Abstract
Using proteins in a therapeutic context often requires engineering to modify functionality and enhance efficacy. We have previously reported that the therapeutic antileukemic protein macromolecule Escherichia coli L-asparaginase is degraded by leukemic lysosomal cysteine proteases. In the present study, we successfully engineered L-asparaginaseto resist proteolytic cleavage and at the same time improve activity. We employed a novel combination of mutant sampling using a genetic algorithm in tandem with flexibility studies using molecular dynamics to investigate the impact of lid-loop and mutations on drug activity. Applying these methods, we successfully predicted the more active L-asparaginase mutants N24T and N24A. For the latter, a unique hydrogen bond network contributes to higher activity. Furthermore, interface mutations controlling secondary glutaminase activity demonstrated the importance of this enzymatic activity for drug cytotoxicity. All selected mutants were expressed, purified, and tested for activity and for their ability to form the active tetrameric form. By introducing the N24A and N24A R195S mutations to the drug L-asparaginase, we are a step closer to individualized drug design.
Introduction
Proteins can be used as efficient drugs with high specificity and limited side effects. However, compared with the total number of known small-molecule drugs, the number of protein drugs is relatively low.1 Nevertheless, recent advances in protein engineering and design2-5 have facilitated the development of new protein drugs that are useful when small molecules do not provide sufficient specificity or cause significant side effects.6
In the present study, we focus on the engineering of Escherichia coli L-asparaginase (L-ASN), a protein drug widely used in the hematological malignancy acute lymphoblastic leukemia (ALL).7 The bacterial enzyme L-ASN, which is active as a homotetramer with 4 nonallosteric active sites, primarily hydrolyzes the amino acid asparagine to aspartic acid and ammonia.8-10 Additionally L-ASN hydrolyzes glutamine,11 although this activity varies according to the source of the enzyme.12 With one exception,8 a steep drop in both serum asparagine and glutamine amino acid levels has been observed 2 weeks after treatment with L-ASN preparations.13-15 When asparagine is depleted in the blood, protein synthesis in malignant lymphoblasts is compromised, leading to the failure of cellular functions and, eventually, apoptosis.16
L-ASN is a key component of chemotherapeutic regimens for childhood ALL. Unlike most other cytotoxic agents, side effects from L-ASN treatment are relatively mild. Nevertheless, because L-ASN is a bacterial protein, a major side effect is the development of hypersensitivity ranging from mild allergic reactions to potentially fatal anaphylaxis.17 The underlying development of antibodies is also known to be responsible for the so-called “silent inactivation,” which leads to a rapid decline in L-ASN activity due to the presence of neutralizing antibodies.17 In such cases, the enzyme Erwinase, derived from the bacterium Erwinia chrysanthemi, is often used as a substitute. However, Erwinase has different pharmacokinetic and immunogenic properties and a shorter half-life than L-ASN.18 L-ASN use is also associated with neurotoxicity,19,20 which can be attributed to its secondary glutaminase activity.11,21,22
We have previously reported that the lysosomal proteases cathepsin B and asparagine endopeptidase (AEP), which are produced by lymphoblasts, hydrolyze L-ASN, resulting in inactivation and exposure of epitopes implicated in the immune response. We determined that the stepwise cleavage of L-ASN by AEP originates at Asn 24 (N24). An N24G point mutant resisted AEP-mediated cleavage but showed decreased asparaginase activity.23
The aim of the present study was to create an AEP-resistant L-ASN mutant with enzymatic activity comparable to the native compound. We also examined the glutaminase/asparaginase activity ratio of the enzyme and explored mutations to modify this property. To achieve this, we used a unique combination of computational protein-engineering tools. High-scoring mutants from the in silico analysis were then experimentally verified.
Methods
Genetic algorithm: mutant sampling and selection
A genetic algorithm (GA) previously used for protein modeling24 was adapted to sample the sequence space of the lid-loop and interfaces of the protein tetramer complex. All operators within our original GA software were kept and an additional sequence point mutation operator plus the DCOMPLEX25 energy score were added. Molecular dynamics (MD) simulations were performed on the wild-type (WT) complex, providing several different starting conformations of the interface residues to be sampled using the GA. In total, 24 000 mutated complexes were created and a subset was defined for further investigation. To show a stable distribution of mutants, the complete GA-sampling protocol was repeated multiple times (for details, see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
MD preparation and simulation
In addition to the negative reference N24G and the WT, the 4 highest-scoring mutations were selected from GA sampling to study the proteins' dynamic and conformational features by MD simulations. These 4 mutants included N24A, N24T, N24H, and N24S D281S. The crystal structure of L-ASN was taken from the Protein Data Bank (entry 3eca).26 Each system was energy minimized and equilibrated for 50 ps at constant volume, and consequently at constant pressure, with backbone atoms restrained to their original positions in both cases. After the equilibration, each system was simulated for 10 ns at room temperature (298K) (for details, see supplemental Methods).
Analysis of MD trajectories
All MD simulations and analyses were performed with the Amber suite Version 10.27 The root mean square fluctuation (RMSF) was calculated for residues 6-38, which contain the flexible lid-loop. The χ1 angle of T12 was monitored to map the orientation of the nucleophilic hydroxyl group of T12 within the active site. The T12 orientation along each MD trajectory was analyzed and a time series plotted (Figure 2B). To generate Figure 2B, we averaged T12 orientations over complete trajectories and for all 4 monomers. In addition to standard MD analysis methods, we used internal coordinate fluctuations as a measure of flexibility. Furthermore, key distances and hydrogen bond counts within the active site were calculated. The free energy of binding was calculated using the MM-PBSA protocol28 (for details, see supplemental Methods). The cavity volume of all 4 monomers was measured along each trajectory using the software suite CAVER.29 Additionally, the compactness of the tetramer was calculated for each snapshot of the trajectory using the method described in the context of the GA.24
Creation and purification of L-ASN recombinants
L-ASN recombinants were created as described in Patel et al.23 Preparation of purified recombinant proteins and estimation of protein molecular weight and aggregation by SEC-MALLS (size exclusion chromatography coupled with multi-angle laser light scattering) were carried out by the Protein Production Division of the Technology Facility in the Department of Biology at the University of York, United Kingdom.
