Abstract
RARA (retinoic acid receptor alpha) haploinsufficiency is an invariable consequence of t(15;17)(q22;q21) translocations in acute promyelocytic leukemia (APL). Retinoids and RARA activity have been implicated in hematopoietic self-renewal and neutrophil maturation. We and others therefore predicted that RARA haploinsufficiency would contribute to APL pathogenesis. To test this hypothesis, we crossed Rara+/− mice with mice expressing PML (promyelocytic leukemia)-RARA from the cathepsin G locus (mCG-PR). We found that Rara haploinsufficiency cooperated with PML-RARA, but only modestly influenced the preleukemic and leukemic phenotype. Bone marrow from mCG-PR+/− × Rara+/− mice had decreased numbers of mature myeloid cells, increased ex vivo myeloid cell proliferation, and increased competitive advantage after transplantation. Rara haploinsufficiency did not alter mCG-PR–dependent leukemic latency or penetrance, but did influence the distribution of leukemic cells; leukemia in mCG-PR+/− × Rara+/− mice presented more commonly with low to normal white blood cell counts and with myeloid infiltration of lymph nodes. APL cells from these mice were responsive to all-trans retinoic acid and had virtually no differences in expression profiling compared with tumors arising in mCG-PR+/− × Rara+/+ mice. These data show that Rara haploinsufficiency (like Pml haploinsufficiency and RARA-PML) can cooperate with PML-RARA to influence the pathogenesis of APL in mice, but that PML-RARA is the t(15;17) disease-initiating mutation.
Introduction
Acute promyelocytic leukemia (APL) is caused by translocations involving RARA (retinoic acid receptor alpha).1 Approximately 95% of APL cases have a reciprocal translocation involving 15q22 and 17q21, resulting in 4 acquired genetic events: expression of the novel fusion protein PML (promyelocytic leukemia)-RARA, inconsistent expression of the reciprocal protein RARA-PML, and haploinsufficiency for both RARA and PML.1 Whereas alternative translocations and fusion proteins are occasionally associated with APL, RARA appears to be a necessary member of each (eg, PLZF-RARA, NPM-RARA, STAT5B-RARA, NUMA1-RARA). These translocations invariably occur in the second intron of RARA, which preserves the DNA-binding domain and ligand-binding domain, but eliminates most of the transcriptional activating (AF1) domain.1-3 Although there is a high degree of homology within the retinoid receptor family, fusion proteins involving RARB, RARG, or any of the RXRs have not been identified in acute leukemia or any other malignancies.
RARA transcriptional activity has been implicated in myeloid development and maturation. RARA is a member of the nuclear receptor superfamily of ligand-dependent transcription factors.4 Retinoids are derived from vitamin A, act as RARA-activating ligands, and have been shown to facilitate hematopoietic stem cell self-renewal, maturation, and lineage commitment.5-8 All-trans retinoic acid (ATRA; a naturally occurring RARA ligand) is a treatment for APL, resulting in differentiation of promyelocytes and transient complete remissions in most patients.1,9,10 Similarly, vitamin A deficiency leads to an expansion of immature myeloid cells, whereas RARA dominant-negative mutations and RARA antagonists block myeloid maturation, leading to increased numbers of myeloid precursors.11-13 ATRA promotes granulopoiesis from common myeloid precursors, and enhances terminal maturation and loss of self-renewal in human and mouse myeloid cells.7,14-17 These effects are essentially lost in Rara-deficient mice, implicating Rara as an important positive mediator of neutrophil maturation.16,18 Finally, RARA expression is decreased in both APL and non-APL acute myelogenous leukemia (AML) compared with normal cord blood CD133+ and CD33+ cells.19 For these reasons, RARA haploinsufficiency has been suggested to be a relevant consequence of t(15;17).20,21
Multiple transgene and knock-in strategies have been used to develop mouse models of APL by targeting PML-RARA expression to the hematopoietic compartment.22-26 These model systems have been used to identify multiple requirements of PML-RARA for leukemogenesis (eg, PML sumoylation, neutrophil elastase, heterodimerization with RXRA, and AF2-dependent PML-RARA transcriptional activity).10,27-30 Integration of additional mutations caused by t(15;17) into these model systems has shown that Pml haploinsufficiency leads to a shortened disease latency, and that expression of the reciprocal bcr3 RARA-PML isoform (but not the shorter bcr1 isoform) increases APL penetrance.30-32 However, the role of RARA haploinsufficiency in APL pathogenesis has not yet been examined.
In the present study, we crossed Rara-deficient mice (Rara+/−) with mice expressing PML-RARA from the cathepsin G (Ctsg) locus (mCG-PR+/−).25,33 Surprisingly, we observed that Rara haploinsufficiency did not alter the latency or penetrance of mCG-PR-associated leukemia. However, Rara haploinsufficiency did cooperate with PML-RARA to shape preleukemic myeloid development and the subsequent distribution of APL cells in the adult mouse; we did not observe either phenotype in mice that were simply haploinsufficient for Rara. These data confirm RARA haploinsufficiency as a relevant mutation caused by the t(15;17) translocation, and further validate the critical importance of PML-RARA as the disease-initiating event.30,31
Methods
Mice
Generation of the Rara L− allele has been described previously.33 LoxP sequences flank exon 3, which contains most of the Rara DNA–binding domain; removal of this exon places the remaining sequence out of frame, generating a null allele. Rara-floxed mice were mated with CMV-Cre mice (The Jackson Laboratory) to generate a germline-null allele (L−, referred to hereafter as Rara+/−). CMV-Cre was then bred out of the founder males, and 4 independently derived Rara+/− mice were mated to mCG-PR+/− mice. All mice were of the C57Bl/6 background.
Competitive repopulation was performed by retro-orbital injection of 1 × 106 bone marrow cells into CD45.1 recipient mice (Ly5.1) 24 hours after receiving 1000 cGy of total body irradiation (Mark1 cesum137 irradiator; J. L. Shepard).
