Figure 3). Furthermore, CCR2 protein could not be detected in HMCLs with an absent Affymetrix call (supplemental Figure 3). CCR2 expression was not expressed in normal memory B cells in agreement with its reported suppression by Pax5 B-cell transcription factor.31 CCR2 was expressed by normal plasmablasts and BM plasma cells and was significantly increased in MMCs compared with normal BMPCs (P ≤ .05, Figure 1B). We then investigated whether CCR2 expression in MMCs could be linked with bone lesions. Using the Maxstat function of R package, a maximum difference in the proportion of patients with major bone structural damage or more than 3 osteolyses was obtained splitting patients into 2 groups with a CCR2 cutoff = 3735. Major bone structural damage or more than 3 osteolyses were observed in 55% of the patients with CCR2 signal in MMCs more than 3735 versus 36% of the patients with CCR2 signal less than or equal to 3735 (P < .05, Tables 1, 2). This CCR2 signal cutoff is 2.7-fold the median value of CCR2 signal in normal plasma cells. Of note, because osteoclasts specifically express the gene coding for CCL23 that activates CCR1 weakly, we also studied whether CCR1 expression in MMC could be linked with bone lesion extent. Contrarily to CCR2, no cutoff of CCR1 gene expression in MMCs could identify a group of newly diagnosed patients with increased bone lesions. An explanation is that various cells in the BM environment could produce other CCR1-targeting chemokines. This is the case for MIP1-α/CCL3 whose gene is expressed by osteoclasts but also by monocytes, stromal cells, and MMCs as mentioned in Supplemental Figure 2. These data suggest these chemokines could be important to recruit MMCs to the BM but not specifically close to osteoclasts.
Chemokine signature of osteoclasts. (A) Affymetrix CCL2, CCL7, CCL8, CCL13, CCL23, CXCL5, and CCL18 gene expression in BM CD34 cells (n = 5), BM stromal cells (n = 5), purified BM CD15 (n = 5), CD14 (n = 5), and CD3 cells (n = 5), osteoclasts (n = 7), normal memory B cells (n = 6), normal polyclonal plasmablasts (n = 7), normal BMPCs (n = 7), and purified myeloma cells from consecutive patients with MM (n = 206). (B) CCR2 expression in memory B cells, normal plasmablasts, and normal BM plasma cells from 7 healthy donors, 206 with MM, and 20 HMCL. The Affymetrix signal of the CCR2 probe set 206978_at is indicated on the y-axis. CCR2 expression in each sample is indicated by the height of the bar. Samples are ordered from the highest to lowest expression of CCR2 gene from left to right on the x-axis. The attached table shows the median value and range of Affymetrix signal and the percentage of samples with a present call.
Chemokine signature of osteoclasts. (A) Affymetrix CCL2, CCL7, CCL8, CCL13, CCL23, CXCL5, and CCL18 gene expression in BM CD34 cells (n = 5), BM stromal cells (n = 5), purified BM CD15 (n = 5), CD14 (n = 5), and CD3 cells (n = 5), osteoclasts (n = 7), normal memory B cells (n = 6), normal polyclonal plasmablasts (n = 7), normal BMPCs (n = 7), and purified myeloma cells from consecutive patients with MM (n = 206). (B) CCR2 expression in memory B cells, normal plasmablasts, and normal BM plasma cells from 7 healthy donors, 206 with MM, and 20 HMCL. The Affymetrix signal of the CCR2 probe set 206978_at is indicated on the y-axis. CCR2 expression in each sample is indicated by the height of the bar. Samples are ordered from the highest to lowest expression of CCR2 gene from left to right on the x-axis. The attached table shows the median value and range of Affymetrix signal and the percentage of samples with a present call.