Estimation of asparaginase and glutaminase enzyme activity
Asparaginase enzyme activity of the L-ASN recombinants was determined using a quantitative chromogenic assay (Medac Asparaginase Activity Test [MAAT], Medac) as reported by Rizzari et al.30 Glutaminase enzyme activity of the L-ASN recombinants was assessed by continuous coupled spectrophotometry based on L-glutamine hydrolysis and L-glutamate oxidation, with L-glutamate dehydrogenase as an auxiliary enzyme and β-nicotinamide adenine dinucleotide as a redox reagent.
Cell viability-proliferation assays
Cell viability was analyzed by using the CellTiter96 AQueous One Solution Cell Proliferation Assay kit (MTS tetrazolium assay; Promega) following the manufacturer's protocol. The half-maximal inhibitory concentrations (IC50s) of WT and mutant L-ASN recombinants were tested at equivalent asparaginase activities of the compounds in the ALL cell lines REH, SupB15, and MV4:11. IC50 values were calculated using the ED50 Plus v1.0 online software. All experiments were performed in triplicate (for details, see supplemental Methods).
Statistical analysis
To demonstrate that the combined usage of the X-ray and 5 key structures derived from MD enriches the structural search space for possible point mutations, we compared several subsets of mutations generated by the GA, and investigated their frequency distributions with a t-distribution test. For each of 2 possible scenarios, X-ray only and X-ray + MD, 3 independent subsets, each consisting of 120 parallel GA runs, were generated. By comparing these subsets in pairs for each scenario, we were able to ascertain whether these were significantly different from each other, thus pointing to the depth of the search space (supplemental Table 4). Correlations for the 12···25 and 25···283 bonds were examined using Pearson χ2 tests with Yates correction. The statistical significance of differences in asparaginase and glutaminase activities and IC50 values of WT and mutant L-ASN compounds were evaluated using a one-sided independent samples Student t test. A P value of < .05 was considered statistically significant. The Pearson correlation coefficient was calculated to examine the correlation between glutaminase activity and drug cytotoxicity, as well as the correlation between computational predictors and asparaginase/glutaminase activity.
Results
Sampling of mutation space
A novel GA-based protocol, described in supplemental Methods, was used to preselect potential amino-acid substitutions for residue 24 and its main interacting partner, D281 (Figure 1A). A combination of different molecular force fields and conformational search techniques was used to find mutants maintaining the fragile network of the active site, as observed in the WT X-ray structure. Conformations exhibiting a sufficiently stable hydrogen bond network are shown in Figure 1B; 2000 new conformations/mutations were created in total. Due to limited computational resources, we chose the top 4 mutants (N24A, N24T, N24H, and N24S D281E), together with the WT and the N24G mutant, for extensive MD analysis.
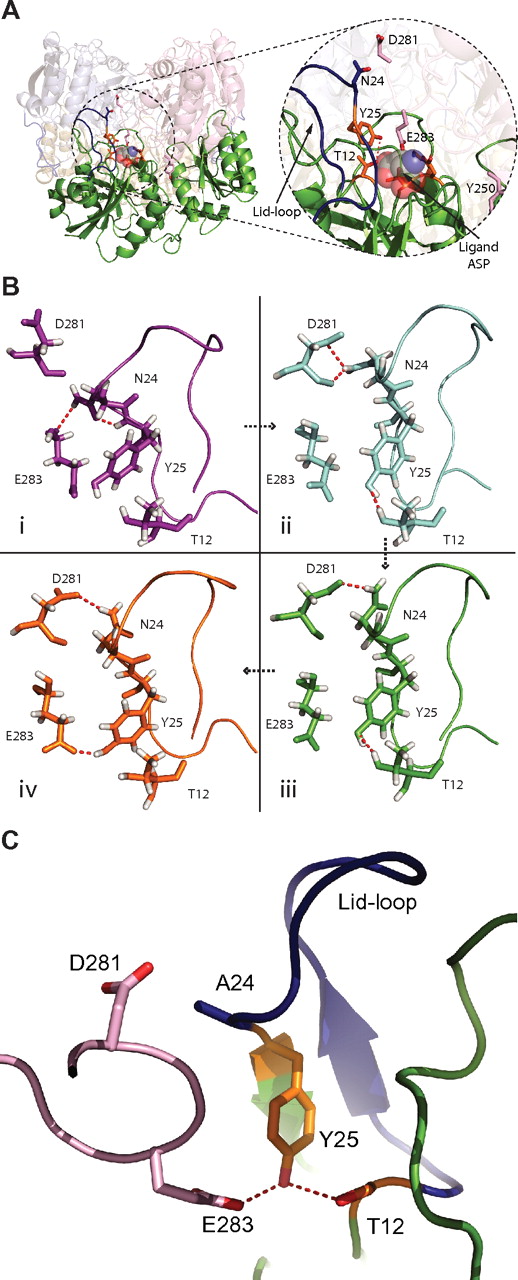
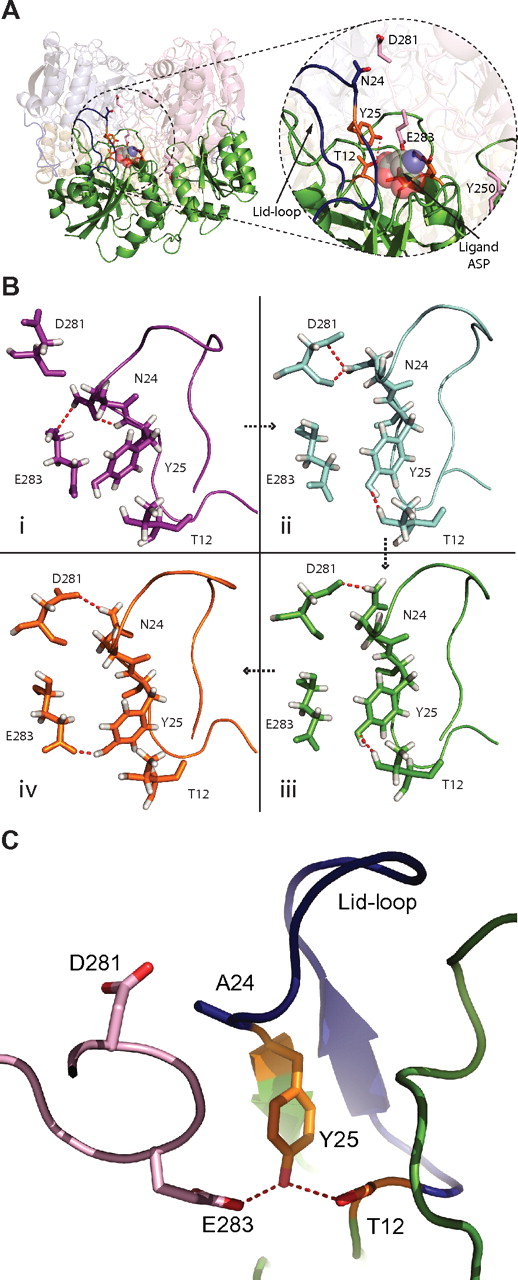
Close-up of the active site and high-occupancy hydrogen bonds during MD simulations. (A) Overall view of the L-ASN tetramer with focus on a single monomer (green). In the close-up, the active site (orange sticks) and lid-loop (blue) are shown, as well as important residues from the interacting second monomer (light pink). An aspartic acid molecule, in space fill, is shown within the active site. (B) Residues of the active site are shown in different conformations and con-figurations found throughout the MD simulation. Simulations I through IV are ordered in a time-dependent fashion denoted by arrows. (I) A more closed conformation for the active site lid-loop is shown, where a hydrogen bond (red) between N24 and E283 is found. (II) One of the main hydrogen bonds in the active site of the WT enzyme is found between D281 and N24. Furthermore, the very important stabilization of T12 by Y25 can be seen. (III) The hydrogen bond between D281 and N24 fluctuates and can be found in different conformations. (IV) Y25 can also form a stabilizing hydrogen bond to residue E283. (C) The active site and surrounding is shown for mutant N24A. The lid-loop is colored blue. D281 and E283 are colored light pink. Y25 (orange) makes a hydrogen bond (red dashed line) to E283 and the nucleophile T12 (orange). This conformation is found more often for the mutant N24A.