Tumor watches were prospectively established, and mice were followed with serial blood counts (Hemavet 950; Drew Scientific Group). Moribund mice were euthanized; peripheral blood counts assessed; and spleen, bone marrow, and lymph node cells cryopreserved in 10% dimethylsulfoxide.
Secondary transplantation and in vivo response to ATRA was performed by retro-orbital injection of 1 × 106 spleen cells (harvested from leukemic mice) into CD45.1 recipient mice without prior irradiation. Five days later, engraftment was confirmed by flow cytometry of peripheral blood (peridinin chlorophyll A protein [PerCP]-Cy5.5-αCD45.2; eBioscience 104), and mice were treated with ATRA (10 mg, 21-day sustained-release pellets, Innovative Research of America), as described previously.34 The Washington University Animal Studies Committee approved all animal experiments.
Expression array profiling
Total cellular RNA was purified using TRIzol reagent, quantified using UV spectroscopy (Nanodrop Technologies), and qualitatively assessed using an Experion Bioanalyzer. Amplified cDNA was prepared from 20 ng of total RNA using the whole transcript WT-Ovation RNA amplification system and biotin labeled using the Encore biotin module (NuGen Technologies), according to the manufacturer's instructions. Labeled targets were then hybridized to Mouse Exon 1.0ST arrays (Affymetrix), washed, stained, and scanned using standard protocols from the Alvin J. Siteman Cancer Center Molecular and Genomic Analysis Core Facility (St Louis, MO). Affymetrix Expression Console software was used to process array images, export signal data, and evaluate image and data quality relative to standard Affymetrix quality-control metrics.35 These data have been deposited into a public database (Gene Expression Omnibus: GSE23291).
Hematopoietic progenitor assay
After red cell lysis in ACK buffer (0.15M NH4Cl, 10mM KHCO3, 0.1mM Na2EDTA [Na2-ethylenediaminetetraacetic acid]) of red blood cells, bone marrow, or spleen cells from individual mice, cells were counted using a hemocytometer and plated (in duplicate) in 1.1 mL of methylcellulose medium containing interleukin-3 (IL-3), IL-6, and stem cell factor (SCF) (MethoCult M3534; StemCell Technologies).36 Cells were plated at 8.3 × 103/mL. Colonies with > 30 cells were counted on day 7. Total cells were collected from the methylcellulose in warm Dulbecco modified Eagle medium with 2% fetal calf serum (FCS), washed, and counted. Cells were replated at the same density. This process was repeated for 4 weeks or until colony formation failed.
ATRA sensitivity assay
Cryopreserved leukemic spleen cells were quickly thawed at 37°C and washed in phosphate-buffered saline with 50μM β-mercaptoethanol and 10% FCS. Cells were plated at 2 × 106/mL in RPMI buffer with 15% FCS, 100 ng/mL of SCF, 6 ng/mL of IL-3, 10 ng/mL of IL-6 (PeproTech) ± 1μM ATRA (Sigma), and maintained at 3% O2 and 5% CO2 in a humidified chamber (Billups-Rothenberg) for 48 hours. Cells were plated at 8.3 × 103/mL (MethoCult M3534; StemCell Technologies) and maintained in 3% O2 and 5% CO2. After 7 days, colonies were counted and assessed for immunophenotypes.
Flow cytometry
After lysis of red blood cells in ACK buffer, peripheral blood, bone marrow, spleen, or lymph node cells were treated with anti–mouse CD16/32 (clone 93; eBioscience) and stained with the indicated combinations of the following antibodies: fluorescein isothiocyanate (FITC)-αGr1 (BD Pharmingen RB6-8C5); phycoerythrin (PE)-αCD11b (eBioscience M1/70); PE-αCD115 (eBioscience AFS98); FITC-αCD34 (BD Pharmingen RAM34); allophycocyanin (APC)-αcKit (eBioscience 2B8); PE-αCD3 (BD Pharmingen 145-2C11); PE-αCD19 (eBioscience MB19-1); PE-αB220 (BD Pharmingen RA3-6B2), biotin-αCD34 (eBioscience RAM34), APC-αSca (eBioscience D7), APC-Cy7-αCD117 (eBioscience 2B8), PerCP-Cy5.5-αGr1 (eBioscience RB6-8C5), PE-αCD11c (BD Pharmingen HL3), FITC-αCD11a (eBioscience M17/4), FITC-αCD29 (BD Pharmingen Ha2/5), FITC-αvascular cell adhesion molecule (VCAM) (BD Pharmingen MVCAM.A), PE-αCD49d (BD Pharmingen R1-2), FITC-αCD61 (BD Pharmingen 01864D), PerCP-Cy5.5-αCD45.2 (eBioscience 104), APC-αCD45.1 (eBioscience A20) or lineage cocktail (αCD3 (CD3e), αCD8 (53.6.7), αCD4 (L3T4), αCD19 (1D3), αB220 (RA3-6B2), αGr1 (RB6-8C5), and αTerr-119 (Ly-76) (all PE-Cy7 conjugated antibodies; eBioscience). Analysis was performed using a FACScan (Beckman Coulter) and data were analyzed using FlowJo Version 8.8.7 (TreeStar), Excel Version 2007 (Microsoft), and Prism Version 5 (GraphPad) software.
Statistical analysis
Exon-level summary data were generated using the RMA algorithm in the Affymetrix expression console. Only core probe sets were used to limit the analysis within well-annotated exons. The ratio of average signal intensity for mCG-PR+/− × Rara+/+ and mCG-PR+/− × Rara+/− samples was calculated as -fold change, and a one-way analysis of variance (ANOVA) was used. Probe sets having an expression signal less than 200 in all samples were removed before the analysis. Candidate probe sets with average signal intensity ≥ 200 in at least one of the groups were retained if they showed at least 2.0-fold dysregulation at significance level of P ≤ .05 (ANOVA) between the mCG-PR+/− × Rara+/+ and mCGPR+/− × Rara+/− samples. Only those mRNAs represented by a minimum of 3 probe sets were chosen for further analysis.