Role of CCR2 chemokines in the osteoclast chemoattractant activity for myeloma cells
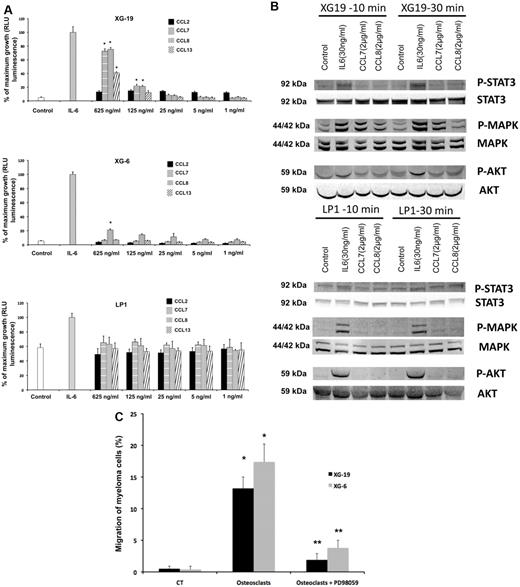
The chemoattractant activity of osteoclasts for MMCs was evaluated using the XG-6, XG-19, and LP1 HMCLs. XG-19 and XG-6 myeloma cells express CCR2 gene and protein, unlike LP1 myeloma cells (Figure 2). Osteoclasts could efficiently attract XG-19 and XG-6 cells, unlike LP1 cells in a transwell assay. This chemoattractant activity was inhibited by a neutralizing mAb to CCR2 by 95% and 81% for the XG-19 and XG-6 cells, respectively (P ≤ .05, Figure 2B). As osteoclasts express MIP1-α/CCL3 chemokine gene together with monocytes or BM stromal cells and specifically CCL23 gene, we also investigated a role of CCR1-targeting chemokines in the osteoclast chemoattractant activity. XG-19 and XG-6 cells expressed CCR1 and a mAb to CCR1 inhibited by 80% and 87% the osteoclast-induced migration of XG-19 and XG-6 cells, respectively (P ≤ .05, Figure 2B). Addition of both anti-CCR1 and anti-CCR2 mAbs further inhibited the osteoclast-mediated migration of myeloma cells (Figure 2B). Similar data were obtained with primary myeloma cells of 5 patients (Figure 2C-D). Primary myeloma cells variably expressed CCR2 or CCR1 assayed by flow cytometry depending on patients' sample. Osteoclasts could attract more than 4% primary myeloma cells for 3 of the 5 patients, the highest chemoattraction being observed with primary myeloma cells with the highest CCR2 and CCR1 expression (Figure 2C-D). The osteoclast chemotactic activity was inhibited by a neutralizing anti-CCR2 mAb (P < .05, Figure 2D). It was also inhibited by a neutralizing anti-CCR1 mAb (P < .05, Figure 2D)
Gene and protein expression of CCR1 and CCR2 chemokine receptors. (A) Data are Affymetrix signals for CCR1 (probe set 205098_at) and CCR2 (probe set 206978_at) in LP1, XG-19, and XG-6 HMCLs. Membrane expression of CCR1 or CCR2 was evaluated by flow cytometry using PE-conjugated anti-CCR1 or anti-CCR2 mAbs or isotype-related PE-conjugated control mAbs. The numbers in the panels are the RFI of the PE-conjugated anti-CCR1 or anti-CCR2 mAbs compared with the PE-conjugated control mAbs. (B) The chemoattractant activity of osteoclasts to myeloma cells was assayed using XG-19, XG-6, and LP1 HMCLs. Data are the fraction of MMC in the upper chamber of the transwell that could migrate to the lower chamber. Results are the mean values plus or minus SD of 3 experiments. *Significant difference of MMC migration in MMC/osteoclast cocultures compared with control using a paired Student t test (P ≤ .05). **Significant difference of MMC migration in MMC/osteoclast cocultures with anti-CCR1 and/or anti-CCR2 mAb compared with MMC/osteoclast cocultures using a paired Student t test (P ≤ .05). (C) The expression of CCR1 and CCR2 by CD138+ primary myeloma cells of patients was evaluated by flow cytometry using FITC-conjugated anti-CD138 mAb and PE-conjugated anti-CCR1 or anti-CCR2 mAbs labeling. Isotype-matched FITC-conjugated or PE-conjugated mAbs recognizing no human antigens were used as control mAb. The numbers in the panels are the RFI of the PE-conjugated anti-CCR1 or anti-CCR2 mAbs compared with the PE-conjugated control mAbs. (D) The chemoattractant activity of osteoclasts to myeloma cells was assayed using primary myeloma cells. Data are the fraction of primary MMC in the upper chamber of the transwell that could migrate to the lower chamber.