Close-up of the active site and high-occupancy hydrogen bonds during MD simulations. (A) Overall view of the L-ASN tetramer with focus on a single monomer (green). In the close-up, the active site (orange sticks) and lid-loop (blue) are shown, as well as important residues from the interacting second monomer (light pink). An aspartic acid molecule, in space fill, is shown within the active site. (B) Residues of the active site are shown in different conformations and con-figurations found throughout the MD simulation. Simulations I through IV are ordered in a time-dependent fashion denoted by arrows. (I) A more closed conformation for the active site lid-loop is shown, where a hydrogen bond (red) between N24 and E283 is found. (II) One of the main hydrogen bonds in the active site of the WT enzyme is found between D281 and N24. Furthermore, the very important stabilization of T12 by Y25 can be seen. (III) The hydrogen bond between D281 and N24 fluctuates and can be found in different conformations. (IV) Y25 can also form a stabilizing hydrogen bond to residue E283. (C) The active site and surrounding is shown for mutant N24A. The lid-loop is colored blue. D281 and E283 are colored light pink. Y25 (orange) makes a hydrogen bond (red dashed line) to E283 and the nucleophile T12 (orange). This conformation is found more often for the mutant N24A.
The GA was also used to sample potential mutations at the interfaces of the monomers that form the active tetramer. For this interface sampling, 24 000 new structures were produced. The mutant distribution of these structures can be seen in supplemental Figure 1, where the most populated residue can be identified as Y250. For this position, the most acceptable mutant was Y250L; Y250N and Y250Q were not applicable, because they would create new potential AEP-cleavage sites. For further analysis, we chose mutant Y250L and the next most frequent non-tyrosine–substituting mutant, R195S.
The calculated position-specific scoring matrix (PSSM) derived from a standard PSI-BLAST (position-specific iterative basic local alignment search tool)31 run using the WT L-ASN sequence is shown in supplemental Table 1. Using the PSSM as a reference for the naturally occurring amino-acid substitutions, the evolutionary likelihood of the mutants selected by our GA approach can be estimated. For example, it can be seen that for residue N24, the most conserved mutation was N24G, with a score of 49% in the calculated PSSM; N24A, the mutation selected by the GA, accounted for only 8% and N24T for only 3%. For D281, the second residue in the double mutant D281G had the highest conservation with 39%; D281E accounted for 13%. R195 had the highest conservation for itself, R (26%), and the score for R195S was only 4%. Finally, for Y250 the most common mutant according to the PSSM would be Y250P, with 42%; Y250L was not observed in homologous sequences at all (0%).
MD simulations
To understand the effect of these mutations on the stability of the lid-loop and the conformation of the active site, we performed MD simulations on WT L-ASN and 5 mutant proteins with variations in position 24 (N24G, N24A, N24T, N24H, and N24S D281E). The influence of interface mutants on glutaminase activity was studied, simulating the WT, N24A Y250L, and N24A R195S. The stability of the loop was measured by RMSF of the loop residues. Furthermore, conformations of key residues and networks in the region of the loop and active site were analyzed in the course of the MD simulations. For the interface mutants, we analyzed the active-site volume and the compactness of the tetramers. These analyses helped to identify structural features that are important for enzyme activity; details are given in the next 4 paragraphs.
Flexibility of the lid-loop
Residue N24 is part of a lid-loop sitting above the active site and serving as a closing lid. This loop is found in an open, helical and a closed, strand-like conformation.32 Furthermore, the region directly preceding the flexible loop includes residue T12, which is a member of the active site and initiates the nucleophilic attack on the substrate.32 The stable, closed conformation of the lid-loop is observed when a substrate molecule is present in the active site of the enzyme; T12 is only positioned correctly in this conformation. In the absence of substrate molecule in the active site, the lid-loop is highly mobile and is observed only rarely in crystal structures. Therefore, the correct balance between stability and flexibility must be maintained for correct functioning of the enzyme. If the lid-loop is too flexible or unstable, it may not be able to close properly and disrupt the orientation of T12, causing significant loss of enzymatic activity. In contrast, an excessively rigid lid-loop may hinder substrate entry into the active site.