All probe sets for the genes Ddx3y, Eif2s3y, and Kdm5d were removed from the analysis, and an unsupervised hierarchical cluster analysis was performed using z-score normalized expression values in DecisionSite software Version 9.9.1 (Spotfire).
Results
Rara haploinsufficiency cooperates with PML-RARA to alter myelopoiesis in young, nonleukemic mice
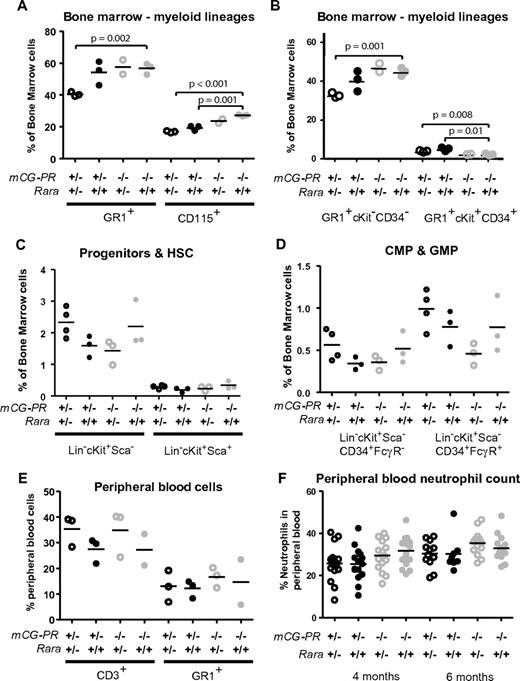
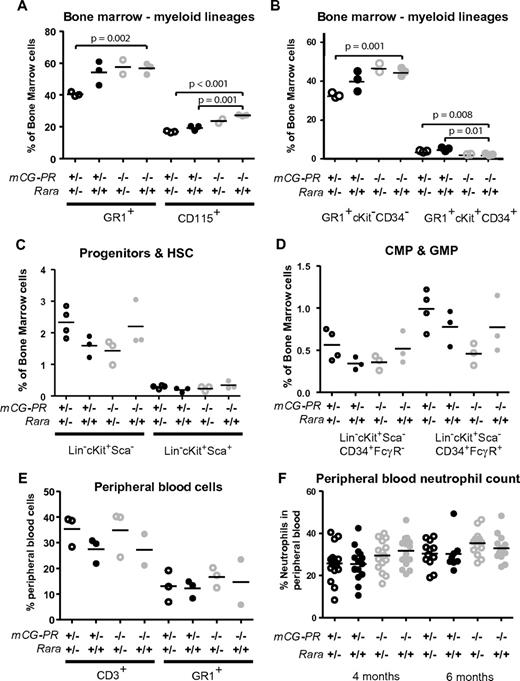
To address the role of RARA haploinsufficiency caused by the t(15;17)(q22;q21) translocation, we crossed Rara+/− mice with our previously described mCG-PR mice.25 Both the mCG-PR and the null Rara alleles were observed at normal Mendelian ratios. At 8 weeks, the bone marrow of mCG-PR+/− × Rara+/− mice exhibited a decreased number of mature myeloid cells (Gr1+ and CD115+; Figure 1A). This loss appeared predominantly within the mature neutrophil compartment (Gr1+CD34−cKit−), with a slight increase in the number of immature myeloid cells (Gr1+CD34+cKit+; Figure 1B). This shift in myeloid maturation was neither associated with a change in bone marrow–progenitor compartments nor with neutropenia or myeloproliferation. In fact, these mice maintained normal cKit+lineage−Sca1+, common myeloid precursor, and granulocyte/macrophage-progenitor compartment sizes, as well as normal spleen sizes, white blood cell (WBC) counts, and peripheral blood neutrophil counts (Figure 1C-F and data not shown). However, the effects of Rara haploinsufficiency appeared to be entirely dependent on cooperation with PML-RARA, because these phenotypes were absent in littermate-matched Rara+/− mice.
Preleukemic cooperation of Rara haploinsufficiency and PML-RARA. Immunophenotype of bone marrow cells from the indicated littermate-matched mice at 8 weeks. (A-B) Myeloid immunophenotype by Gr1, CD115, cKit, and CD34. (C-D) Progenitor immunophenotype by lineage (CD3, CD8, CD4, CD19, B220, Gr1, and Terr-119), cKit, Sca, CD34, and FcγRIII-R. (E) Peripheral blood immunophenotype by CD3 and Gr1. (F) Neutrophil counts assessed by Coulter counter at 4 and 6 months of age.
Preleukemic cooperation of Rara haploinsufficiency and PML-RARA. Immunophenotype of bone marrow cells from the indicated littermate-matched mice at 8 weeks. (A-B) Myeloid immunophenotype by Gr1, CD115, cKit, and CD34. (C-D) Progenitor immunophenotype by lineage (CD3, CD8, CD4, CD19, B220, Gr1, and Terr-119), cKit, Sca, CD34, and FcγRIII-R. (E) Peripheral blood immunophenotype by CD3 and Gr1. (F) Neutrophil counts assessed by Coulter counter at 4 and 6 months of age.