Gene and protein expression of CCR1 and CCR2 chemokine receptors. (A) Data are Affymetrix signals for CCR1 (probe set 205098_at) and CCR2 (probe set 206978_at) in LP1, XG-19, and XG-6 HMCLs. Membrane expression of CCR1 or CCR2 was evaluated by flow cytometry using PE-conjugated anti-CCR1 or anti-CCR2 mAbs or isotype-related PE-conjugated control mAbs. The numbers in the panels are the RFI of the PE-conjugated anti-CCR1 or anti-CCR2 mAbs compared with the PE-conjugated control mAbs. (B) The chemoattractant activity of osteoclasts to myeloma cells was assayed using XG-19, XG-6, and LP1 HMCLs. Data are the fraction of MMC in the upper chamber of the transwell that could migrate to the lower chamber. Results are the mean values plus or minus SD of 3 experiments. *Significant difference of MMC migration in MMC/osteoclast cocultures compared with control using a paired Student t test (P ≤ .05). **Significant difference of MMC migration in MMC/osteoclast cocultures with anti-CCR1 and/or anti-CCR2 mAb compared with MMC/osteoclast cocultures using a paired Student t test (P ≤ .05). (C) The expression of CCR1 and CCR2 by CD138+ primary myeloma cells of patients was evaluated by flow cytometry using FITC-conjugated anti-CD138 mAb and PE-conjugated anti-CCR1 or anti-CCR2 mAbs labeling. Isotype-matched FITC-conjugated or PE-conjugated mAbs recognizing no human antigens were used as control mAb. The numbers in the panels are the RFI of the PE-conjugated anti-CCR1 or anti-CCR2 mAbs compared with the PE-conjugated control mAbs. (D) The chemoattractant activity of osteoclasts to myeloma cells was assayed using primary myeloma cells. Data are the fraction of primary MMC in the upper chamber of the transwell that could migrate to the lower chamber.
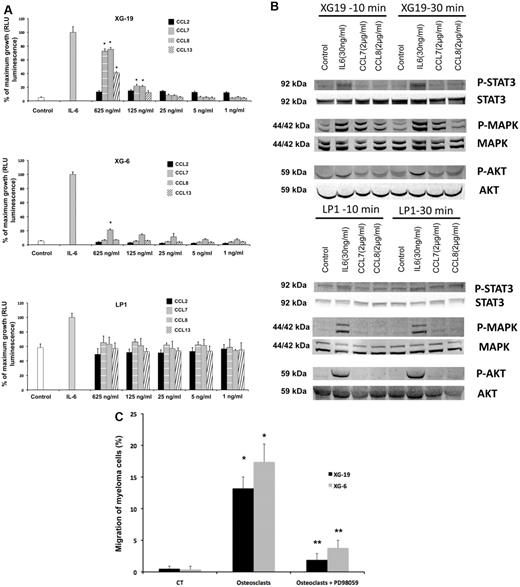
CCR2 chemokines activate MAP kinase pathway in myeloma cells and promote myeloma cell growth, and a MAP kinase inhibitor abrogates the chemoattractant activity of osteoclasts to myeloma cells
The recombinant CCL7, CCL8, and CCL13 CCR2 chemokines significantly increased the growth of CCR2+ XG-19 cells and only CCL8 that of XG-6 cells (P ≤ .05, Figure 3A). These chemokines did not stimulate the growth of CCR2− LP1 cells (Figure 3A). CCL7 and CCL8 chemokines, which had the highest effects on myeloma cell growth, were assayed for signal transduction. The 2 chemokines induced MAPK phosphorylation in XG-19 cells. They did not induce STAT3 or AKT phosphorylations, whereas IL-6 induced the phosphorylation of STAT3, MAPK, and AKT, in agreement with previous data.32 CCL7 or CCL8 did not induce MAPK phosphorylation in CCR1−CCR2− LP1 cells (Figure 3B). As the osteoclast chemoattractant activity for MMC was inhibited by anti-CCR2 mAbs and as CCR2 chemokines activated MAPK pathway in MMC, we looked for the effect of a MAP kinase inhibitor on osteoclast chemoattractant activity for MMC. The PD98059 MAP kinase inhibitor dramatically inhibited the migration of myeloma cells toward osteoclasts (Figure 3C).