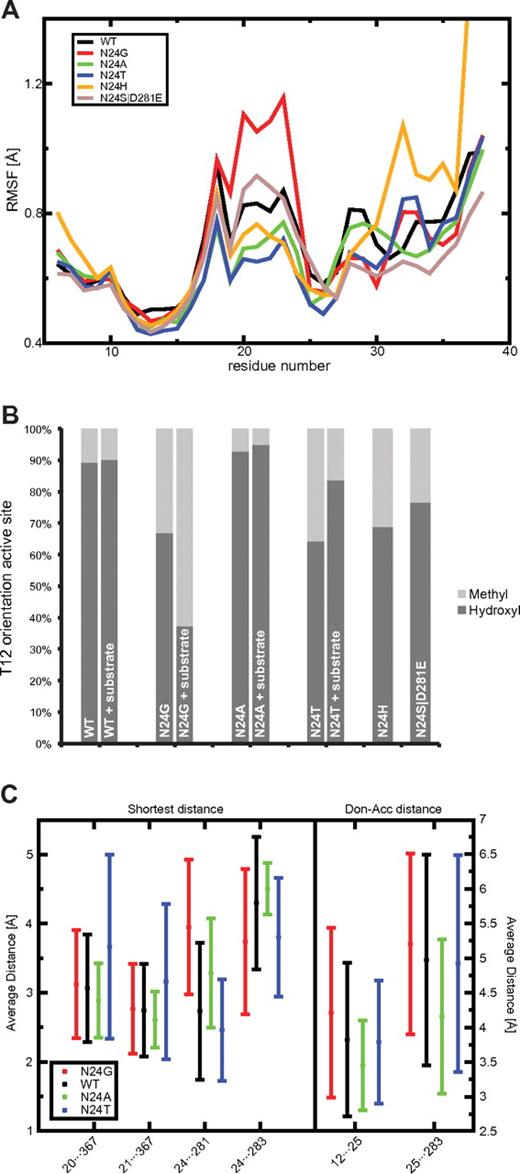
Calculating RMSF profiles from the MD simulations allowed the analysis of the lid-loop in WT L-ASN and the 5 N24 mutants. These RMSF profiles, averaged over the 4 monomers of each protein, are shown in Figure 2A. The N24G mutant had a much higher loop flexibility compared with those of WT and the other mutants, which clearly is one of the reasons for its decreased catalytic activity (Figure 3). On the contrary, N24A and N24T had very stable lid-loops, resulting in a tightly locked substrate molecule in the active site, stabilized for the catalytic reaction. The N24H mutant also displayed low flexibility in the central part of the loop; however, the C-terminal region of the loop showed high RMSF values that are likely to cause stability problems. The double mutant (N24S D281E) had a RMSF profile similar to that of WT, with a slight increase in flexibility for residues 20-24. These observations were further confirmed by the analysis of the Φ/Ψ angle deviations of the lid-loop (supplemental Figure 2).
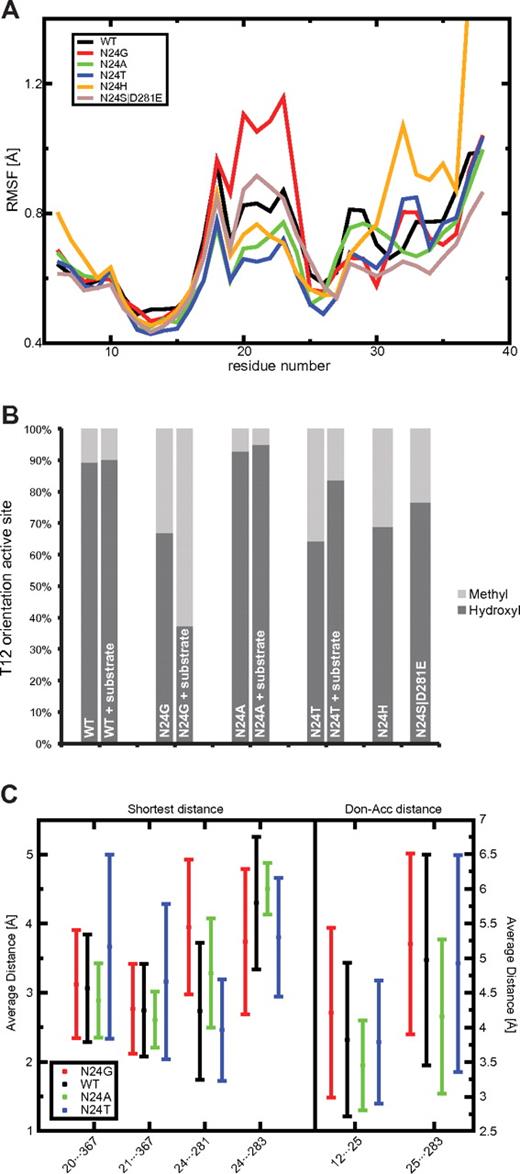
Lid-loop flexibility (RMSF), THR12 side-chain orientation, and distance analysis for important residues of the active site. (A) The RMSF for L-ASN WT (black) and 5 mutants are depicted in different colors calculated for the residue range 6-38. The lowest RMSF was found for N24T (blue) and N24A (green). N24H (yellow) shows a very high c-terminal RMSF. N24G (red) showed the highest RMSF of all L-ASN mutants. The double mutant N24S D281E (light brown) had a slightly increased RMSF compared with the WT. (B) The fraction of correctly placed hydroxyl groups toward the active site compared with the incorrect conformation involving the methyl group is shown. The highest fraction can be seen for N24A, the lowest for N24T and N24G. (C) Key distances measured in Å. The average and SDs are calculated for all snapshots found in the MD simulations of the WT (black) and the mutants N24G (red), N24A (green), and N24T (blue). Hydrogen bonds can be formed for distances around 2.8Å.
Lid-loop flexibility (RMSF), THR12 side-chain orientation, and distance analysis for important residues of the active site. (A) The RMSF for L-ASN WT (black) and 5 mutants are depicted in different colors calculated for the residue range 6-38. The lowest RMSF was found for N24T (blue) and N24A (green). N24H (yellow) shows a very high c-terminal RMSF. N24G (red) showed the highest RMSF of all L-ASN mutants. The double mutant N24S D281E (light brown) had a slightly increased RMSF compared with the WT. (B) The fraction of correctly placed hydroxyl groups toward the active site compared with the incorrect conformation involving the methyl group is shown. The highest fraction can be seen for N24A, the lowest for N24T and N24G. (C) Key distances measured in Å. The average and SDs are calculated for all snapshots found in the MD simulations of the WT (black) and the mutants N24G (red), N24A (green), and N24T (blue). Hydrogen bonds can be formed for distances around 2.8Å.