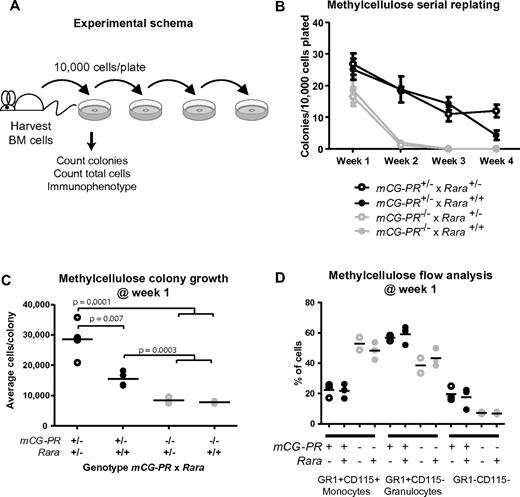
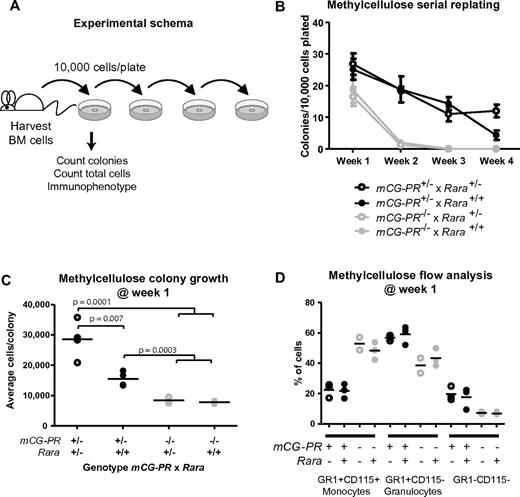
Rara haploinsufficiency cooperated with PML-RARA to increase cell growth during myeloid colony formation. Bone marrow cells were plated in methylcellulose containing IL-3, IL-6, and SCF. After 1 week, we observed a small increase in the total number of colonies in mCG-PR+/− bone marrow cells (Figure 2A-B). This phenotype was not altered by Rara haploinsufficiency. However, Rara haploinsufficiency did cooperate with mCG-PR to increase the average number of cells per colony (Figure 2C). mCG-PR altered myeloid maturation in methylcellulose, shifting the dominant immunophenotype from Gr1+CD115+ monocytes to Gr1+CD115− granulocytes; however, this phenotype was neither altered by Rara haploinsufficiency nor detected in littermate-matched Rara+/− mice (Figure 2D). Cytomorphology of these cells confirmed populations of monocytes and granulocytes with corresponding Gr1+CD115+ and Gr1+CD115− immunophenotypes, respectively, with a minor population of mast cells corresponding to the Gr1−CD115− population (data not shown).
Ex vivo growth of myeloid bone marrow cells. (A) Experimental schema. (B) Colony formation and serial replating. Bone marrow cells from littermate-matched mice at 8 weeks of age were plated in duplicate in methylcellulose containing IL-3, IL-6, and SCF, and after 7 days, colonies and total cells per duplicate plates were counted. Cells were then replated in duplicate and 7 days later, colonies were counted and replated. Replating continued for 4 weeks or until colony formation failed. (C) Methylcellulose colony growth. On day 7 of week 1, total colonies and total cells on duplicate were counted to assess the average number of cells per colony. (D) Immunophenotype of day-7, week-1 total cells from duplicate methylcellulose plates.
Ex vivo growth of myeloid bone marrow cells. (A) Experimental schema. (B) Colony formation and serial replating. Bone marrow cells from littermate-matched mice at 8 weeks of age were plated in duplicate in methylcellulose containing IL-3, IL-6, and SCF, and after 7 days, colonies and total cells per duplicate plates were counted. Cells were then replated in duplicate and 7 days later, colonies were counted and replated. Replating continued for 4 weeks or until colony formation failed. (C) Methylcellulose colony growth. On day 7 of week 1, total colonies and total cells on duplicate were counted to assess the average number of cells per colony. (D) Immunophenotype of day-7, week-1 total cells from duplicate methylcellulose plates.
Rara haploinsufficiency cooperates with PML-RARA to increase competitive repopulation efficiency
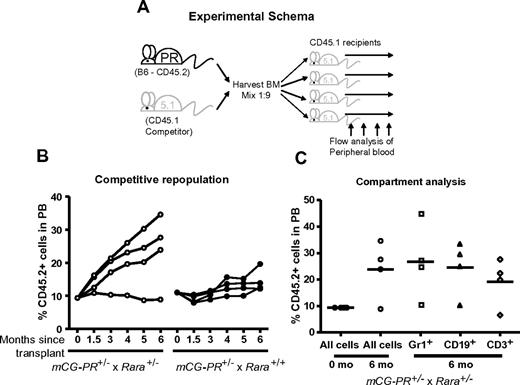
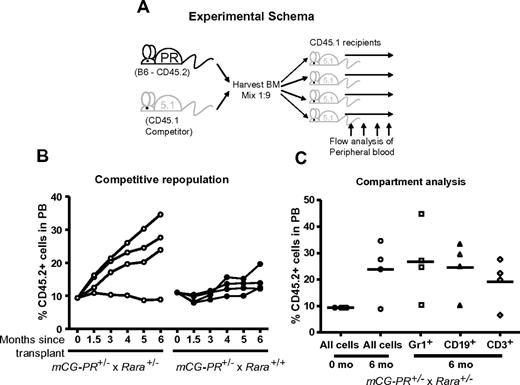
Bone marrow cells from mCG-PR+/− × Rara+/− and littermate mCG-PR+/− × Rara+/+ mice were transplanted at a ratio of 1 to 9 into age- and sex-matched CD45.1 recipients (Figure 3A). We observed a small, consistent competitive advantage in mCG-PR+/− × Rara+/+ bone marrow cells (Figure 3B). During 6 months of observation, CD45.2+ donor cells expanded from 10% of the peripheral blood to ∼ 15%. Rara haploinsufficiency cooperated with PML-RARA to enhance this phenotype, expanding CD45.2+ peripheral blood cells from 10% to ∼25% during 6 months of observation (Figure 3B). This phenotype was not observed in Rara+/− bone marrow cells (data not shown). During these 6 months of observation, recipient mice maintained normal peripheral blood counts. At 6 months, the percentage of CD45.2+ cells was assessed in both myeloid (Gr1+) and lymphoid (CD3+, CD19+) compartments. Surprisingly, the marked expansion of CD45.2+ cells observed from mCG-PR+/− × Rara+/− donor cells was detected in both the myeloid and lymphoid compartments, suggesting that mCG-PR+/− may act at a multipotent progenitor stage (Figure 3C).