CCL7, CCL8, and CCL13 support the growth of CCR2+ HMCLs. (A) XG-19, LP1, and XG-6 were IL-6 starved for 3 hours and cultured either with no cytokine, or in the presence of IL-6 (3 ng/mL) or in the presence of increasing concentrations of CCL2, CCL7, CCL8, or CCL13. Results are the mean plus or minus SD values of the RLU fluorescence determined on sextuplet culture wells. Results are those of one experiment representative of 5. *Mean value is significantly different from that obtained without adding cytokine using a Student t test (P ≤ .05). (B) XG-19 or LP1 cells were starved overnight and cultured without cytokine, or with IL-6 (30 ng/mL), CCL7 (2 μg/mL), or CCL8 (2 μg/mL) for 10 and 30 minutes at 37°C. Cell lysates were probed by Western blotting with antibodies against phospho-STAT3 (pSTAT3), phospho-ERK1/2 (pMAPK), and phospho-AKT (pAKT). Blots were reprobed with antibodies to STAT3, MAPK, and AKT proteins to quantitate protein loading. Western blots are of one experiment representative of 3. (C) The chemoattractant activity of osteoclasts to XG-19 or XG-6 myeloma cells was assayed with or without a MAPK inhibitor (PD98059). Data are the fraction of primary MMC in the upper chamber of the transwell that could migrate to the lower chamber and are mean values of 3 experiments. *Significant increase in MMC migration in MMC/osteoclast cocultures compared with MMC alone using a paired Student t test (P ≤ .05). **Significant decrease of MMC migration in MMC/osteoclast cocultures with PD98059 (10μM) compared with MMC/osteoclast cocultures using a paired Student t test (P ≤ .05).
CCL7, CCL8, and CCL13 support the growth of CCR2+ HMCLs. (A) XG-19, LP1, and XG-6 were IL-6 starved for 3 hours and cultured either with no cytokine, or in the presence of IL-6 (3 ng/mL) or in the presence of increasing concentrations of CCL2, CCL7, CCL8, or CCL13. Results are the mean plus or minus SD values of the RLU fluorescence determined on sextuplet culture wells. Results are those of one experiment representative of 5. *Mean value is significantly different from that obtained without adding cytokine using a Student t test (P ≤ .05). (B) XG-19 or LP1 cells were starved overnight and cultured without cytokine, or with IL-6 (30 ng/mL), CCL7 (2 μg/mL), or CCL8 (2 μg/mL) for 10 and 30 minutes at 37°C. Cell lysates were probed by Western blotting with antibodies against phospho-STAT3 (pSTAT3), phospho-ERK1/2 (pMAPK), and phospho-AKT (pAKT). Blots were reprobed with antibodies to STAT3, MAPK, and AKT proteins to quantitate protein loading. Western blots are of one experiment representative of 3. (C) The chemoattractant activity of osteoclasts to XG-19 or XG-6 myeloma cells was assayed with or without a MAPK inhibitor (PD98059). Data are the fraction of primary MMC in the upper chamber of the transwell that could migrate to the lower chamber and are mean values of 3 experiments. *Significant increase in MMC migration in MMC/osteoclast cocultures compared with MMC alone using a paired Student t test (P ≤ .05). **Significant decrease of MMC migration in MMC/osteoclast cocultures with PD98059 (10μM) compared with MMC/osteoclast cocultures using a paired Student t test (P ≤ .05).