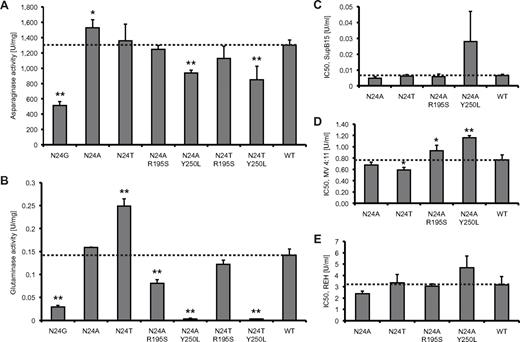
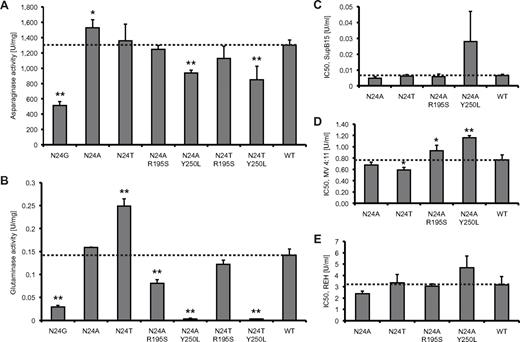
L-ASN activity and cell-kill assays. All experiments were conducted 3 times (n = 3). SDs were calculated for each protein. (A) Asparaginase activity measured for WT and mutant L-ASN. (B) Glutaminase activity for WT and mutant L-ASN. IC50 values were estimated for WT and mutants at asparaginase-equivalent doses of the compounds, thus reflecting the sole impact of modification of glutaminase activity on drug cytotoxicity. (C) Cytotoxicity assays on drug-sensitive SupB15 cells. (D) Cytotoxicity assays on intermediate drug-sensitive MV4:11 cells. (E) Cytotoxicity assays on drug-resistant REH cells. *P < .05; **P < .01.
L-ASN activity and cell-kill assays. All experiments were conducted 3 times (n = 3). SDs were calculated for each protein. (A) Asparaginase activity measured for WT and mutant L-ASN. (B) Glutaminase activity for WT and mutant L-ASN. IC50 values were estimated for WT and mutants at asparaginase-equivalent doses of the compounds, thus reflecting the sole impact of modification of glutaminase activity on drug cytotoxicity. (C) Cytotoxicity assays on drug-sensitive SupB15 cells. (D) Cytotoxicity assays on intermediate drug-sensitive MV4:11 cells. (E) Cytotoxicity assays on drug-resistant REH cells. *P < .05; **P < .01.
Orientation of T12
MD simulations also showed that mutations in position 24 changed the hydrogen bond network in the area around the active site and influenced the conformation of Y25, which has the important function of stabilizing the conformation of T12. Visual analysis of the T12···Y25 pair revealed that for the WT and the N24A mutant, the correct orientation of T12 was maintained, with the hydroxyl group pointing to the center of the active site. In contrast, for the N24G mutant (and to some extent N24T), Y25 was shifted and was not able to form hydrogen bonds with T12. As a result, the hydroxyl group of T12 was often replaced by the methyl group in the active site, which cannot perform a nucleophilic attack.
To describe the orientation of the T12 hydroxyl and methyl groups in the active site, we monitored the value of the χ1 dihedral angle along the MD trajectory for each mutant. The resulting time series, converted to binary plots (see “Analysis of MD trajectories”) are shown in Figure 2B (details in supplemental Figure 2C). Our results show that WT and the N24A mutant had the hydroxyl group present in the active site for the majority of the simulations. Conversely, the N24G and N24T mutants showed a lower occupancy of the correct hydroxyl group orientation, suggesting a lower stability of the active site. This explains why the N24T mutant, although it had the most stable lid-loop among all of the studied mutants, had a lower activity compared with the N24A mutant. For the N24G mutant, the low occupancy of the hydroxyl group and the decreased stability of the lid-loop explain its substantial reduction of activity. Interestingly, with the substrate bound, N24T was able to recover some of the correct T12 orientation, whereas this was not the case for N24G. The T12 occupancy of N24H was intermediate between N24T and WT. However, given the fact that N24H displayed significant lid-loop instability and that N24S D281E also showed suboptimal results, we decided to exclude these lid-loop mutants from further analysis. We therefore decided to only focus on WT, the N24G mutant as a negative reference, and the N24T and N24A mutant proteins.
Distance and analysis of active site residues
The stability of the active site is monitored by calculating the number of hydrogen bonds and the distances between several key residues in and around the active site along each MD trajectory. These residues include T12 and Y25, and Y25 is important for the correct orientation of T12.32 Distances between residues A20/T21 and Q367 describe the openness of the lid-loop. Interactions between N24···D281, N24···E283, and Y25···E283 stabilize the active site. Figure 2C shows the shortest side-chain distances for all of these pairs and the donor-acceptor distances for T12···Y25 and Y25···E283 (details in supplemental Figure 3) in all of the proteins studied. The hydrogen bond counts for T12···Y25, N24···Y25, N24···D281, N24···E283, and Y25···E283 are indicated in supplemental Table 2, and a graphical illustration is provided in Figure 1B. Calculating both the distances and counts allows the analysis of weak interactions that do not fall within the hydrogen-bond distance and angle cutoff (see “Analysis of MD trajectories”).
Regarding the distance analysis for the amino-acid pair T12 and Y25, the lowest distance was found for N24A, followed by N24T and then the others. WT showed a less stable T12···Y25 interaction, and the largest distance and lowest hydrogen bond count (supplemental Table 2) were observed for N24G. These observations were further confirmed by the results shown in supplemental Figure 3C. Lid-loop openness monitoring distances, A20···Q367 and T21···Q367, were smallest for N24A and showed the highest average value and deviation for N24T; WT and N24G behaved similarly.
