Abstract
The stromal cell–derived factor-1 (SDF-1)/chemokine C-X-C receptor 4 (CXCR4) axis plays a critical role in homing and engraftment of hematopoietic stem/progenitor cells (HSCs) during bone marrow transplantation. To investigate the transcriptional regulation provided by this axis, we performed the first differential transcriptome profiling of human cord blood CD34+ cells in response to short-term exposure to SDF-1 and identified a panel of genes with putative homing functions. We demonstrated that CD9, a member of the tetraspanin family of proteins, was expressed in CD34+CD38−/lo and CD34+CD38+ cells. CD9 levels were enhanced by SDF-1, which simultaneously down-regulated CXCR4 membrane expression. Using specific inhibitors and activators, we demonstrated that CD9 expression was modulated via CXCR4, G-protein, protein kinase C, phospholipase C, extracellular signal-regulated kinase, and Janus kinase 2 signals. Pretreatment of CD34+ cells with the anti-CD9 monoclonal antibody ALB6 significantly inhibited SDF-1–mediated transendothelial migration and calcium mobilization, whereas adhesion to fibronectin and endothelial cells was enhanced. Pretreatment of CD34+ cells with ALB6 significantly impaired their homing to bone marrow and spleen of sublethally irradiated NOD/SCID (nonobese diabetic/severe combined immune-deficient) mice. Sorted CD34+CD9− cells displayed lower bone marrow homing capacity compared with that of total CD34+ cells. CD9 expression on homed CD34+ cells was significantly up-regulated in vivo. Our results indicate that CD9 might possess specific functions in HSC homing.
Introduction
Homing of hematopoietic stem/progenitor cells (HSCs) to their bone marrow niches is crucial to successful transplantation. This multistep process starts with rolling and tethering of HSCs to bone marrow sinusoidal endothelial cells, followed by migration through the endothelium and extracellular matrix barrier to engage their bone marrow niches.1,2 The molecular mechanism controlling HSC homing is still not fully understood. Experimental evidence suggests that it requires the orchestrated action of chemokines,3,4 adhesion molecules,5,6 and proteolytic enzymes.7,8 Signaling provided by the interaction of stromal cell-derived factor-1 (SDF-1) with its receptor, chemokine CXC receptor 4 (CXCR4), plays essential roles in regulating HSC homing. Mice lacking SDF-1 or CXCR4 are severely defective in seeding of stem cells in the bone marrow and in the establishment of B-lymphopoiesis and myelopoiesis during development.9-11 In vitro, SDF-1 induces chemotactic and transendothelial migration,12 adhesion to extracellular matrix proteins under static or shear stress conditions,5 actin polymerization,13 and calcium flux12 in human CD34+ cells. Moreover, homing and engraftment of transplanted CD34+ cells in NOD/SCID (nonobese diabetic/severe combined immune-deficient) mice are greatly impaired by neutralization of CXCR4 or desensitization by high doses of SDF-1.3,14 However, CXCR4−/− fetal liver cells are capable (albeit at a lower level) of engraftment in the bone marrow of wild-type mice,15,16 suggesting that HSC homing and repopulation might not be exclusively controlled by the SDF-1/CXCR4 axis.
We previously demonstrated that a short exposure of human cord blood–derived CD34+ cells to a peptide analog of SDF-1 enhances their engraftment in the NOD/SCID mice model.17 Similarly, others have reported that homing of human or murine HSCs could be significantly improved by pretreatment with the sphingosine 1-phosphate receptor agonist FTY720,18 extracellular nucleotides,19 prostaglandin E2,20 or CD26 inhibitors.21 Thus, the identification of molecular mediators governing stem cell homing may lead to new strategies for improving the efficacy of stem cell transplantation. This application will be particularly beneficial for cord blood transplantation, in which cell dose remains a major limiting factor.
CD9, a member of the tetraspanin superfamily, has been implicated in regulating various physiological processes, including cell motility, adhesion, and fusion.22 In several types of cancer, the loss of CD9 expression is associated with tumor progression and metastasis.23-26 In human dendritic cells27 and mast cells,28 CD9 is involved in chemokine-induced migration. In the hematopoietic progenitor compartment, CD9 was shown to be heterogeneously expressed on human bone marrow–derived CD34+ cells,29 and was reported to play a role in murine myeloid differentiation30 and human megakaryocytic differentiation.29 However, to our knowledge, there has been no evidence on the role of CD9 in the homing-related functions of primary CD34+ cells. In general, tetraspanins have the ability to recruit transmembrane and cytosolic proteins to form structures known as tetraspanin-enriched microdomains in which the functions of the associated partners are modulated.22 Some of the known CD9-associating molecules,31 including β1 integrin,5 MT1-MMP (membrane type 1–matrix metalloproteinase),8 and CD26,21 are known to play crucial roles in HSC motility and homing.
In the present study, we identified CD9 as one of the SDF-1–responsive genes in cord blood CD34+ cells using the microarray approach. We further characterized the signaling pathway along the SDF-1/CXCR4 axis that led to CD9 expression, and examined its roles in in vitro and in vivo functional assays of migration and homing of CD34+ cells.
Methods
Cells
Human cord blood samples from full-term deliveries were obtained with informed written consent and in accordance with procedures approved by the Ethics Committee for Clinical Research of The Chinese University of Hong Kong. Low-density mononuclear cells were isolated by density gradient centrifugation over Ficoll-Paque Plus (Amersham). CD34+ cells were enriched using the indirect microbead kit (MiniMACS, Miltenyi Biotec) according to the manufacturer's instructions. The purity of enriched CD34+ cells was 93.7% ± 0.5% (range, 84.4%-98.4%; n = 45).
Human umbilical vein endothelial cells (HUVECs) were purchased from Cascade Biologics and grown in Medium 200 supplemented with low serum growth supplement (Cascade Biologics).
Microarray analysis
Highly purified CD34+ cells (> 95% purity, n = 4) were cultured for 4 hours in the absence or presence of 100 ng/mL of recombinant human SDF-1α (R&D Systems). Total RNA was isolated using TRIzol reagent (Invitrogen) and was further purified with the RNeasy Micro Kit (QIAGEN). RNA integrity was determined using the RNA 6000 Pico LabChip kit (Agilent Technologies) and quantified with an ND-1000 spectrophotometer (Nanodrop Technologies).
Amplification and labeling of RNA, hybridization to the HG-U133 Plus 2.0 GeneChip, staining, and scanning were performed according to manufacturer's protocols (Affymetrix), using 100 ng of total RNA from each sample.
The GeneChip Operating Software absolute analysis algorithm was applied to determine the amount of transcripts (signal) using the global scaling option. The algorithm-generated data were then subjected to per-chip and per-gene normalization using GeneSpring v10.0 (Agilent Technologies) normalization algorithms. Probe sets showing an absent call in all conditions were excluded. Additional filtering procedures were performed to remove poorly and inconsistently changed genes using GeneSpring software (fold change cutoff = 1.5 between untreated and SDF-1–treated samples; paired t test, P value cutoff = .05).
DAVID 2.0 (National Institute of Allergy and Infectious Diseases, http://david.abcc.ncifcrf.gov/) was used for functional annotation of differentially expressed genes, and TIGR MeV (MultiExperiment Viewer version 2.2, http://www.jcvi.org/) was used for hierarchical clustering analysis.
Quantitative polymerase chain reaction
Total RNA (20-100 ng) was reverse transcribed using the High Capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative polymerase chain reaction (qPCR) reactions were performed with Taqman universal PCR master mix and Taqman gene expression assays (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) on the Prism 7300 real-time PCR System according to the manufacturer's instructions (Applied Biosystems). The relative expression level of each target gene was calculated by the comparative CT method and was normalized to glyceraldehyde 3-phosphate dehydrogenase expression.