Growth factors produced by osteoclasts
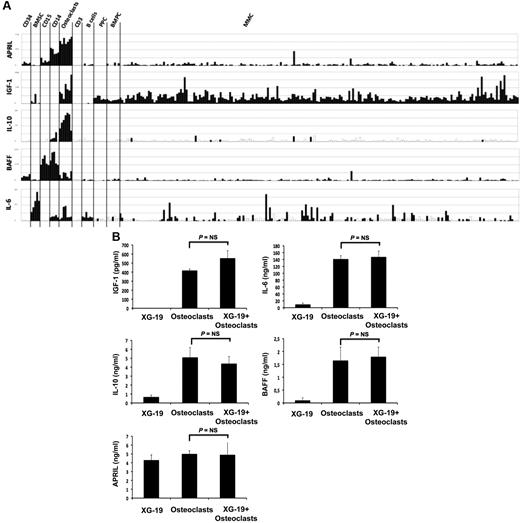
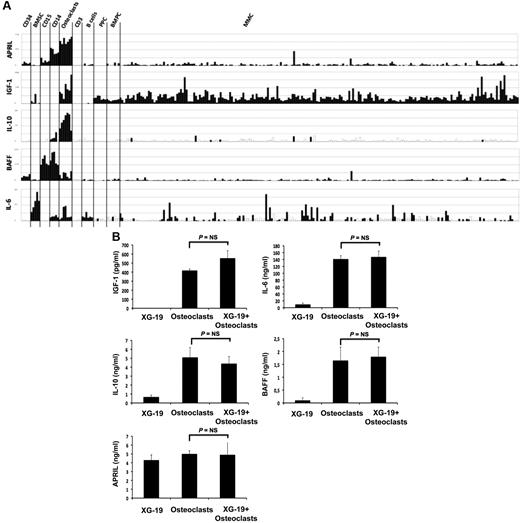
Osteoclasts overexpress genes encoding for previously reported MGFs, including IGF-1,33 IL-10,34 and APRIL32 (Figure 4A). IL-6 and BAFF genes are also highly expressed by osteoclasts but were not picked in the osteoclast gene list because they are also highly expressed by BM stromal cells (IL-6), monocytes, or polymorphonuclear cells (BAFF; Figure 4A). Osteoclasts produced IGF-1 or APRIL proteins. A total of 418 plus or minus 16 pg/mL of IGF-1 and 5 plus or minus 0.3 ng/mL of APRIL were detected in 3-day culture supernatants of osteoclasts (Figure 4B), concentrations that are active on myeloma cells.32,33 Osteoclasts also produced IL-6 (141 ± 13 pg/mL) and BAFF (1.6 ± 0.5 ng/mL) and produced a low amount of IL-10 (5.1 ± 1.1 pg/mL) compared with the known biologically active concentrations.35 A coincubation of osteoclasts with XG-19 MMC did not significantly affect the production of IGF-1, IL-6, APRIL, BAFF, or IL-10 (Figure 4B).
Gene expression of several myeloma cell growth factors. (A) Affymetrix APRIL, IGF-1, IL-10, BAFF, and IL-6 gene expression in BM CD34 cells (n = 5), BM stromal cells (n = 5), purified BM CD15 (n = 5), CD14 (n = 5) and CD3 cells (n = 5), osteoclasts (n = 7), normal memory B cells (n = 6), normal polyclonal plasmablasts (n = 7), normal BMPCs (n = 7), and purified myeloma cells from patients with MM (n = 131). (B) The concentrations of BAFF, APRIL, IL-10, IL-6, or IGF-1 were assayed with an enzyme-linked immunosorbent assay in 3-day culture supernatant of XG-1 and XG19 cells, osteoclasts, and XG-1/osteoclasts and XG-19/osteoclast cocultures. Results are the mean value of 3 independent experiments.
Gene expression of several myeloma cell growth factors. (A) Affymetrix APRIL, IGF-1, IL-10, BAFF, and IL-6 gene expression in BM CD34 cells (n = 5), BM stromal cells (n = 5), purified BM CD15 (n = 5), CD14 (n = 5) and CD3 cells (n = 5), osteoclasts (n = 7), normal memory B cells (n = 6), normal polyclonal plasmablasts (n = 7), normal BMPCs (n = 7), and purified myeloma cells from patients with MM (n = 131). (B) The concentrations of BAFF, APRIL, IL-10, IL-6, or IGF-1 were assayed with an enzyme-linked immunosorbent assay in 3-day culture supernatant of XG-1 and XG19 cells, osteoclasts, and XG-1/osteoclasts and XG-19/osteoclast cocultures. Results are the mean value of 3 independent experiments.