As shown in Figure 2C and supplemental Table 2, the hydrogen bond between residues N24 and D281 was only maintained for WT and N24T; N24G and N24A lacked the donors and acceptors in the side chain of residue 24. In absolute numbers, compared with WT, N24T showed an increase of ∼37% in the capacity of the N24···D281 distance to maintain a formal hydrogen bond.
When using the hydrogen bond counts to calculate the correlation between T12···Y25, the important bond for correct orientation of T12, and Y25···E283, the bond compensating the loss of N24···D281 in N24A, we made several interesting observations. For WT and N24T we observed a significant anticorrelation (WT: P < .002, N24T: P < .006): it was more likely that the bonds were observed separately. There was no significant level of correlation associated with the 2 bonds found for N24G. Interestingly, a significant correlation was found for N24A (P < .001), which means that the 2 bonds are more likely to occur together. This analysis reveals a special configuration in N24A, which was locally more stable than in any of the other 3 cases, and likely contributed to the increased activity. This stabilizing network between T12, Y25, and E283 can be seen in Figure 1C.
Active-site cavity volume, tetramer compactness, and free energy of binding as a measure of glutaminase activity
Using the GA, 2 interface mutants were selected to increase the stability of the active tetramer: Y250L and R195S. These 2 mutants were combined with the lid-loop mutant N24A. The active site of L-ASN lies partly at the monomer-monomer interface, and therefore, changes in the interface packing will likely affect the activity of the enzyme. Because each active site is composed of 2 monomers and the entrance to the binding pocket lies within the interface, we assumed that tighter packing might decrease the cavity size and thus specifically prevent the entering of the larger Gln molecule, whereas access for the smaller Asn is not interrupted. Therefore, this optimization should specifically reduce glutaminase activity, while asparaginase activity may be affected to a lesser degree. Increased tetramer stability was assessed by monitoring the free energy of binding (supplemental Figure 4A) and compactness of the tetramer (supplemental Figure 4B), while altered properties of the active site likely to influence the glutaminase/asparaginase activity ratio were assessed by monitoring cavity size and flexibility (SD; supplemental Figure 4C). N24T resulted in the least compact tetramer; however, the cavity volume was comparable to WT, providing an explanation as to why N24T is predicted to have a higher glutaminase activity. In contrast, N24A Y250L had the smallest cavity with the least flexibility and the lowest tetramer compactness. This result was also partly confirmed by the free energy of binding, for which N24A Y250L had the lowest value of the 2 interface mutants, indicating that this mutant is likely to have substantially decreased glutaminase activity (supplemental Figure 4A). The cavity volume of the N24A R195S mutant was also smaller than in WT, indicating that this mutant might also have a decreased glutaminase activity, although to a lesser extent than that seen for the N24A Y250L mutant. These computational predictions were experimentally confirmed (see next paragraph).
Experimental confirmation
To confirm the computationally derived findings, the WT and mutant L-ASN proteins were expressed and the state of oligomerization and activity measured (supplemental Table 3, Figure 3). As we previously reported,23 N24G activity was reduced to approximately 40% of the WT; however, the oligomerization ability was not compromised in this mutant, with 95% of the compound occurring as tetramers, similar to that in the N24A mutant. The activity of N24A was higher than that of the WT, with a ratio of 1:1.17. However, N24T showed no significant change in activity compared with WT. As shown in Figure 3, only N24T had a significantly increased glutaminase activity; the double mutants N24A Y250L and N24T Y250L had barely detectable glutaminase activity. These double mutants also had reduced asparaginase activity (N24A Y250L, 72%, and N24T Y250L, 65%, compared with WT). In contrast, the N24A R195S double mutant had close to half of the WT glutaminase activity but a similar asparaginase activity. These experimental findings correlate well with modeling predictions; a correlation coefficient of 0.96 was observed between measured asparaginase activity and T12 hydroxyl group positioning, and 0.83 between the glutaminase activity and our compactness measure.
To investigate the effect of altered glutaminase activity on drug cytotoxicity, IC50 concentrations for WT and mutant L-ASN were estimated in leukemia cell lines (Figure 3). These IC50 values were determined using equivalent asparaginase activities of the compounds (Figure 4). Therefore, all IC50 values have been weighted by the measured asparaginase activities. Three leukemia cell lines were studied, SupB15, MV4:11, and REH, with, respectively, high, intermediate, and low sensitivity to L-ASN. The N24A mutant was the compound with the lowest average and most consistent IC50 values (on average, 79% of WT with ratios of 75%, 88%, and 75% in SupB15, MV4:11, and REH, respectively) and therefore the best cell-kill properties. Overall, the N24T and N24A R195S mutants were equivalent to WT L-ASN (averaged estimated IC50 compared with WT, 93% and 103%, respectively); however, both showed inconsistent results between the different cell lines and higher SDs than seen for N24A. N24T, with the highest glutaminase activity, had lower IC50 values compared with WT, but the average value was still higher than that of N24A. Halving the glutaminase activity did not adversely affect cytotoxicity, with IC50 values of the N24A R195S mutant similar to that of the WT compound. Importantly, the N24A Y250L mutant, with minimal glutaminase activity, was also the least potent cytotoxic L-ASN compound, with an overall IC50 of 243% compared with WT L-ASN. Accordingly, near-complete elimination of glutaminase activity was associated with reduced cytotoxicity. An overall Pearson correlation coefficient of 0.73 was observed between the modification in glutaminase activity and drug cytotoxicity, with the highest correlation observed in the intermediate-sensitive MV4:11 cells (MV4:11, 0.99; SupB15, 0.7; REH, 0.5). Thus, in MV4:11 cells, L-ASN mutants that varied significantly in glutaminase activity (N24T, N24A R195S, and N24A Y250L) also demonstrated significantly different cytotoxic efficacy.