Characterization of CD9 and CXCR4 expression
Enriched CD34+ cells were stimulated with SDF-1 (100 ng/mL) for 1-24 hours or with protein kinase C (PKC) activators for 4 hours in Iscove modified Dulbecco medium (IMDM; Invitrogen) with 20% fetal calf serum (FCS; StemCell Technologies) at 37°C. In some experiments, cells were preincubated with pharmacological inhibitors at the indicated concentrations for 1 hour at 37°C prior to SDF-1 stimulation. AMD3100, pertussis toxin, and chelerythrine chloride were obtained from Sigma-Aldrich. GF 109203X, PD 98 059, D609, U-73 122, AG490, and wortmannin were products of Calbiochem. Phorbol 12-myristate 13-acetate (PMA), mezerein (MEZ), and ingenol 3,20-dibenzoate (IDB) were purchased from Enzo Life Sciences. The cell-surface expression of CD9 and CXCR4 was assessed by staining with anti-CD9–fluorescein isothiocyanate (anti-CD9–FITC; M-L13) and anti-CXCR4–phycoerythrin (anti-CXCR4–PE; 12G5), together with anti-CD34–PerCP-Cy5.5 (8G12) and anti-CD38–APC (HB7) antibodies for 30 minutes at 4°C. All antibodies were products of BD Biosciences unless specified otherwise. Cells were washed and acquired on a FACSCalibur (BD Biosciences). Data were analyzed by CellQuest Version 3.1 (BD Biosciences) or FlowJo Version 7 software (TreeStar).
Detection of phosphorylated extracellular signal-regulated kinase and Akt
CD34+ cells were stimulated with SDF-1 (200 ng/mL) in IMDM with 20% FCS at 37°C for 1 minute, and then fixed and permeabilized using the Phosflow fixation/permeabilization kit (BD Biosciences) according to the manufacturer's protocols. Cells were stained with anti-extracellular signal-regulated kinase 1/2 (anti-ERK1/2)–PE (pT202/pY204) or anti-Akt–Alexa Fluor 488 (pS473) for 30 minutes at room temperature, washed, and analyzed by flow cytometry.
Migration assay
Chemotaxis and transendothelial migration experiments were performed using Transwells (6.5-mm diameter; 5-μm pore; Costar, Corning). Enriched CD34+ cells were cultured for 4 hours in the presence of 10 μg/mL of neutralizing anti-CD9 monoclonal antibody (mAb; clone ALB6; Beckman Coulter) or an isotypic control antibody (mouse immunoglobulin 1 [IgG1]; R&D Systems), washed, and resuspended in IMDM with 1% FCS. These cells (6 × 104 to 1 × 105 in 0.1 mL of assay medium) were seeded in the upper chamber, and 0.6 mL of medium containing 100 ng/mL of SDF-1 was added to the bottom chamber. After 4 hours at 37°C, migrated cells were enumerated with a hematocytometer. For transendothelial migration, HUVECs (2 × 104 cells) were grown to confluence on the upper chamber precoated overnight with 10 μg/mL of fibronectin (Sigma-Aldrich). HUVECs were preactivated with 20 ng/mL of tumor necrosis factor-α (TNF-α; Peprotech) for 6 hours, and washed before being subjected to migration assay. A pilot study using annexin V/7-amino-actinomycin D (7-AAD; BD Biosciences) staining indicated that the anti-CD9 mAb did not induce apoptotic cell death.
Adhesion assay
Adhesion of CD34+ cells to fibronectin was performed in high-binding 96-well plates (Costar) as described by Basu and Broxmeyer32 with minor modifications. Briefly, plates were coated overnight with 20 μg/mL of fibronectin at 4°C, followed by incubation for 2 hours at 37°C. Nonspecific binding was blocked by 2.5% bovine serum albumin (BSA; Sigma-Aldrich) for 1 hour. CD34+ cells were labeled with 5μM 5-(6)-carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes) for 15 minutes at 37°C, and allowed to adhere to fibronectin for 45 minutes at 37°C (1 × 105 cells/well) in the absence or presence of SDF-1 (100 ng/mL). Nonadherent cells were removed by 3 washes with prewarmed PBS (Invitrogen). Adherent cells from 5 random fields were counted by fluorescence microscopy (model IX71; Olympus). Cellular adhesion assays were performed on monolayers of HUVECs. Briefly, HUVECs (1-2 × 105) were seeded on 96-well plates and allowed to grow to confluence. Adhesion assays of CD34+ cells were performed as described above. HUVECs were activated by 20 ng/mL of TNF-α for 6 hours, and washed before the addition of CD34+ cells.
Calcium flux
Intracellular free Ca2+ was measured by flow cytometry as described in Aiuti et al12 with minor modifications. Briefly, CD34+ cells were resuspended in calcium assay buffer (Hanks buffer containing 20mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 0.2% BSA, pH 7.4; Invitrogen) and loaded with 4μM Fluo-3AM (Molecular Probes) in the presence of 0.04% pluronic acid (Molecular Probes) for 30 minutes at 37°C. After a 1:5 dilution with assay buffer followed by incubation for 40 minutes, cells were washed twice, and further incubated for 15 minutes at 37°C. Basal fluorescence in each sample was measured by flow cytometry. After stimulation with SDF-1 (100 ng/mL), the change in Fluo-3 fluorescence levels was monitored for 3 minutes.
Actin polymerization
CD34+ cells were stimulated with SDF-1 (100 ng/mL) in IMDM with 0.2% BSA at 37°C for 15-60 seconds. Reactions were stopped by adding 3 volumes of fixation/permeabilization solution (BD Biosciences), and washed twice with Perm/Wash buffer (BD Biosciences). Cells were stained with phalloidin-FITC (0.4μM; Sigma-Aldrich) for 30 minutes at 37°C, washed, and analyzed by flow cytometry.
In vivo homing assay
NOD.CB17-Prkdcscid/J (NOD/SCID) mice were purchased from The Jackson Laboratory and bred and maintained in the Laboratory Animal Services Center at The Chinese University of Hong Kong. All experiments were approved by the Animal Research Ethics Committee of The Chinese University of Hong Kong. Eight- to 11-week-old mice were sublethally irradiated (137Cs; 375 cGy; Gammacell 1000 Elite Irradiator, MDS Nordion), and intravenously injected with CD34+ cells (1.9 × 105 to 5 × 105 cells/mouse) 24 hours after irradiation. Prior to transplantation, cells were incubated with ALB6 or mouse IgG1 for 4 hours at 37°C. In some experiments, mice were intraperitoneally administered with 200 μg of anti-mouse CD122 mAb (clone TM-β1; Serotec) immediately after irradiation,33 to deplete natural killer cells and macrophages.34 Single-cell suspensions were prepared from bone marrow (2 femurs) and spleen 20 hours after cell infusion, and red blood cells were lysed in buffer EL (QIAGEN). Human cells were detected by flow cytometry using human-specific antibodies (anti-CD34–FITC, anti-CD9–PE, and anti-CD45–APC). Fc receptors were blocked by anti–mouse CD16/CD32 mAb (clone 2.4G2, BD Biosciences) and human serum (Sigma-Aldrich). Dead cells were identified by 7-AAD staining and were excluded from analysis. For enrichment of subpopulations, CD34+ cells were stained with anti-CD34–FITC and anti-CD9–PE or isotypic IgG1-PE antibodies. They were sorted for CD34+CD9− cells or total CD34+ cells, respectively, by FACSVantage (BD Biosciences), obtaining a purity of > 98%. Sorted CD34+CD9− or total CD34+ cells (8.7 × 104 to 2.1 × 105 cells/mouse) were transplanted into NOD/SCID mice. A pilot study demonstrated that the anti-CD9–PE antibody used for identification of the CD34+CD9+ subpopulation possessed neutralizing effects on homing functions (data not shown). Therefore, this subpopulation was not included in the in vivo homing assay.
Statistics
The paired t test was used for determination of statistical significance between groups, in which P values of .05 or less were considered significant. All data were expressed as means ± SEM.