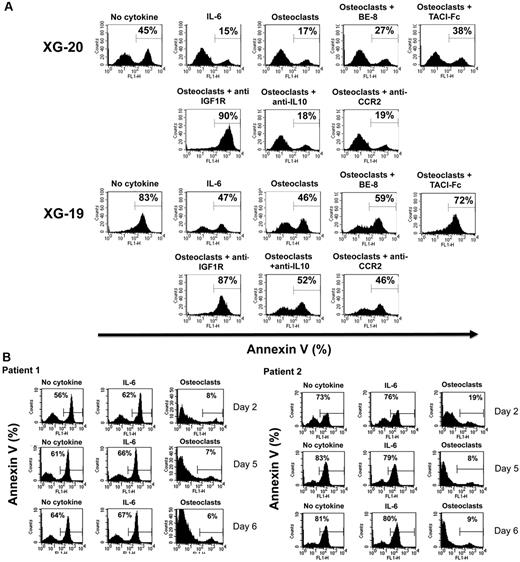
To investigate the role of these various growth factors in the survival activity of MMC induced by osteoclasts, we used the XG-19 and XG-20 HMCLs that expressed IL-6R, IL-10R, IGF1R, TACI, and BCMA BAFF/APRIL receptors. On IL-6 deprivation, XG-19 and XG-20 cells rapidly apoptosed, and a coculture with osteoclasts protected them from apoptosis (P = .01). Fluorescence-activated cell sorter data from one representative experiment are shown in Figure 5A and the mean values plus or minus SD of 5 experiments in Table 3. The survival-promoting activity of osteoclasts was partially inhibited blocking IL-6 using the B-E8 anti-IL-6 mAb (P = .05 for XG-19 and P = .01 for XG-20) or BAFF/APRIL using TACI-Fc fusion protein (P = .01 for XG-19 and P = .02 for XG-20). Inhibiting IGF-1 using an anti-IGF-1R mAb completely blocked MMC survival supported by osteoclasts in serum-free culture medium (P = .003 for XG19 and P = .0005 for XG20; Figure 5A). The anti-IGF-1R mAb further increased the apoptosis rate observed without osteoclasts, probably by inhibiting the autocrine IGF-1/IGF-1R loop occurring in some HMCLs.33 A neutralizing anti-IL-10 mAb had no significant effect. The anti-CCR2 mAb did not either inhibit the MMC growth activity of osteoclasts, but this anti-CCR2 mAb partially blocked the growth activity of recombinant CCL7 or CCL8 (supplemental Figure 4), whereas it blocked efficiently their chemoattractant activity (Figure 2B).
Osteoclasts promote the survival of cytokine-dependent myeloma cell lines. (A) XG-19 and XG-20 myeloma cells were cultured at 105 cells/mL without cytokine, or with IL-6 (2 ng/mL), or with 104 osteoclasts with or without anti–IL-6 (BE8) mAb (10 μg/mL), TACI-Fc (20 μg/mL), anti–IGF-1R mAb (4 μg/mL), anti–IL-10 mAb (10 μg/mL), or anti-CCR2 mAb (10 μg/mL). Cells were recovered after 3 days of culture, and apoptotic cells were detected by annexin V staining. Results are those of one experiment representative of 5. (B) Purified MMCs of patients were cultured at 105 cells/mL without cytokine or in the presence of osteoclasts (1 osteoclast for 4 MMC) or IL-6 (2 ng/mL). Cells were recovered after 2, 5, and 6 days of culture, and apoptotic cells were detected by annexin V staining.
Osteoclasts promote the survival of cytokine-dependent myeloma cell lines. (A) XG-19 and XG-20 myeloma cells were cultured at 105 cells/mL without cytokine, or with IL-6 (2 ng/mL), or with 104 osteoclasts with or without anti–IL-6 (BE8) mAb (10 μg/mL), TACI-Fc (20 μg/mL), anti–IGF-1R mAb (4 μg/mL), anti–IL-10 mAb (10 μg/mL), or anti-CCR2 mAb (10 μg/mL). Cells were recovered after 3 days of culture, and apoptotic cells were detected by annexin V staining. Results are those of one experiment representative of 5. (B) Purified MMCs of patients were cultured at 105 cells/mL without cytokine or in the presence of osteoclasts (1 osteoclast for 4 MMC) or IL-6 (2 ng/mL). Cells were recovered after 2, 5, and 6 days of culture, and apoptotic cells were detected by annexin V staining.
Osteoclasts promote the survival of primary MMC
Purified primary MMC rapidly apoptosed in vitro within 2 days36 and adding recombinant IL-6 did not prevent apoptosis. A coculture with osteoclasts completely prevented primary MMC from apoptosis on day 2, and this protective effect lasted until day 6 (Figure 5B). Given the rarity of primary MMC, it was not possible to add the inhibitors to the various growth factors produced by osteoclasts to identify those that are critical.
Discussion
The aim of this study was to identify some major cell communication signals involved in recruitment of MMC close to osteoclasts and subsequent support of MMC survival.4,37
We first identified 552 genes that are significantly overexpressed in osteoclasts compared with various cell components of the BM microenvironment and to MMC. Comparing our results with previous gene array analyses of osteoclasts, 22 common genes coding for secreted or extracellular matrix proteins were identified in the Pederson et al study38 and in our current study, in particular CCL7, CCL8, and CXCL5 chemokine genes and RANK, IGF-1, and IL-10 genes (supplemental Table 2). This stringent method was initially chosen to limit the size of the osteoclast gene list, being aware that some genes that are highly expressed both in osteoclasts and in another cell population could be eliminated. Our data show that this method is of interest to find the signals that may attract MMC specifically to osteoclasts and promote MMC survival.