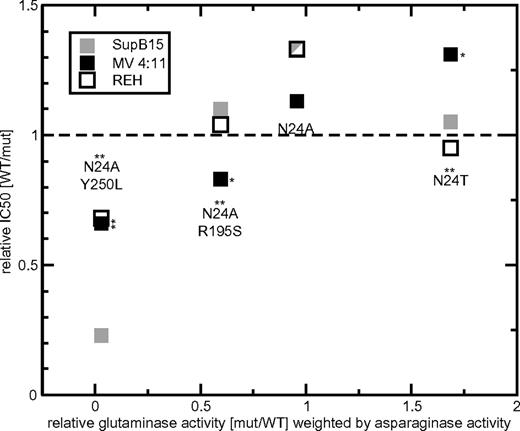
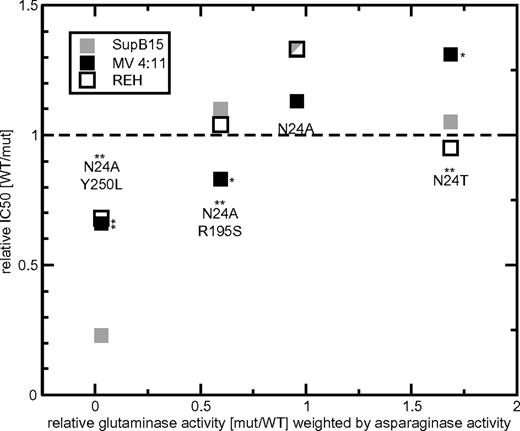
Relative glutaminase activity in the context of leukemic cell kill. In these cell-kill assays, the asparaginase activity was kept constant. For N24A, N24T, N24A R195S, and N24A Y250L, the relative glutaminase activity (x-axis) weighted by the asparaginase activity and relative IC50 (y-axis) values are shown separately. The cytotoxic activity of each mutant L-ASN is indicated for the leukemic cell lines SupB15 (very drug sensitive, ▩), MV 4:11 (intermediate drug sensitivity, ■), and REH (resistant, □). Values > 1 (dashed line) indicate IC50 values lower than WT compound, values equal to 1 indicate cytotoxicity equivalent to WT L-ASN, and values < 1 indicate a higher IC50 than WT. For each mutant, an asterisk above the identifier indicates significant differences in glutaminase activity. Significant relative differences in cytotoxicity of mutant L-ASN (compared with WT) are indicated by an asterisk on the right side of the data point. *P < .05; **P < .01.
Relative glutaminase activity in the context of leukemic cell kill. In these cell-kill assays, the asparaginase activity was kept constant. For N24A, N24T, N24A R195S, and N24A Y250L, the relative glutaminase activity (x-axis) weighted by the asparaginase activity and relative IC50 (y-axis) values are shown separately. The cytotoxic activity of each mutant L-ASN is indicated for the leukemic cell lines SupB15 (very drug sensitive, ▩), MV 4:11 (intermediate drug sensitivity, ■), and REH (resistant, □). Values > 1 (dashed line) indicate IC50 values lower than WT compound, values equal to 1 indicate cytotoxicity equivalent to WT L-ASN, and values < 1 indicate a higher IC50 than WT. For each mutant, an asterisk above the identifier indicates significant differences in glutaminase activity. Significant relative differences in cytotoxicity of mutant L-ASN (compared with WT) are indicated by an asterisk on the right side of the data point. *P < .05; **P < .01.
Discussion
We previously showed that L-ASN is degraded by 2 proteases, one of which, AEP, is predominantly expressed in lymphoblasts of high-risk patients. Furthermore, we were able to show that the stepwise cleavage of L-ASN by AEP originates at N24 in the L-ASN lid-loop, and can be blocked by a single amino-acid substitution in this position.23 In the present study, we identified the most suitable amino-acid substitution in the L-ASN lid-loop that maintained resistance to AEP cleavage while preserving or even improving L-ASN activity. Combining this substitution with a second mutation at the monomer interface of the active tetrameric L-ASN molecule allowed control over hydrolysis of the secondary substrate, glutamine. All mutants were identified using a GA to sample possible amino-acid substitutions; none of the described mutants could have been identified using an approach based on evolutionary amino-acid conservation. Flexibility and structural properties of WT and mutant L-ASN were investigated using MD simulations. Applying a combination of several structural descriptors to interpret the results of MD simulations helped to link the impact of mutations to asparaginase/glutaminase activity. Based on the descriptors, our predictions of mutant activities correlated well with experimental measurements of asparaginase and glutaminase activities, and also to a large extent with the results of cytotoxicity assays. From these observations, we propose that the L-ASN mutants N24A and N24A R195S show the best potential for further drug development.
A clear correlation between the flexibility of the lid-loop and the activity of different mutants was observed. For example, N24G had the highest flexibility observed among the mutations, but the lowest asparaginase activity. The 2 mutants with the most stable lid-loop, N24A and N24T, showed the highest asparaginase and glutaminase activities compared with the WT. Measuring the independent flexibility of the lid-loop in internal coordinate space, averaged over the 4 monomeric subunits, revealed that N24A had the lowest SD. Experiments showed that N24A was more active than N24T, although N24T was more stable in the lid-loop region than N24A. As an additional, complementing measure, we used the correct orientation of the nucleophile in the active site to help in predicting which of these 2 mutants had the higher asparaginase activity. This measure revealed that, indeed, for N24A the hydroxyl group of T12 was in the correct substrate-attack conformation most of the time, similar to WT—a trend not observed for N24T or the other lid-loop mutants.
In the context of the analysis, the N24A mutant had certain distinctive characteristics. N24A had the most stable hydrogen bond for the pair 25···283 and, unique to the N24A mutation, a significant direct correlation was observed between the hydrogen bonds of 25···283 and 12···25. These features markedly rearranged the active site in N24A. In contrast, there was a significant negative correlation between the occurrences of these pairs in WT and N24T, probably caused by a very stable 24···281 hydrogen bond. Indeed, the network associated with the mechanism can be described as a double lever, where hydrogen bonds between residues 24 and 281 on top of the active site slightly pulled out the residues below (which include Y25 and E283) and thus prevented a hydrogen bond between these residues. Consequently, N24T and WT could rarely simultaneously form 25···283 and 12···25 hydrogen bonds. N24A, on the other hand, formed both bonds, which imparted an improved, stabilizing network in the most crucial part of the active site. In support of these findings, we note the importance of the interaction between Y25 and T12, also demonstrated by Aung et al,32 who showed that any mutation of Y25 leads to a complete loss of enzyme activity.