Results
SDF-1 induces functionally relevant transcriptional changes in cord blood CD34+ cells
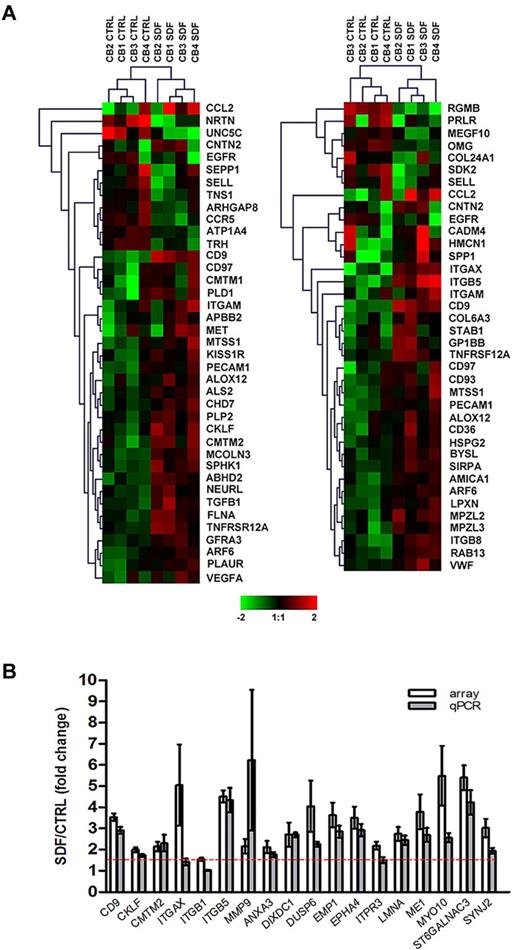
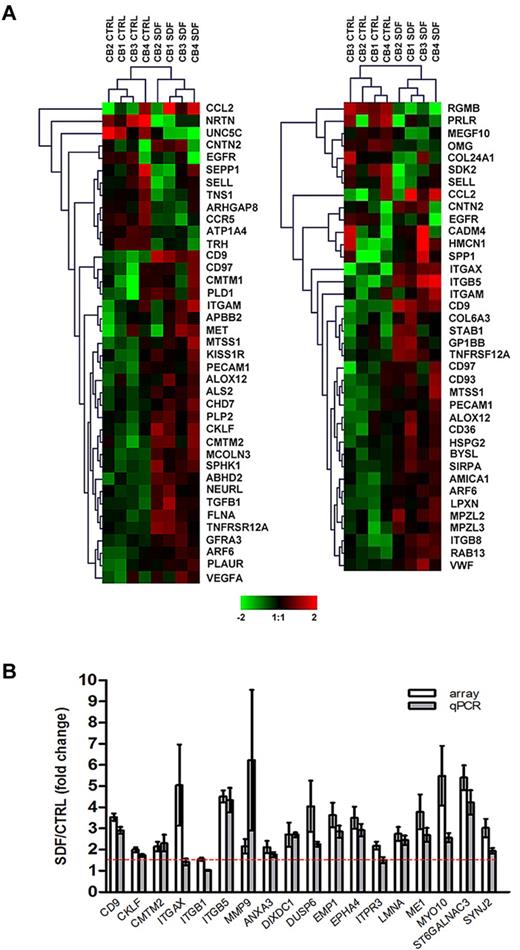
Of over 26 000 probe sets, exposure of CD34+ cells to SDF-1 resulted in the differential expression of 745 transcripts (P < .05), corresponding to 477 well-characterized genes with a minimum of 1.5-fold change compared with control, untreated cells. Consistent with observations that SDF-1 regulates motility12 and adhesion5 of CD34+ cells, functional annotation of the SDF-1–modulated gene set identified 39 and 38 differentially expressed genes in these categories, respectively (P < .001; supplemental Table 2; Figure 1A). Genes involved in other functions such as cell proliferation, hematopoiesis, and actin cytoskeleton organization were also significantly modulated by SDF-1 (P < .05; supplemental Table 2). To validate the microarray data, 18 genes were selected from the SDF-1–modulated gene set based either on their putative role in HSC homing (first 7 genes in Figure 1B) or the magnitude of expressional changes for further assessment by qPCR. Using a 1.5-fold cutoff, differential expressions of 16 genes (89%) were confirmed (P < .01; Figure 1B). The microarray data have been deposited in a public functional data repository (Gene Expression Omnibus, National Center for Biotechnology Information, accession no. GSE20891; supplemental Table 3).
Microarray analysis and validation. (A) Differential gene expression of cord blood CD34+ cells (n = 4) in response to SDF-1 (100 ng/mL) was analyzed by Affymetrix HG-U133 Plus 2.0 GeneChip software. Hierarchical analysis of the SDF-1–modulated genes involved in cell motility (left panel) and cell adhesion (right panel) were performed using the Euclidean distance metric. The color scale represents log2 normalized signal intensity. CTRL, control. (B) Differential expression of 18 SDF-1–induced genes was validated by qPCR in CD34+ cells derived from the same cord blood samples used in microarray experiments (n = 4) plus CD34+ cells from an independent cohort of cord blood (n = 4-6). Gene expression levels were calculated by the comparative CT method and are represented as the fold change of gene expression level in SDF-1–treated cells compared with control untreated cells. The red dashed line represents the 1.5-fold cutoff. Results are depicted as means ± SEM.
Microarray analysis and validation. (A) Differential gene expression of cord blood CD34+ cells (n = 4) in response to SDF-1 (100 ng/mL) was analyzed by Affymetrix HG-U133 Plus 2.0 GeneChip software. Hierarchical analysis of the SDF-1–modulated genes involved in cell motility (left panel) and cell adhesion (right panel) were performed using the Euclidean distance metric. The color scale represents log2 normalized signal intensity. CTRL, control. (B) Differential expression of 18 SDF-1–induced genes was validated by qPCR in CD34+ cells derived from the same cord blood samples used in microarray experiments (n = 4) plus CD34+ cells from an independent cohort of cord blood (n = 4-6). Gene expression levels were calculated by the comparative CT method and are represented as the fold change of gene expression level in SDF-1–treated cells compared with control untreated cells. The red dashed line represents the 1.5-fold cutoff. Results are depicted as means ± SEM.
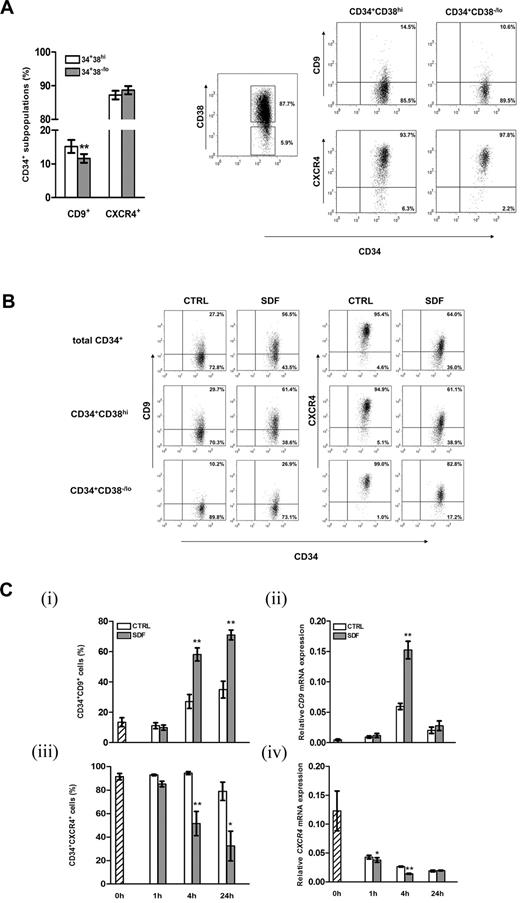
CD9 is expressed in CD34+ cells and is regulated by SDF-1 via specific signaling pathways
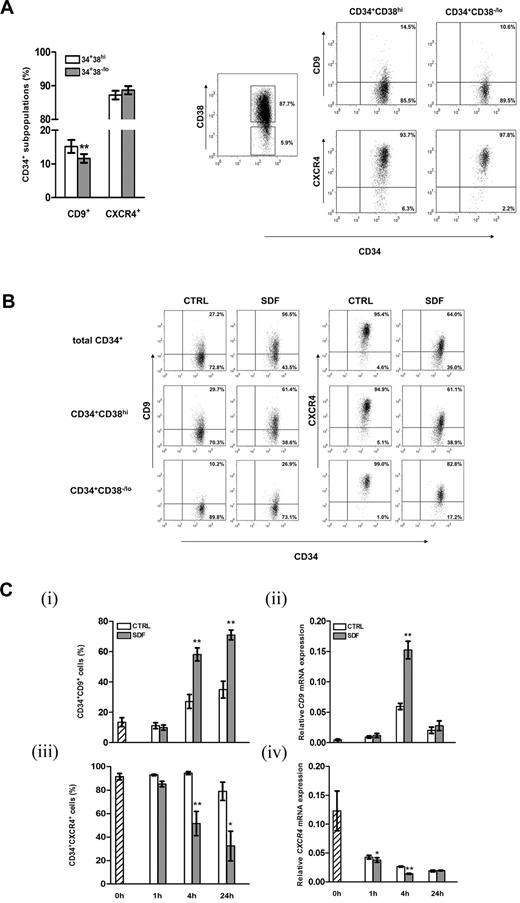
From the list of regulatory genes responsive to SDF-1, we focused on the characterization of CD9, which has been reported to modulate motility and adhesion of various cell types.22,27,28,35 Flow cytometric analysis showed that 14.2% ± 1.6% of cord blood CD34+ cells expressed CD9 on their surfaces. The more primitive CD34+CD38−/lo subpopulation did express CD9, with the level being slightly but significantly lower than the more lineage-committed CD34+CD38hi cells (P = .002; Figure 2A). Simultaneous analysis demonstrated similar levels of CXCR4 expressions in the CD34+CD38−/lo and CD34+CD38hi subpopulations (Figure 2A). Exposure of CD34+ cells to SDF-1 (100 ng/mL) for 4 hours resulted in a > 2-fold increase in CD9 expression in total CD34+ cells or their CD38−/lo or CD38hi subpopulations (Figure 2B), confirming the microarray data at protein level (Figure 1B). A 5-fold increase in SDF-1 concentration did not lead to further increases in CD9 expression level (data not shown). We then investigated the kinetics of SDF-1–induced CD9 expression in CD34+ cells. Treatment with SDF-1 for 1 hour did not change the percentage of CD34+CD9+ cells (Figure 2Ci). Subsequently, a time-dependent increase in CD9+ cells was observed at 4 hours and stayed at high levels at 24 hours (P < .01; Figure 2Ci). At the mRNA level, induction of CD9 was noted at 4 hours of SDF-1 treatment (2.6-fold increase; P = .003; Figure 2Cii), but its expression declined to the basal level at 24 hours. Consistent with other reports,3,32,36 SDF-1 decreased cell-surface CXCR4 expressions on both CD34+CD38−/lo and CD34+CD38hi cells (Figure 2B), which was noted at 4 hours and 24 hours post-SDF-1 treatment (Figure 2Ciii). In line with membrane protein expression, exposure of CD34+ cells to SDF-1 reduced CXCR4 mRNA levels at 1 hour and 4 hours (Figure 2Civ). In general, CXCR4 mRNA expression declined with duration of culture (Figure 2Civ).