Competitive repopulation. (A) Experimental schema. (B) Bone marrow cells from the indicated mice at 8 weeks of age were mixed at a ratio of 1 to 9 with competitor CD45.1 bone marrow cells from sex- and age-matched mice. These cells were transplanted into sex-matched, 6-week-old, lethally irradiated CD45.1 recipients. At the indicated time points, peripheral blood was assessed for ratios of CD45.2+ and CD45.1+ WBCs. (C) At 6 months of age, peripheral blood from mCG-PR+/− × Rara+/− recipients was assessed for ratios of CD45.2+ vs CD45.1+ cells in Gr1+, CD19+, and CD3+ compartments.
Competitive repopulation. (A) Experimental schema. (B) Bone marrow cells from the indicated mice at 8 weeks of age were mixed at a ratio of 1 to 9 with competitor CD45.1 bone marrow cells from sex- and age-matched mice. These cells were transplanted into sex-matched, 6-week-old, lethally irradiated CD45.1 recipients. At the indicated time points, peripheral blood was assessed for ratios of CD45.2+ and CD45.1+ WBCs. (C) At 6 months of age, peripheral blood from mCG-PR+/− × Rara+/− recipients was assessed for ratios of CD45.2+ vs CD45.1+ cells in Gr1+, CD19+, and CD3+ compartments.
Rara haploinsufficiency does not alter the phenotype of mCG-PR–associated leukemia
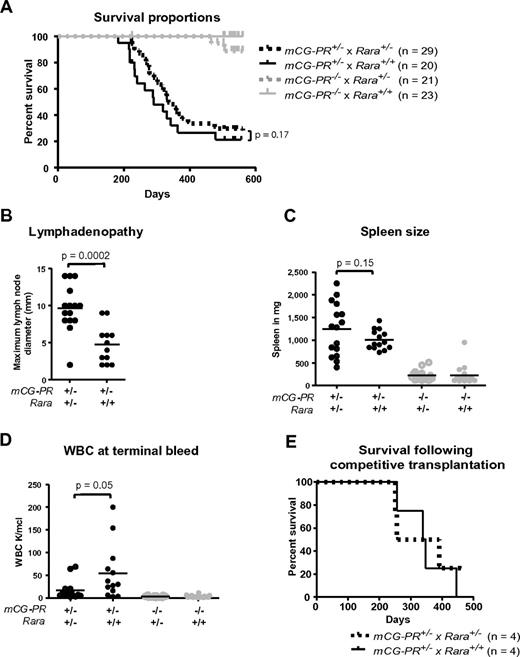
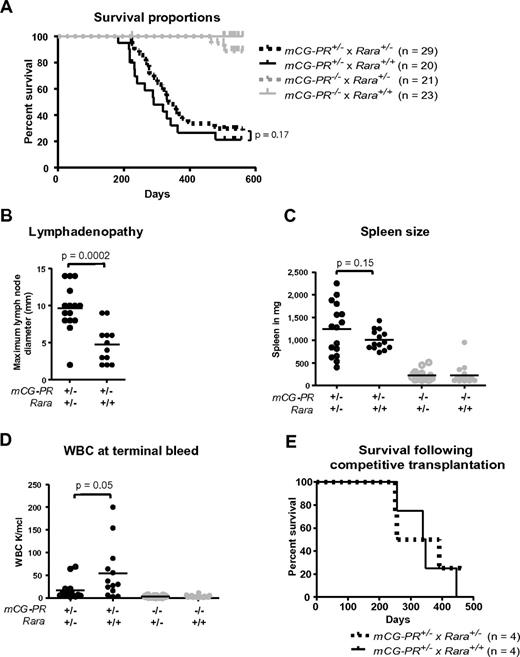
We established a tumor watch consisting of 29 mCG-PR+/− × Rara+/−, 20 mCG-PR+/− × Rara+/+, 21 mCG-PR−/− × Rara+/−, and 23 mCG-PR−/− × Rara+/+ littermate control mice. After 18 months of observation, we observed a leukemia penetrance of 88% and 84% in mCG-PR+/− × Rara+/− and mCG-PR+/− × Rara+/+ mice, respectively (Figure 4A). Median survival times of the 2 cohorts were 335 and 290 days, respectively, which were not statistically different.
Tumor watch. (A) The indicated cohorts of littermate-matched mice were prospectively established in a tumor watch. Mice were followed for 18 months and moribund animals were killed. Leukemia occurred in mCG-PR+/− mice, but not in littermate controls. There was no significant difference in latency or penetrance in mCG-PR+/− × Rara+/− vs mCG-PR+/− × Rara+/+ mice. (B) Maximum diameter of largest cervical lymph node at the time of killing. (C) Spleen size in the indicated mice at the time of killing or at 18 months of age, whichever came first. (D) Peripheral WBC counts at the time of killing or at 18 months of age. (E) Mice shown in Figure 3 were followed, and moribund mice were killed. There was no significant difference in survival by genotype of transplanted bone marrow cells.
Tumor watch. (A) The indicated cohorts of littermate-matched mice were prospectively established in a tumor watch. Mice were followed for 18 months and moribund animals were killed. Leukemia occurred in mCG-PR+/− mice, but not in littermate controls. There was no significant difference in latency or penetrance in mCG-PR+/− × Rara+/− vs mCG-PR+/− × Rara+/+ mice. (B) Maximum diameter of largest cervical lymph node at the time of killing. (C) Spleen size in the indicated mice at the time of killing or at 18 months of age, whichever came first. (D) Peripheral WBC counts at the time of killing or at 18 months of age. (E) Mice shown in Figure 3 were followed, and moribund mice were killed. There was no significant difference in survival by genotype of transplanted bone marrow cells.