There are approximately 50 chemokines,39 some of them being already found to be produced in the BM milieu of patients with MM: mainly CXCL12/SDF-1 (targeting CXCR4 and produced by stromal cells, endothelial cells, and myeloma cells) and MIP1α/CCL3 (targeting CCR1 and CCR5 and produced by osteoclasts, BM stromal cells, osteoblasts, monocytes, and myeloma cells29 ). These chemokines are important to recruit circulating MMCs into the BM as illustrated for CXCL12/SDF-1.40 They could be also critical to trigger the fate of a myeloma cell in the presence of gradients of multiple chemokines produced by various environment cells. The current data provide some answer regarding the chemokines targeting CCR1 or CCR2, both receptors being expressed by myeloma cells.28 These CCR1 or CCR2 chemokines are produced by osteoclasts, and we show that CCR1 and CCR2 chemokines contribute to osteoclast chemoattractant activity for myeloma cells in vitro, in agreement with previous data for CCR1 chemokines.29 In addition, we show that the expression of CCR2 chemokines is very specific to osteoclasts, whereas it was previously shown that various cells, including osteoclasts, produce the MIP-1α/CCL3 CCR1 chemokine.13,14 This suggests that a myeloma cell that expresses CCR1 could be recruited close to the various cells producing MIP-1α/CCL3 (ie, osteoclasts, stromal cells, monocytes, or myeloma cells). But if the myeloma cell expresses CCR2, it could be attracted by osteoclasts specifically. Given that osteoclast can recruit and promote survival of CCR2+ myeloma cells, and in turn, myeloma cells promote osteoclast recruitment and activation, it is hardly surprising that the bone lesion number in patients with newly diagnosed MM was associated with high CCR2 expression in myeloma cells, unlike CCR1 expression. This hypothesis should be further tested injecting CCR2+ or CCR2− myeloma cell lines in immunodeficient mice. If the specificity for human CCR2-targeting chemokine expression is kept by murine osteoclasts, one should expect that CCR2+ myeloma cell lines could create more bone lesions than CCR2− ones and that anti-CCR2 antibodies could prevent bone lesions and even tumor growth. Alternatively, this could be tested in the murine 5T2 MM model because these murine myeloma cells express CCR2.41
Chemokine receptors are G-protein-coupled receptors that can activate various signaling pathways depending on chemokine receptor and cell type: MAPK, PI-3/AKT, Src, JNK, and Rho.42,43 The CCR2 chemokines induced MAPK phosphorylation in myeloma cells and did not activate Akt or Stat3 pathways, in agreement with the inhibition of osteoclast chemoattractant activity for myeloma cells by the PD98059 MAPK inhibitor. High concentrations of recombinant CCR2 chemokines (≥ 125 ng/mL) can trigger the survival of CCR2+ MMC unlike CCR2− MMC.
Osteoclasts also highly expressed genes encoding for some major MGFs and produced high levels of these MGFs: IGF-1, APRIL, and IL-6 and produced the corresponding proteins. A high expression of IL-10 gene was found, but no protein production. In agreement with growth factor production, the survival of MMC induced by osteoclast was partly inhibited by IL-6 and APRIL inhibitors, indicating their contribution to osteoclast-induced MMC survival. An interesting finding is the complete inhibition of osteoclast-induced survival of MMC by the anti-IGF-1R mAb. This may be for 2 reasons. First, IGF-1 is a critical factor for the generation and survival of osteoclasts44 and the neutralizing anti-IGF-1R decreased osteoclast survival in our culture system (results not shown). Second, IGF-1 is a major MGF, both as a paracrine and autocrine growth factor. In particular, the MGF activities of IL-6, HGF, or EGF-family members are inhibited in part by IGF-1R inhibitors, being dependent of an autocrine IGF-1 loop produced by MMCs.33
Bone lesions in MM are the result of an abnormal bone remodeling with increased osteoclast and decreased osteoblast activities.37,45 In healthy persons, bone remodeling occurs in closed bone remodeling compartments (BRCs) containing osteoblasts and osteoclasts.46 These BRCs are protected from BM cells by a canopy of flat cells with osteoblastic markers.46-48 Red blood cells are found in the BRCs, and radiolabeled ferritin is detected in BRCs 5 minutes after injection.47,48 This indicated that BRCs are linked to the vasculature through capillaries that are directly connected to the BRC canopy.48 This connection of closed BRCs with the vasculature should ensure the recruitment of osteoblasts or osteoclast progenitors while protecting BRC cells from other BM cells. In patients with MM, the canopy of BRCs is frequently disrupted in association with the extent of bone lesions.48 This canopy disruption makes a direct contact possible between osteoclasts and MMC.