Expression of CD9 on cord blood CD34+ cells and regulation by SDF-1. (A) Cell-surface expression of CD9 and CXCR4 on CD34+CD38hi and the more primitive CD34+CD38−/lo cells were analyzed by flow cytometry (n = 41). Right panel, gating strategy of cord blood CD34+ subsets based on CD38 expression and dot plots of CD9 and CXCR4 expression of a representative experiment. Quadrants were set on the basis of staining with respective isotypic antibodies, and the percentage of cells in each quadrant is depicted. (B) Expressions of CD9 and CXCR4 on total cord blood CD34+, CD34+CD38hi, and CD34+CD38−/lo cells after 4 hours of culture in medium alone (CTRL) or in the presence of SDF-1 (100 ng/mL). A representative of 4 independent experiments is shown. (C) Kinetics of SDF-1–regulated CD9 and CXCR4 expression in cord blood CD34+ cells at the (i,iii) protein (n = 3-5) and (ii,iv) mRNA levels (n = 3-4). Results in subpanels ii and iv are gene expression levels relative to GAPDH (glyceraldehyde 3-phosphate dehydrogenase) expression. *P < .05; **P < .01. Data are means ± SEM.
Expression of CD9 on cord blood CD34+ cells and regulation by SDF-1. (A) Cell-surface expression of CD9 and CXCR4 on CD34+CD38hi and the more primitive CD34+CD38−/lo cells were analyzed by flow cytometry (n = 41). Right panel, gating strategy of cord blood CD34+ subsets based on CD38 expression and dot plots of CD9 and CXCR4 expression of a representative experiment. Quadrants were set on the basis of staining with respective isotypic antibodies, and the percentage of cells in each quadrant is depicted. (B) Expressions of CD9 and CXCR4 on total cord blood CD34+, CD34+CD38hi, and CD34+CD38−/lo cells after 4 hours of culture in medium alone (CTRL) or in the presence of SDF-1 (100 ng/mL). A representative of 4 independent experiments is shown. (C) Kinetics of SDF-1–regulated CD9 and CXCR4 expression in cord blood CD34+ cells at the (i,iii) protein (n = 3-5) and (ii,iv) mRNA levels (n = 3-4). Results in subpanels ii and iv are gene expression levels relative to GAPDH (glyceraldehyde 3-phosphate dehydrogenase) expression. *P < .05; **P < .01. Data are means ± SEM.
We then used a panel of pharmacological inhibitors of specific SDF-1 signal transducers37-41 to dissect the pathways leading to CD9 induction. As shown in Figure 3A, SDF-1–induced CD9 expression was totally blocked by the CXCR4 antagonist AMD3100, whereas pre-exposure of CD34+ cells to specific inhibitors of the G-protein α subunit (Gα; pertussis toxin) and Janus kinase 2 (JAK2; AG 490) partially inhibited CD9 induction (Figure 3A). Interestingly, although SDF-1 activated Akt (Figure 3Bi) and ERK (Figure 3Bii) in CD34+ cells, an inhibitor of MAP kinase kinase (MEK; PD 98 059), but not the inhibitor of phosphatidylinositol 3-kinase (PI3K; wortmannin) suppressed SDF-1–induced CD9 up-regulation (Figure 3A). We also found that CD9 expression was partially blocked by GF 109203X, a PKC inhibitor, and completely blocked by a broader-range PKC inhibitor chelerythrine chloride (Figure 3A). U-73 122, a selective inhibitor of phosphatidylinositol-specific phospholipase C (PI-PLC), completely abolished SDF-1–induced CD9 expression, whereas the inhibitor of phosphatidylcholine-specific PLC (D609) had no effect (Figure 3A). In parallel, we investigated the effect of these inhibitors on SDF-1–mediated CXCR4 down-regulation. As shown in Figure 3A, the decrease in CD34+CXCR4+ cells was completely inhibited by pretreatment with AMD3100 and U-73 122, and to a lesser extent reduced by pertussis toxin, PD 98 059, wortmannin, and chelerythrine chloride. Conversely, AG490 slightly promoted CXCR4 down-regulation. In addition, effects of signal inhibitors on SDF-1–induced CD9 expression were observed in both CD34+CD38hi and CD34+CD38−/lo subpopulations (data not shown).
Signaling pathways leading to SDF-1–induced CD9 expression. (A) Cord blood CD34+ cells, either untreated (IMDM or dimethylsulfoxide [DMSO]; ie, solvent control), or pretreated with AMD3100 (AMD; 10 μg/mL), pertussis toxin (PTX; 1 μg/mL), PD 98 059 (PD; 40μM), wortmannin (W; 50 nM), GF 109203X (GF; 2μM), chelerythrine chloride (CC; 5μM), AG490 (AG; 50μM), U-73 122 (U; 2.5μM), or D609 (D; 10μM) for 1 hour, were stimulated with or without SDF-1 (100 ng/mL) for 4 hours. CD9 and CXCR4 expressions were detected by flow cytometry (n = 4-8). Results are represented as the percentage changes in CD9+ or CXCR4+ cells after SDF-1 stimulation in the presence of each inhibitor. AMD and PTX were dissolved in IMDM, while other inhibitors were dissolved in DMSO. (B) Enriched CD34+ cells were left untreated (filled histogram) or stimulated with SDF-1 (200 ng/mL; open histogram) for 1 minute. (i) Phosphorylated Akt (pAkt) and (ii) phosphorylated ERK (pERK) levels were measured by flow cytometry. Representative histograms and the mean fluorescence intensity are shown. (C) Cells were stimulated with SDF-1 (200 ng/mL) for 1 minute after pretreatment with wortmannin (W; 50nM), chelerythrine chloride (CC; 5μM), AG490 (AG; 50μM) or U-73 122 (U; 2.5μM) for 1 hour. Phosphorylated ERK (p-ERK) level was detected by flow cytometry (n = 3). (D) CD34+ cells were cultured for 4 hours in the absence (DMSO) or presence of PKC activators PMA (200 ng/mL), MEZ (200 ng/mL), or IDB (200 ng/mL). Cell-surface CD9 and CXCR4 expressions were detected by flow cytometry (n = 3). (E) Proposed model for the signaling pathways of SDF-1–induced CD9 expression. Results are depicted as means ± SEM. *P ≤ .05; **P < .01.