Although Rara haploinsufficiency did not alter the latency or penetrance of mCG-PR–associated leukemia, it did subtly shape the leukemic tissue distribution. Leukemic mCG-PR+/− × Rara+/− mice generally presented with enlarged and matted cervical lymph nodes and occasionally with abdominal lymphadenopathy (Figure 4B and data not shown). Flow cytometry of lymph node cells revealed an infiltration of Gr1+/cKit+/CD11b+ granulocytes (a similar immunophenotype to the splenic leukemia in all animals; supplemental Figures 1 and 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Enlarged cervical lymph nodes were occasionally observed in mCG-PR+/− × Rara+/+ leukemic mice, but these were consistently smaller than those arising in mCG-PR+/− × Rara+/− littermates, and only once presented with multiple, matted lymph nodes. Fourteen of 15 evaluable mCG-PR+/− × Rara+/− mice had at least one cervical lymph node > 5 mm in size versus 5 of 12 evaluable mCG-PR+/− × Rara+/+ mice; the average maximum diameter of the largest lymph node was 9.7 mm in mCG-PR+/− × Rara+/− mice versus 4.5 mm in mCG-PR+/− × Rara+/+ mice (P = .0001). Leukemia arising in mCG-PR+/− × Rara+/− mice also presented with lower peripheral blood WBC counts (16 000/μL vs 56 000/μL, P = .045) and a nonsignificant trend toward larger spleen sizes (average 1238 mg vs 1000 mg, P = .15; Figure 3B-D).
Because the Rara haploinsufficiency–associated competitive advantage contrasted with the lack of influence on leukemic latency, the mice generated in the competitive repopulation study were followed for the development of leukemia (Figure 3). Surprisingly, the marked competitive advantage and difference in the percentage of PML-RARA+ hematopoietic cells at 6 months did not alter the latency or penetrance of the subsequent leukemia (Figure 4E). These 2 separate experiments suggest that Rara haploinsufficiency does not influence the occurrence of secondary mutations required for leukemia progression.
Because Rara has been implicated in neutrophil maturation, we assessed the effect of Rara haploinsufficiency on the maturation arrest of leukemic cells in mCG-PR mice. We performed blinded, 200-cell-count differentials on spleen cells from 7 independent mCG-PR+/− × Rara+/− leukemic samples and 7 mCG-PR+/− × Rara+/+ samples. We observed variable maturation ranging from 1% to 72% promyelocytes, but Rara haploinsufficiency did not consistently alter the fraction of promyelocytes in the tumors (average 18% vs 27%, P = .46; supplemental Figure 3A-B). We assessed the same 14 samples using flow-based immunophenotyping for Gr1, cKit, and CD11b expression. We observed a correlation between samples with a high percentage of promyelocytes by cytomorphology and those samples with large Gr1+cKit+CD11b− populations, but there was no immunophenotypic correlation by genotype (data not shown). Thus, Rara haploinsufficiency does not appear to influence the degree of myeloid maturation in mCG-PR–associated APL.
To define possible molecular mechanisms related to differences in tissue distribution, we used both candidate gene and unbiased approaches. We assessed the immunophenotype of spleen leukemia cells from 3 mCG-PR+/− × Rara+/− mice with marked lymphadenopathy and 3 mCG-PR+/− × Rara+/+ mice with no lymphadenopathy. We observed expression of Gr1, CD11b, CD34, cKit, CD29, VCAM, VLA4, CD11a, and CD44, but little or no expression of CD11c, CD61, CD62L, and CXCR4. We observed no differences based on the genotype or phenotypic presentation (supplemental Figure 1 and data not shown). We observed similar immunophenotypes in cells prepared from spleens and lymph nodes, although the percentage of Gr1+ leukemic cells was lower in lymph nodes compared with spleen (supplemental Figures 1-2 and data not shown).