The current data suggest that osteoclasts can directly recruit MMC by producing CCR2 chemokines, promote MMC survival, growth, and drug resistance by producing various growth factors. Vice versa, MMC will further promote osteoclast progenitor recruitment and differentiation producing MIP1-α/CCL3, MIP-1β, and CXCL12 chemokines, IGF-1, and increasing RANKL production by stromal cells.4,37,45,49,50 Thus, osteoclasts and MMC can recruit each other and mutually promote their survival through various mechanisms. It will be interesting to investigate whether this strong cooperation between MMC and osteoclasts is a reflection of a cooperation occurring between osteoclasts and normal plasma cells. Indeed, CCR2 is a plasma cell marker, which is not expressed by B cells. The reason is that CCR2 gene expression is repressed by the Pax5 B-cell transcription factor.31 This negative control by Pax5 is removed when B cells differentiate into plasma cells because of the repression of Pax5 gene expression by the Blimp1 plasma cell transcription factor.23,31 After generation in the lymph node and exit into the circulation, plasma cells have to find a niche either in the BM or in mucosa. CXCR4 is one of the critical homing molecules for normal plasma cells to the BM by SDF-1-producing stromal cells.28 CCR2 could be another critical homing molecule of normal plasma cells toward osteoclasts producing CCR2 chemokines, providing a physiologic explanation of its specific induction in plasma cell differentiation. The concentration of CCR2 chemokines as well as plasma cell survival factors produced by osteoclasts (IL-6 and APRIL) should be high in the closed BRCs whose canopy is directly linked to a capillary, making the recruitment and survival of these normal plasma cells possible. Using an in vitro model of normal plasma cell generation,23 we have found that osteoclasts can promote the survival of normal plasma cells (Michel Jourdan, unpublished observations, March 2010).
In conclusion, we have identified an additional mechanism involved in the interaction of osteoclasts and MMC. Osteoclasts are the main cells in the BM environment that produce various CCR2 chemokines enabling malignant plasma cells attraction. Osteoclasts also produce the major growth factors for MMC (IGF-1, IL-6, and APRIL). Targeting the osteoclast/MMC interaction through CCR2 and/or IGF-1 appears to be a promising therapeutic approach in myeloma.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Ligue Nationale Contre le Cancer (équipe labellisée 2009), Paris, France, Institut National du Cancer (no. RPT09001FFA), and European Myeloma Stem Cell Network European strep (no. E06005FF), and in part by grants from the Hopp-Foundation (Germany); the University of Heidelberg (Heidelberg, Germany); the National Center for Tumor Diseases (Heidelberg, Germany); the Tumorzentrum Heidelberg/Mannheim, Germany; the Deutsche Krebshilfe (Bonn, Germany); the Deutsche Forschungsgemeinschaft Transregio TRR79 (Bonn, Germany); and Novartis Pharma, Nürnberg, Germany. Microarray experiments were performed at the Institute of Research in Biotherapy (http://irb.chu-montpellier.fr/en/laboratories_microarray.html) in the Montpellier University Hospital.
Authorship
Contribution: J.M. performed research and participated in the writing of the paper; A.K. completed Western blot experiments; D.H. and H.G. collected bone marrow samples and clinical data and participated in the writing of the paper; T.R. participated in the bioinformatics studies and participated in the writing of the paper; P.M. and G.R. provided with technical assistance; and B.K. participated in the design of the research and the writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bernard Klein, Inserm U847, Institute of Research in Biotherapy, Centre Hospitalier Universitaire Montpellier, Av Augustin Fliche, 34285 Montpellier cedex, France; e-mail: bernard.klein@montp.inserm.fr.