Signaling pathways leading to SDF-1–induced CD9 expression. (A) Cord blood CD34+ cells, either untreated (IMDM or dimethylsulfoxide [DMSO]; ie, solvent control), or pretreated with AMD3100 (AMD; 10 μg/mL), pertussis toxin (PTX; 1 μg/mL), PD 98 059 (PD; 40μM), wortmannin (W; 50 nM), GF 109203X (GF; 2μM), chelerythrine chloride (CC; 5μM), AG490 (AG; 50μM), U-73 122 (U; 2.5μM), or D609 (D; 10μM) for 1 hour, were stimulated with or without SDF-1 (100 ng/mL) for 4 hours. CD9 and CXCR4 expressions were detected by flow cytometry (n = 4-8). Results are represented as the percentage changes in CD9+ or CXCR4+ cells after SDF-1 stimulation in the presence of each inhibitor. AMD and PTX were dissolved in IMDM, while other inhibitors were dissolved in DMSO. (B) Enriched CD34+ cells were left untreated (filled histogram) or stimulated with SDF-1 (200 ng/mL; open histogram) for 1 minute. (i) Phosphorylated Akt (pAkt) and (ii) phosphorylated ERK (pERK) levels were measured by flow cytometry. Representative histograms and the mean fluorescence intensity are shown. (C) Cells were stimulated with SDF-1 (200 ng/mL) for 1 minute after pretreatment with wortmannin (W; 50nM), chelerythrine chloride (CC; 5μM), AG490 (AG; 50μM) or U-73 122 (U; 2.5μM) for 1 hour. Phosphorylated ERK (p-ERK) level was detected by flow cytometry (n = 3). (D) CD34+ cells were cultured for 4 hours in the absence (DMSO) or presence of PKC activators PMA (200 ng/mL), MEZ (200 ng/mL), or IDB (200 ng/mL). Cell-surface CD9 and CXCR4 expressions were detected by flow cytometry (n = 3). (E) Proposed model for the signaling pathways of SDF-1–induced CD9 expression. Results are depicted as means ± SEM. *P ≤ .05; **P < .01.
We next studied the effect of specific inhibitors on SDF-1–induced ERK activation (an immediate downstream target of MEK). Exposure of CD34+ cells to SDF-1 resulted in a 2.1-fold increase in the phosphorylated ERK level (Figure 3C). Pretreatment with chelerythrine chloride or U-73 122 significantly blocked ERK activation, whereas wortmannin and AG490 had no effect (Figure 3C). These data indicated that SDF-1–induced ERK activation was PKC- and PI-PLC–dependent, but PI3K- and JAK2-independent.
We also tested whether direct activation of the identified signal transducers can enhance CD9 expression. As shown in Figure 3D, exposure of CD34+ cells to the PKC activators PMA, MEZ, and IDB significantly increased CD34+CD9+ cells by > 2.4-fold (P < .05). Consistent with other reports,3,36 these PKC activators markedly reduced CXCR4 expression (Figure 3D).
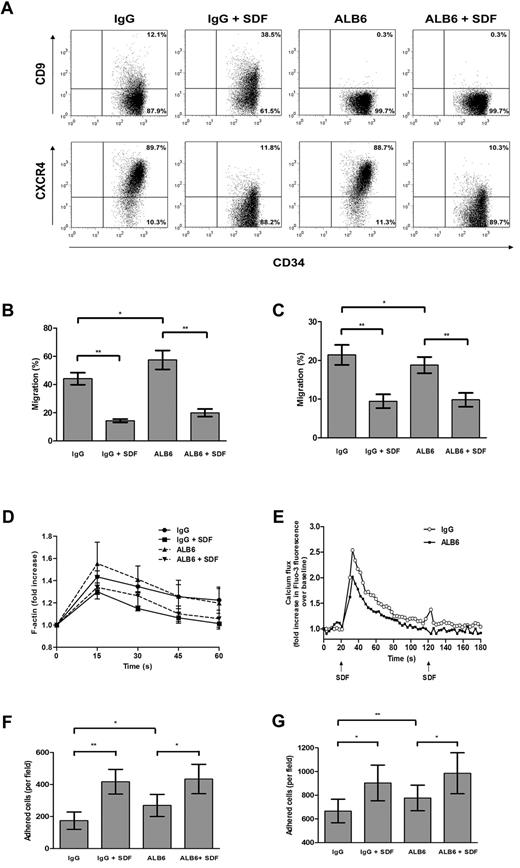
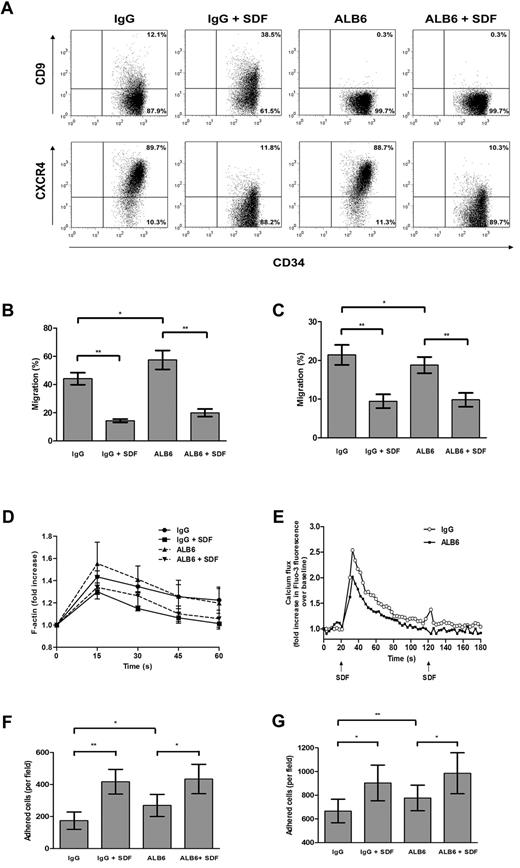
Anti-CD9 antibody alters migratory and adhesive functions of cord blood CD34+ cells in vitro
To address the possible involvement of CD9 in SDF-1–mediated motility, CD34+ cells were cultured for 4 hours in the presence of a neutralizing anti-CD9 mAb (clone ALB6)42,43 or an isotypic control antibody. Without changing CXCR4 expression (Figure 4A), pretreatment with ALB6 enhanced Transwell chemotactic migration of CD34+ cells to a gradient of SDF-1 by 30.2% (P = .033; Figure 4B). To investigate whether migration can be improved under a situation in which CXCR4 expression is suppressed, we pretreated CD34+ cells with SDF-1 (100 ng/mL) in the absence or presence of ALB6. Exposure of CD34+ cells to SDF-1 decreased cell-surface CXCR4 expression (Figure 4A), which coincided with a significant inhibition of SDF-1–mediated migration (P = .004; Figure 4B). However, the presence of ALB6 did not affect the migratory potential of SDF-1–pretreated cells (Figure 4A).
Effects of neutralizing anti-CD9 mAb on in vitro homing-related functions of cord blood CD34+ cells. (A) Cells were cultured in the presence of an anti-CD9 mAb (ALB6; 10 μg/mL) or an irrelevant isotypic control mAb (IgG; 10 μg/mL), or pretreated with SDF-1 (100 ng/mL) for 4 hours in the presence of the anti-CD9 mAb (ALB + SDF-1) or the isotypic control mAb (IgG + SDF-1). Cell-surface expressions of CD9 and CXCR4 were detected by flow cytometry. Representative dot plots of CD9 and CXCR4 expression and the percentage of cells in each quadrant are shown. Cell migration through (B) bare filters (n = 4) or across (C) HUVEC monolayers (n = 4) to a gradient of SDF-1 (100 ng/mL) was determined using Transwells. The percentage migration was calculated as follows: (number of migrated cells/input cell number) × 100. (D) Actin polymerization assay: cells were stimulated with SDF-1 (100 ng/mL) for the indicated times, and the mean phalloidin-FITC fluo-rescence intensity was determined by flow cytometry (n = 3-4). (E) Cells were loaded with the Fluo-3AM, and calcium flux following SDF-1 stimulation (100 ng/mL) was monitored by flow cytometry (n = 4). (F,G) Adhesion assay: CFSE-labeled cells were pretreated with IgG or ALB6 for 4 hours and plated on (F) fibronectin-coated (n = 4) or (G) HUVEC-coated microwell plates (n = 4). Cells were either left unstimulated or stimulated with SDF-1 (100 ng/mL) for 45 minutes. Adherent cells from 5 random 20 × fields were counted under the Olympus IX71 fluorescence microscope. Data represent means ± SEM. *P ≤ .05; **P < .01.