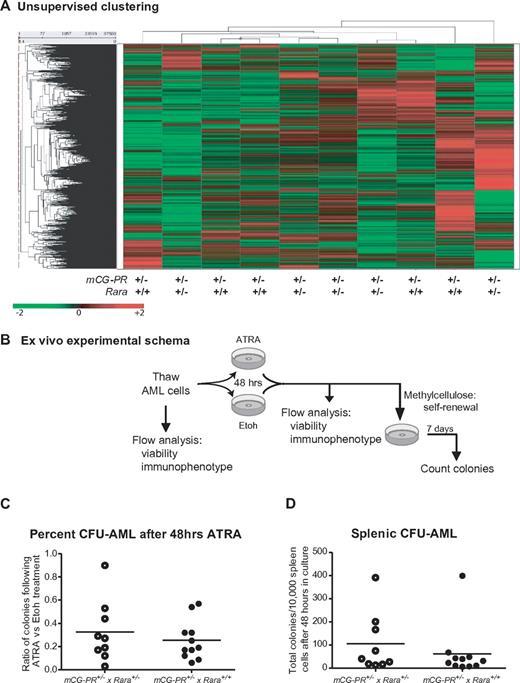
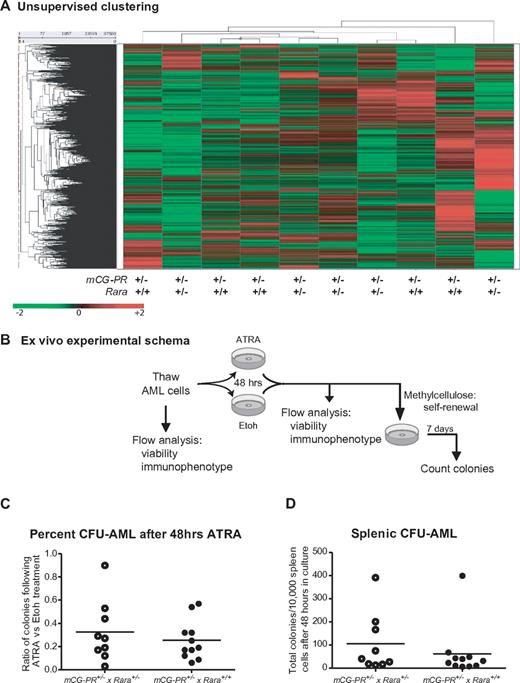
We further assessed expression differences between mCG-PR+/− × Rara+/− and mCG-PR+/− × Rara+/+ leukemias using expression array profiling on Affymetrix Exon 1.0 arrays. We observed almost no differences between spleen cells from 5 individual mCG-PR+/− × Rara+/− and 5 mCG-PR+/− × Rara+/+ leukemias. These samples did not genotypically segregate during unsupervised clustering (Figure 5A). Using one-way ANOVA, along with multiple-filter criteria (-fold change, ANOVA P value, average signal intensity > = 200), we identified only 7 mRNAs that were differentially expressed between mCG-PR+/− × Rara+/− and mCG-PR+/− × Rara+/+ leukemic spleen cells (Ddx3y, Eif2s3y, Kdm5d, IL1r2, Fn1, Hook1, and Myef2; supplemental Figure 4A). Of these, 3 (Ddx3y, Eif2s3y, and Kdm5d) are encoded by genes located on the Y chromosome, and correlate with male sex. Of the other 4, none had P values < .001. The expression of these mRNAs was compared with the expression of their human counterparts in 111 cases of human AML using our previously published dataset.38 Two of these transcripts (HOOK1 and FN1) are expressed at very low to undetectable levels in human APL; the other 2 (IL1R2 and MYEF2) are regulated during myeloid maturation and do not appear to be uniquely altered by t(15;17) compared with FAB-M4 and FAB-M5 AML (supplemental Figures 4B-G). These data suggest that Rara haploinsufficiency leads to a subtle shift in the tissue distribution of leukemic cells, but does not significantly alter the latency, penetrance, cytomorphology, immunophenotype, or expression signature of this leukemia.
Analysis of mCG-PR+/− × Rara+/− and mCG-PR+/− × Rara+/+ leukemia cells. (A) Expression array profiling of leukemic spleen cells from 5 mCG-PR+/− × Rara+/− and 5 mCG-PR+/− × Rara+/+ mice using Affymetrix Exon 1.0 arrays. Tumors did not cluster by genotype using an unsupervised analysis. (B-D) Ex vivo ATRA sensitivity. (B) Experimental schema. (C) Cryopreserved spleen cells from 9 mCG-PR+/− × Rara+/− and 9 mCG-PR+/− × Rara+/+ independent tumors were thawed and cultured for 48 hours in liquid medium containing IL-3, IL-6, and SCF ± 1μM ATRA. Viable cells were counted and plated in methylcellulose containing IL-3, IL-6, and SCF. After 7 days, colonies (C) were counted and pairwise assessed for ATRA sensitivity (D).
Analysis of mCG-PR+/− × Rara+/− and mCG-PR+/− × Rara+/+ leukemia cells. (A) Expression array profiling of leukemic spleen cells from 5 mCG-PR+/− × Rara+/− and 5 mCG-PR+/− × Rara+/+ mice using Affymetrix Exon 1.0 arrays. Tumors did not cluster by genotype using an unsupervised analysis. (B-D) Ex vivo ATRA sensitivity. (B) Experimental schema. (C) Cryopreserved spleen cells from 9 mCG-PR+/− × Rara+/− and 9 mCG-PR+/− × Rara+/+ independent tumors were thawed and cultured for 48 hours in liquid medium containing IL-3, IL-6, and SCF ± 1μM ATRA. Viable cells were counted and plated in methylcellulose containing IL-3, IL-6, and SCF. After 7 days, colonies (C) were counted and pairwise assessed for ATRA sensitivity (D).
ATRA responsiveness of Rara-haploinsufficient APL
We assessed the response of mCG-PR+/− × Rara+/− splenic leukemia cells to ATRA in vivo and ex vivo. Spleen cells from 2 independent mCG-PR+/− × Rara+/− and 2 independent mCG-PR+/− × Rara+/+ leukemias were transplanted into 6 nonirradiated CD45.1 recipients each. All 4 unique leukemias engrafted and caused a lethal disease. Similar to our prior observations, there was heterogeneity in the latency (6-12 weeks), but very little variability between replicate recipients.34 After confirming engraftment of CD45.2+/Gr1+ peripheral blood cells 5 days after transplant, half of each replicate cohort was treated with sustained-release ATRA pellets. ATRA led to decreased numbers of CD45.2+ peripheral blood cells, evidence of leukemic differentiation with an immunophenotypic shift of CD45.2+ peripheral blood cells from Gr1+/cKit+/CD11blow to Gr1+/cKit−/CD11bhigh, and improved survival regardless of Rara status (data not shown).
The biologic variability in secondary tumor engraftment limited our ability to quantitate Rara haploinsufficiency–dependent effects on ATRA responsiveness. We therefore assessed ATRA responsiveness ex vivo. Cryopreserved spleen cells from 9 independent mCG-PR+/− × Rara+/− and 9 mCG-PR+/− × Rara+/+ leukemias were thawed and cultured in liquid medium containing IL-3, IL-6, and SCF ± ATRA for 48 hours, followed by culture in methylcellulose (containing the same cytokines) for 7 days (Figure 5B). We observed little (if any) ATRA-dependent reduction in viable cells after 48 hours of culture (data not shown). After 7 days in methylcellulose, we observed that ATRA pretreatment led to a 10%-97% decline in the number of CFUs from leukemic spleen cells independent of Rara status (average reduction in CFUs: 71%; Figure 5C-D). Immunophenotyping of these colonies revealed the majority to be Gr1+/CD11b+/cKit− (data not shown).