The second objective of this study was to identify mutants in the interface of the L-ASN monomers that rearrange the packing close to the active site, thereby affecting the relative selectivity of asparagine/glutamine hydrolysis. Instead of random or evolutionary-based selection of potential mutants, we used a GA to identify energetically favorable mutations. For this, we created several thousand mutants with lower or equal energy to the WT and chose the ones with the highest occurrences, a principle that has been successfully used in protein design.33 Surprisingly, by far the residue with the highest count during all of the GA simulations was residue Y250. In this position, the mutant Y250L was most common, a mutation that is not likely to be observed in a PSSM for a stable homotetramer interface, due to the importance of tyrosine for solvation.34 It is important to note that Y250 lies next to N248, a residue for which mutations have been shown to cause a significant reduction of glutaminase and, to a lesser degree, asparaginase activity.35 N248 forms a hydrogen bond with the catalytic residue D90 across the monomer-monomer interface (supplemental Figure 5A) and is involved in the stabilization of the transition state during catalysis. The mutation N248A has been shown to destabilize the catalysis of the weaker glutamine substrate to a much higher degree than asparagine.35 However, there is no direct interaction between N248 and Y250, and no major conformational change in the vicinity of N248 can be seen with the Y250L mutation. Consequently, we suspect that the decreased glutaminase activity of the N24A Y250L mutant comes from small conformational changes at the interface that lead to tighter monomer-monomer binding and a subsequent decrease in volume. The second interface mutant selected was R195S. In L-ASN, 2 monomers form a tightly packed dimer; 2 of these dimers form the final tetrameric structure.36 Four R195 residues, one from each monomer, were situated at the interface of the 2 dimers, each interacting with a double strand on their opposite monomer. These large residues appear to act as spacers, preventing tight packing between monomers (supplemental Figure 5B). Therefore, once the amino acid Arg was replaced by the smaller Ser, this effect was reduced. Indeed, MD simulations revealed that both Y250L and R195S showed increased tetramer compactness and a decreased active site cavity, correlating well with the measured reductions in glutaminase activities.
Calculations for the free energy of binding indicated that the Y250L mutant had tighter monomer-monomer binding. However, it is important to stress that the sole use of free energy changes would neither have correlated particularly strongly with the prediction of mutation effects nor given us useful insights into the structural mechanisms of this protein drug. Thus, we believe that multiple structural descriptors are needed for a clear understanding of the effect of interface mutants in L-ASN, and the descriptors listed in “Active-site cavity volume, tetramer compactness, and free energy of binding as a measure of glutaminase activity” provide a firm base for further improvements.
Although the glutaminase activity of L-ASN has been linked to therapeutic toxicity, its effect on the leukemic cell has been debated.8 Our data show that N24A, with stable glutaminase activity, appears to be the most effective drug. N24A R195S, which had half of the glutaminase activity of the WT, nevertheless had comparable asparaginase activity and cytotoxicity. N24A Y250L, which had no glutaminase activity, had significantly reduced cytotoxicity when compared with WT. Thus, some glutaminase activity is required to induce apoptosis in cells already affected by a substantial lack of asparagine. Although glutamine is a nonessential amino acid, it appears to be essential to cancer cells as a major source of energy, nitrogen, and carbon.37,38 It is possible that glutamine metabolism links with the mitochondrial tricarboxylic acid cycle to maintain the availability of metabolic intermediates critical for cancer cell survival and proliferation. Indeed, glutamine addiction in cancer cells represents a potential therapeutic target.39 This may explain why, at least in vitro, L-ASN depletion of exogenous glutamine enhances cytotoxicity. Because glutaminase activity is also associated with therapeutic toxicity, a compound with a lower enzymatic activity but comparable cytotoxicity has potential clinical application.
We altered L-ASN properties to go beyond evolution and optimize this drug for different treatment scenarios. First, the drugs were engineered to resist degradation and inactivation by AEP produced by leukemic blast cells. We designed 2 mutants that can be exchanged during cancer therapy, according to the needs and observed side effects of the patient, either reducing the dosage and thus the antigenicity (N24A), or reducing the glutaminase activity and thus potentially reducing toxicity (N24A R195S). Also, we show for the first time that secondary glutaminase activity is required for the action of the drug. This was achieved by only looking at one species of L-asparaginase, as opposed to previous studies comparing the activities and cancer cell-kill ability of homologous asparaginase proteins. Our newly introduced L-ASN mutants N24A and N24A R195S show potential for therapeutic use and our results should form a sound basis for future clinical trials.
Future directions include further molecular optimization of the drug, such as humanization of the antigenic epitopes to reduce allergic reactions and modification of subunit aggregation to enable increased drug access in solid tumors. A combination of these different aims will enable us to create a tailored cancer drug with maximum effectiveness and minimum side effects. To target solid tumors, the ultimate aim is to combine the above enhancements of the drug with tissue-specific antibodies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank all of the members of the Biomolecular Modelling Laboratory and the Children's Cancer Group for many useful discussions and insights. We thank the Protein Production division of the Technology Facility in the Department of Biology at the University of York for preparation of purified L-ASN recombinants.
This work was funded by program grants from Cancer Research UK (to P.A.B. and V.S.), the Barbara Mary Hill Memorial Fund, a B'nai B'rith Leo Back Lodge Scholarship, and a Minerva Fellowship awarded to M.N.O.
Authorship
Contribution: M.N.O., M.K., V.S., and P.A.B. designed and devised the study and wrote the manuscript; M.N.O. and M.K. performed and analyzed the computational experiments; N.P., S.K., J.L., and V.S. designed and performed the experimental work; M.N.O. and M.K. made the figures; and all authors read and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for N.P. is Nephro-Urology Unit, University College London Institute of Child Health, London, United Kingdom.
Correspondence: Paul Alan Bates, Biomolecular Modelling Laboratory, Cancer Research UK London Research Institute, Lincoln's Inn Fields Laboratories, London, WC2A 3LY, UK; e-mail: paul.bates@cancer.org.uk; and Vaskar Saha, Cancer Research UK Children's Cancer Group, School of Cancer and Enabling Sciences, Manchester Academic Health Science Centre and Central Manchester University Hospitals, NHS Foundation Trust, University of Manchester, Wilmslow Rd, Manchester M20 4BX, UK; e-mail:vaskar.saha@manchester.ac.uk.
References
Author notes
M.N.O. and M.K. contributed equally to this study.