Effects of neutralizing anti-CD9 mAb on in vitro homing-related functions of cord blood CD34+ cells. (A) Cells were cultured in the presence of an anti-CD9 mAb (ALB6; 10 μg/mL) or an irrelevant isotypic control mAb (IgG; 10 μg/mL), or pretreated with SDF-1 (100 ng/mL) for 4 hours in the presence of the anti-CD9 mAb (ALB + SDF-1) or the isotypic control mAb (IgG + SDF-1). Cell-surface expressions of CD9 and CXCR4 were detected by flow cytometry. Representative dot plots of CD9 and CXCR4 expression and the percentage of cells in each quadrant are shown. Cell migration through (B) bare filters (n = 4) or across (C) HUVEC monolayers (n = 4) to a gradient of SDF-1 (100 ng/mL) was determined using Transwells. The percentage migration was calculated as follows: (number of migrated cells/input cell number) × 100. (D) Actin polymerization assay: cells were stimulated with SDF-1 (100 ng/mL) for the indicated times, and the mean phalloidin-FITC fluo-rescence intensity was determined by flow cytometry (n = 3-4). (E) Cells were loaded with the Fluo-3AM, and calcium flux following SDF-1 stimulation (100 ng/mL) was monitored by flow cytometry (n = 4). (F,G) Adhesion assay: CFSE-labeled cells were pretreated with IgG or ALB6 for 4 hours and plated on (F) fibronectin-coated (n = 4) or (G) HUVEC-coated microwell plates (n = 4). Cells were either left unstimulated or stimulated with SDF-1 (100 ng/mL) for 45 minutes. Adherent cells from 5 random 20 × fields were counted under the Olympus IX71 fluorescence microscope. Data represent means ± SEM. *P ≤ .05; **P < .01.
The functional involvement of CD9 in migration across a HUVEC monolayer was further explored. Unexpectedly, incubation of CD34+ cells with ALB6 resulted in an opposite effect on transendothelial migration compared with migration across a bare Transwell insert. As shown in Figure 4C, ALB6 inhibited transendothelial migration of CD34+ cells toward an SDF-1 gradient by 12.3% (P = .024). However, transmigration of SDF-1–pretreated cells was not affected by ALB6 (Figure 4C).
Actin polymerization and mobilization of intracellular calcium are early cellular responses to SDF-1 in CD34+ cells. In agreement with Voermans et al,13 SDF-1 markedly increased the polymerized actin content after a 15-second stimulation, which was inhibited by 4 hours of SDF-1 pretreatment (Figure 4D). Blocking CD34+ cells with ALB6 did not significantly alter these responses (Figure 4D). However, CD9 neutralization significantly inhibited SDF-induced calcium flux in CD34+ cells (Figure 4E).
We next examined the role of CD9 in CD34+ cell adhesion to fibronectin- or HUVEC-coated surfaces. ALB6 significantly enhanced basal adhesion of CD34+ cells to fibronectin (P = .035; Figure 4F) and the HUVEC monolayer (P = .006; Figure 4G) by 55% and 16.6%, respectively. Consistent with Peled et al,5 the addition of SDF-1 significantly increased adhesion to either fibronectin (P = .004; Figure 4F) or HUVECs (P = .021; Figure 4G). In the presence of SDF-1, ALB6 did not affect the adhesion of CD34+ cells to fibronectin (Figure 4F), but showed a trend of promoting adhesion to HUVECs by 9.1% (P = .054; Figure 4G).
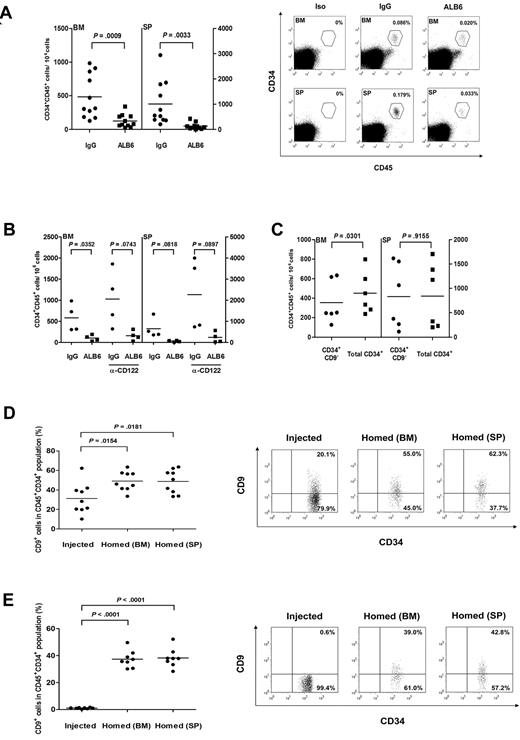
CD9 neutralization impairs homing of transplanted CD34+ cells in NOD/SCID mice
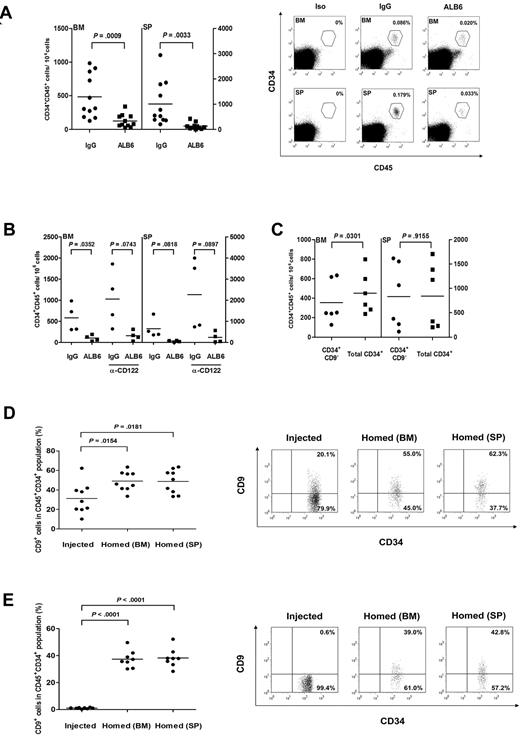
To assess whether CD9 regulates the in vivo homing process, CD34+ cells were cultured with either ALB6 or isotypic control antibody for 4 hours, and infused into irradiated NOD/SCID mice. Preincubation of CD34+ cells with ALB6 significantly reduced homing to the recipient bone marrow and spleen by 73.5% (P < .001) and 86.9%, respectively (P = .003; Figure 5A). Similar results were obtained with a shorter incubation time (30 minutes) of ALB6 treatment (data not shown). The inhibitory effect of ALB6 on homing of CD34+ cells in these animals was comparable with that in mice pretreated with anti-CD122 mAb (84.2% for bone marrow and 89.6% for spleen; Figure 5B). These data ruled out the possibility that the ALB6-bearing cells were cleared by the innate immune system via antibody-dependent cellular cytotoxicity, a phenomenon that has been recently reported for anti-CD38 mAb.44
The role of CD9 on homing of cord blood CD34+ cells in NOD/SCID mice. (A) Cord blood CD34+ cells were cultured in the presence of an anti-CD9 mAb (ALB6; 10 μg/mL) or an isotypic mAb (IgG; 10 μg/mL) for 4 hours, and injected intravenously into NOD/SCID mice (1.9 × 105 to 5 × 105 cells per mouse). Bone marrow (BM) and spleens (SP) of recipient mice were analyzed for the presence of human CD45+CD34+ cells by flow cytometry 20 hours after transplantation (n = 11). P values are indicated. Right panel, representative flow cytometry analysis plots of human cell detection in BM and SP samples of transplanted mice. BM and SP samples from a mouse stained with isotypic antibodies (Iso) are also presented. The gates used for identification of human CD45+CD34+ cells and the frequency of human cells are indicated. (B) CD34+ cells were pretreated with IgG or ALB6, and transplanted into anti-CD122 (α-CD122)-treated or untreated NOD/SCID mice (n = 4). (C) Sorted CD34+CD9− or total CD34+ cells (8.7 × 104 to 2.1 × 105 cells/mouse) were transplanted into NOD/SCID mice (n = 6). Homing level of each population was determined by flow cytometry. (D,E) CD9 expressions in (D) total CD34+ cells (n = 9) or (E) sorted CD34+CD9− cells (n = 8) before injection were compared with those homed to BM and SP in NOD/SCID mice. Right panels, representative dot plots showing CD9 expression on injected and homed CD34+ cells. The percentage of cells in each quadrant is depicted.
The role of CD9 on homing of cord blood CD34+ cells in NOD/SCID mice. (A) Cord blood CD34+ cells were cultured in the presence of an anti-CD9 mAb (ALB6; 10 μg/mL) or an isotypic mAb (IgG; 10 μg/mL) for 4 hours, and injected intravenously into NOD/SCID mice (1.9 × 105 to 5 × 105 cells per mouse). Bone marrow (BM) and spleens (SP) of recipient mice were analyzed for the presence of human CD45+CD34+ cells by flow cytometry 20 hours after transplantation (n = 11). P values are indicated. Right panel, representative flow cytometry analysis plots of human cell detection in BM and SP samples of transplanted mice. BM and SP samples from a mouse stained with isotypic antibodies (Iso) are also presented. The gates used for identification of human CD45+CD34+ cells and the frequency of human cells are indicated. (B) CD34+ cells were pretreated with IgG or ALB6, and transplanted into anti-CD122 (α-CD122)-treated or untreated NOD/SCID mice (n = 4). (C) Sorted CD34+CD9− or total CD34+ cells (8.7 × 104 to 2.1 × 105 cells/mouse) were transplanted into NOD/SCID mice (n = 6). Homing level of each population was determined by flow cytometry. (D,E) CD9 expressions in (D) total CD34+ cells (n = 9) or (E) sorted CD34+CD9− cells (n = 8) before injection were compared with those homed to BM and SP in NOD/SCID mice. Right panels, representative dot plots showing CD9 expression on injected and homed CD34+ cells. The percentage of cells in each quadrant is depicted.
CD9-negative cells have inferior homing capacity to bone marrow
We transplanted sorted cells to NOD/SCID animals and compared homing activities in those receiving enriched CD34+CD9− cells or total CD34+ cells (containing 14.3% of CD9+ cells). Our results showed that animals transplanted with total CD34+ cells had slightly but significantly increased levels of homing to the bone marrow (27.8% increase) compared with those receiving highly enriched CD34+CD9− cells (P = .03; Figure 5C). These data suggested that CD34+CD9+ cells might possess higher homing capacity to their bone marrow niche. Such differences in the homing of sorted cell populations were not observed in the spleens of recipient animals (Figure 5C).
CD9 expression is up-regulated after transplantation
We compared the proportions of CD34+CD9+ cells in the initial injected CD34+ population with those homed to bone marrow and spleen 20 hours after cell infusion. After injection of total CD34+ cells containing 31.3% of the CD9+ subset, we observed a 1.6-fold increase in CD34+CD9+ cells in the bone marrow and spleens of NOD/SCID animals (P < .05; Figure 5D). The up-regulation of CD9 expression was most obvious in mice transplanted with CD34+CD9− cells, because more than 35% of the homed CD34+ cells expressed CD9 membrane protein (P < .001; Figure 5E). These data indicated that CD9 expression in CD34+ cells was subjected to regulation in vivo and could be functionally important for homing/retention of these cells in their niches.
Discussion
In this study, we reported a microarray expression profile of cord blood CD34+ cells in response to a short-term (4-hour) exposure to SDF-1. Among the differentially regulated genes, we identified the tetraspanin CD9 as one of the SDF-1 downstream proteins that was regulated via the CXCR4, G-protein, PKC, PLC, ERK, and JAK2 signals. We also provided the first evidence that CD9 is functionally involved in modulating SDF-1–mediated migration, adhesion, and in vivo homing of CD34+ cells.
In agreement with a recent microarray study using the Affymetrix HG-U133A GeneChip on CD34+ cells derived from granulocyte-colony stimulating factor-mobilized peripheral blood,19 our results revealed that SDF-1 altered the expression of a large number of genes involved in cell motility, cell adhesion, cytoskeleton organization, and cell proliferation. An example is MMP-9, one of the few genes known to be transcriptionally regulated by SDF-1.7 Other SDF-1–modulated genes identified in our profiling experiments only displayed modest overlapping with those reported by Rossi et al.19 This discrepancy could be due to the difference in the source of CD34+ cells (cord blood vs granulocyte-colony stimulating factor–mobilized peripheral blood)45 and the duration of SDF-1 treatment (4 hours vs 24 hours). Although some of the SDF-1–regulated genes in cord blood CD34+ cells have known homing functions (eg, MMP-97 and integrins5 ), we discovered many others (eg, CD9,22 CKLF,46 and EPHA447 ) that have not been characterized in hematopoietic progenitors but were involved in regulating homing-related functions in other cell types. Our microarray data have thus provided a useful platform for the identification of novel intrinsic regulators of HSC homing.
Binding of SDF-1 to CXCR4 has been known to activate several signal transducers, including ERK, JAK2, PI3K, PKC, and PLC, in several types of hematopoietic cells.37-41 In human CD34+ cells, Petit et al41 showed that SDF-1–induced MMP-9 transcription was dependent on ERK, PI3K, PI-PLC, and PKC. Using a panel of inhibitors, we identified G-protein, JAK2, PKC, PLC, and ERK, but not PI3K, as the downstream signals leading to protein expression of CD9 on CD34 cells via the SDF-1/CXCR4 axis. We also showed that the regulatory involvement of PI3K activation on CD9 and CXCR4 induction might be different, and we observed that CD9 expression could be increased by the activation of PKC. Based on these data and reported SDF-1 signals,37-41,48,49 we proposed a model of the SDF-1/CXCR4 signaling cascade leading to CD9 expression in human CD34+ cells (Figure 3E). Upon binding of SDF-1 to CXCR4, uncoupling of the G-protein subunits might activate PI-PLC, which would result in the activation of various PKC isoforms. PKC, in turn, would lead to MEK/ERK activation, followed by JAK2 activation. Finally, JAK2 might mediate CD9 expression via activation of the transcription factor signal transducers and activators of transcription (STAT).
In the in vitro assays of homing-related functions, we showed that effects of the CD9 neutralizing antibody ALB6 on CD34+ cells were specific and could be modulated by pretreatment with SDF-1. While neutralizing CD9 had no effect on actin polymerization, it inhibited transendothelial motility toward a SDF-1 gradient and reduced calcium influx. On the other hand, ALB6 increased CD34+ cell motility through a bare filter and enhanced fibronectin or HUVEC adhesion. In SDF-1–pretreated cells, ALB6 failed to alter migratory responses, suggesting that the migratory function of CD9 might be masked when CXCR4 expression is suppressed. Furthermore, the regulatory role of CD9 in cell motility appears to be complex, and might vary depending on the presence of an endothelial barrier. Such complications have also been reported in a variety of cancer cells.26 While an inverse correlation between CD9 expression and metastastic development was observed in melanoma,50 cervical carcinoma,24 and multiple myeloma,43 strong CD9 expression was found in contact sites between tumor and endothelial cells, where it promoted transendothelial migration and invasion. We also demonstrated the role of CD9 in inhibiting cell adhesion to fibronectin and HUVECs, and to a lesser extent in the presence of SDF-1. Our results are in agreement with studies on some cancer cells. Pretreatment of pre-B–cell acute lymphoblastic leukemia,35 melanoma,50 or colon carcinoma51 with CD9 mAb enhanced their adhesion to fibronectin, endothelial, and bone marrow stromal cells, suggesting that CD9 is a negative regulator of cellular adhesion. Our observations, together with those in metastatic tumors, suggest that the presence of CD9 on CD34+ cells might inhibit SDF-1–mediated cell motility to a certain extent, but that such activities could be modulated by adhesion to endothelial cells. As a result, CD9 neutralization may strengthen heterotypic cell-cell interactions and thus hamper transendothelial migration.
We demonstrated that CD9 was involved in homing of CD45+CD34+ cells in the NOD/SCID mouse transplantation model. While CD9 neutralization mildly inhibited transendothelial migration of CD34+ cells in vitro, we found that the same treatment resulted in more than 70% inhibition of cord blood CD34+ cell homing to murine bone marrow and over 80% inhibition to the spleen. We also observed a slight but significantly higher level of bone marrow homing when NOD/SCID animals were injected with total CD34+ cells (ie, both CD9+ and CD9− cells) compared with those receiving only CD34+CD9− cells. Thus, we speculate that CD9+ cells might possess higher homing potential or are preferentially retained in these niches. A similar observation has been recently demonstrated in human B-acute lymphoblastic leukemia cells.52 Using the NOD/SCID/gcnull mouse model, Nishida et al52 found that transplantation of sorted CD9+, but not CD9−, leukemic cells resulted in tumorigenesis in the bone marrow and spleens of recipients. Another interesting observation was the consistent up-regulation of CD9 expression in CD34+ cells recovered from the bone marrow and spleens of NOD/SCID animals regardless of the CD9 status of the cells originally infused. Such regulation could result from exposure of CD34+ cells to the SDF-1–rich microenvironment,53 and indicates a possible role of CD9 in the retention of CD34+ in these niches. These data might also explain the only slight decrease of bone marrow homing when the sorted CD34+CD9− cell subpopulation was infused.
The differential homing efficiency of CD34+ subpopulations has remained a controversial issue.14,54 In the present study, CD9 expression and induction signals were demonstrated in both CD34+CD38+ and the more primitive CD34+CD38−/lo cells. However, due to cell-dose limitations, we could not specifically address the effect of CD9 expression on the homing capacity of the CD34+CD38+ and CD34+CD38−/lo subpopulations. Such information would be particularly important for determining the role of CD9 in long-term engraftment of CD34+ cells.
In summary, we have demonstrated the wide-coverage involvement of SDF-1 in modulating homing-related gene expressions in cord blood CD34+ cells through differential microarray profiling. We have also provided the first evidence that CD9 mRNA and membrane protein expressions were up-regulated by a short exposure to SDF-1 through various specific CXCR4 downstream signals. Significantly, CD9 was shown to play a role in in vitro homing-related functions and in vivo homing of CD34+ cells to the bone marrow and spleens of NOD/SCID mice. It is anticipated that strategies for modulating CD9 expression and functions might improve homing of CD34+ cells to their hematopoietic niches.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Mr Henry Pong for his assistance in cell sorting, and Prof Chi Chiu Wang, Ms Hanna Fong, and Ms Fiona Leung for cord blood collection.
This study was financially supported by the Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong; by the Research Grant Council, The Government of the Hong Kong Special Administrative Region, Hong Kong (earmarked grant 4509/06M); and by the Cost Center for Marginally Funded Research Postgraduate Places (Molecular Medicine), The Chinese University of Hong Kong.
Authorship
Contribution: K.T.L. conceptualized and designed research, collected and analyzed data, and cowrote the manuscript; K.Y.Y.C. collected and analyzed data and cowrote the manuscript; P.C.N. conceptualized and designed research and contributed to research funding; T.Z.L. provided essential materials and contributed to research funding; W.M.C. collected and analyzed data; K.S.T. analyzed data and provided essential materials; C.K.L. designed research and provided essential materials; C.K.L.K. collected and analyzed data; and K.L. conceptualized, designed and coordinated research, analyzed data, contributed to research funding, and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karen Li, PhD, Department of Paediatrics, Prince of Wales Hospital, The Chinese University of Hong Kong, Shatin, NT, Hong Kong; e-mail: lipang@cuhk.edu.hk.

![Figure 3. Signaling pathways leading to SDF-1–induced CD9 expression. (A) Cord blood CD34+ cells, either untreated (IMDM or dimethylsulfoxide [DMSO]; ie, solvent control), or pretreated with AMD3100 (AMD; 10 μg/mL), pertussis toxin (PTX; 1 μg/mL), PD 98 059 (PD; 40μM), wortmannin (W; 50 nM), GF 109203X (GF; 2μM), chelerythrine chloride (CC; 5μM), AG490 (AG; 50μM), U-73 122 (U; 2.5μM), or D609 (D; 10μM) for 1 hour, were stimulated with or without SDF-1 (100 ng/mL) for 4 hours. CD9 and CXCR4 expressions were detected by flow cytometry (n = 4-8). Results are represented as the percentage changes in CD9+ or CXCR4+ cells after SDF-1 stimulation in the presence of each inhibitor. AMD and PTX were dissolved in IMDM, while other inhibitors were dissolved in DMSO. (B) Enriched CD34+ cells were left untreated (filled histogram) or stimulated with SDF-1 (200 ng/mL; open histogram) for 1 minute. (i) Phosphorylated Akt (pAkt) and (ii) phosphorylated ERK (pERK) levels were measured by flow cytometry. Representative histograms and the mean fluorescence intensity are shown. (C) Cells were stimulated with SDF-1 (200 ng/mL) for 1 minute after pretreatment with wortmannin (W; 50nM), chelerythrine chloride (CC; 5μM), AG490 (AG; 50μM) or U-73 122 (U; 2.5μM) for 1 hour. Phosphorylated ERK (p-ERK) level was detected by flow cytometry (n = 3). (D) CD34+ cells were cultured for 4 hours in the absence (DMSO) or presence of PKC activators PMA (200 ng/mL), MEZ (200 ng/mL), or IDB (200 ng/mL). Cell-surface CD9 and CXCR4 expressions were detected by flow cytometry (n = 3). (E) Proposed model for the signaling pathways of SDF-1–induced CD9 expression. Results are depicted as means ± SEM. *P ≤ .05; **P < .01.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/6/10.1182_blood-2010-04-281329/4/m_zh89991064890003.jpeg?Expires=1771190239&Signature=CJngnreceppNjz8z9BS4bpAFJxCjg~9uKDPko6uVxiL4wPPxMeWYlluPX5Fx~S2nrd3vjTjklcUXmrF9gK5L8xlEZ70jPQFp1ytIn2khtpvd~FhBA9UnT35j-leDGlGzJ2K~fQQfGPyarbcgIQhlxH-sS19Y5bWKPin3zolWu4KGcxu97VjLEYBpsprxPC07mbu3g5umWRvtN2BRooTdLyXTRRvaRr59f4rnsDx93EIhaPLcjMX2s5Rl-XwEuCgeV25RFke2RacAIcVM6FI-Y9Riw1Eo4fE4lz0MG-1Hi4GTZ7B~Ms5pMGYQb3FjBzmDBGtWdSeh1KAZlS23VSDR7Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 3. Signaling pathways leading to SDF-1–induced CD9 expression. (A) Cord blood CD34+ cells, either untreated (IMDM or dimethylsulfoxide [DMSO]; ie, solvent control), or pretreated with AMD3100 (AMD; 10 μg/mL), pertussis toxin (PTX; 1 μg/mL), PD 98 059 (PD; 40μM), wortmannin (W; 50 nM), GF 109203X (GF; 2μM), chelerythrine chloride (CC; 5μM), AG490 (AG; 50μM), U-73 122 (U; 2.5μM), or D609 (D; 10μM) for 1 hour, were stimulated with or without SDF-1 (100 ng/mL) for 4 hours. CD9 and CXCR4 expressions were detected by flow cytometry (n = 4-8). Results are represented as the percentage changes in CD9+ or CXCR4+ cells after SDF-1 stimulation in the presence of each inhibitor. AMD and PTX were dissolved in IMDM, while other inhibitors were dissolved in DMSO. (B) Enriched CD34+ cells were left untreated (filled histogram) or stimulated with SDF-1 (200 ng/mL; open histogram) for 1 minute. (i) Phosphorylated Akt (pAkt) and (ii) phosphorylated ERK (pERK) levels were measured by flow cytometry. Representative histograms and the mean fluorescence intensity are shown. (C) Cells were stimulated with SDF-1 (200 ng/mL) for 1 minute after pretreatment with wortmannin (W; 50nM), chelerythrine chloride (CC; 5μM), AG490 (AG; 50μM) or U-73 122 (U; 2.5μM) for 1 hour. Phosphorylated ERK (p-ERK) level was detected by flow cytometry (n = 3). (D) CD34+ cells were cultured for 4 hours in the absence (DMSO) or presence of PKC activators PMA (200 ng/mL), MEZ (200 ng/mL), or IDB (200 ng/mL). Cell-surface CD9 and CXCR4 expressions were detected by flow cytometry (n = 3). (E) Proposed model for the signaling pathways of SDF-1–induced CD9 expression. Results are depicted as means ± SEM. *P ≤ .05; **P < .01.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/6/10.1182_blood-2010-04-281329/4/m_zh89991064890003.jpeg?Expires=1771190240&Signature=gPQCRroiJlZad706wB84P8T9sUJJdhpmlLa6xLMsEoS-NvAjMrdB435kjpIDSTfAYBD82qAtzZYe7LZHsrxGIQJR3giUy0inhQ6pkZcxz2L3ae17fJJvnKJJEycNhlc0wtQ-1QyPGKClzX6wtNlYHEaFOJdruBXvRHuYYRHRuwNTcVwfVa9RFmVzojaLszlqvoqorp0goJamS7VyWv-ROeCOqSMCug5~npFeMiv3Q-q0PeyRm1t-hTvo5VfEiep2jLpcdJpm0355BF4Z0munlsgkI7HWYKYOzUdiqw5SMLjH-lWGjI5WgiK7LrNRuuo5w6Ehp~fLDKqrF7WXzlp~zg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)