Discussion
The t(15;17)(q22;q21) translocation associated with APL results in 4 genetic events: formation and expression of the fusion transcript PML-RARA, inconsistent formation and/or expression of the reciprocal RARA-PML fusion, and haploinsufficiency for PML and RARA. Because RARA and retinoid activity have been implicated in neutrophil maturation and hematopoietic self-renewal, RARA haploinsufficiency has been suggested to be a leukemic consequence of t(15;17).20,21 Pml haploinsufficiency and RARA-PML have previously been examined in mouse models of APL; with this study, all 4 genetic events of t(15;17) have now been evaluated for their ability to cooperate with PML-RARA and to alter the phenotype of APL.30,31
We assessed the effects of Rara haploinsufficiency on PML-RARA preleukemic and leukemic outcomes. We found that Rara haploinsufficiency cooperated with PML-RARA to shape both its preleukemic and leukemic effects. In conjunction with prior studies of Pml haploinsufficiency and RARA-PML, these data suggest that PML-RARA is the dominant contributor to t(15;17) leukemogenesis, although each of the other 3 elements can uniquely influence APL pathogenesis.30-32
We found that Rara haploinsufficiency cooperates with PML-RARA to cause a decrease in the percentage of mature Gr1+cKit−CD34− bone marrow cells and a subtle increase in the percentage of immature Gr1+cKit+CD34+ bone marrow cells. However, it does not perturb numbers of bone marrow–progenitor cells or lymphoid cells. Rara haploinsufficiency also cooperated with PML-RARA to augment a competitive repopulation advantage. However, neither of these phenotypes was associated with preleukemic myeloproliferation (as measured by flow cytometry in the peripheral blood), again suggesting that multiple “hits” are required for leukemia; PML-RARA initiates APL, but cooperating secondary mutations are required for disease progression. Interestingly, these phenotypes were not observed in Rara-haploinsufficient mice alone, suggesting that they arise from a unique interaction between PML-RARA and Rara haploinsufficiency.
Unexpectedly, Rara haploinsufficiency (unlike Pml haploinsufficiency or bcr3 RARA-PML co-expression) did not alter the latency or penetrance of mCG-PR leukemia.30,31 However, Rara haploinsufficiency altered the tissue distribution of the subsequent leukemia, with myeloid infiltration of lymph nodes, leukopenia, and a trend to greater splenomegaly. Leukopenia in Rara-haploinsufficient tumors may better reflect human APL, which usually presents with low WBC counts; however, tissue infiltration and granulocytic sarcomas are rarely observed in human APL.1 Consistent differences in cytomorphology, immunophenotype, and expression profiles were not observed, suggesting that RARA haploinsufficiency does not contribute to promyelocytic maturation arrest. We further observed that Rara haploinsufficiency did not alter ATRA sensitivity of leukemic spleen cells ex vivo.
The absence of an ATRA-dependent phenotype in the Rara-haploinsufficient tumors correlates with clinical observations that ATRA resistance is associated with acquired mutations in the ligand-binding domain or AF2 domain of PML-RARA, not in the remaining RARA allele.39,40 These data support the hypothesis that at least one PML-RARA leukemic mechanism involves binding to DNA with extended RARA-responsive elements (RAREs) or binding to non-RARE sites, because wild-type RARA does not appear to compete with PML-RARA for binding of critical DNA target sequences.
Rara haploinsufficiency cooperated with PML-RARA to increase a competitive repopulation advantage, but this was not associated with a shortened leukemic latency. This suggests that polyclonal expansion of mCG-PR+/− × Rara+/− bone marrow cells is not a rate-limiting step in leukemogenesis, and again supports the observation that Rara haploinsufficiency does not alter the probability of acquiring secondary mutations that are necessary for leukemic progression.
These data suggest that all 4 elements of t(15;17) can contribute to the pathogenesis of APL. PML-RARA is the initiating event, but other translocation elements can shape the pattern and presentation of APL. In mouse models, Pml haploinsufficiency reduces APL latency, bcl-3 RARA-PML increases penetrance (and the acquisition of an interstitial deletion of chromosome 2 that includes Spi1/Pu.1), and Rara haploinsufficiency shapes preleukemic hematopoiesis and leukemia tissue distribution.30,31
The mechanisms responsible for the t(15;17)-associated promyelocytic maturation arrest and the myeloid lineage exclusivity of t(15;17)-associated leukemia remain uncertain. Because retinoids and RARA have been implicated in neutrophil maturation, we tested the hypothesis that RARA haploinsufficiency contributes to the promyelocytic maturation arrest. However, we did not observe a more pronounced degree of promyelocytic maturation arrest in mCG-PR+/− × Rara+/− leukemias.
Differences in APL mouse model targeting strategies (although using exactly the same PML-RARA cDNA) appear to influence penetrance and promyelocytic maturation arrest, suggesting that dose and timing of PML-RARA expression influence leukemic latency, penetrance, and promyelocytic arrest.10,41 Environmental factors (eg, contemporaneous expression of neutrophil elastase and PML-RARA) have also been shown to shape APL penetrance.29 Thus, the unique morphologic presentation of t(15;17)-associated leukemia may result from a combination of factors within the microenvironment of the promyelocyte and from the lineage- and dose-specific expression of PML-RARA. In contrast, Rara haploinsufficiency per se does not appear to influence the promyelocytic maturation arrest.
Many other recurrent translocations have been modeled to varying extents in mice (eg, core bind factor translocations; non-t(15;17) APL fusion proteins; and ETV6, NUP98, and MLL translocations42 ). All 4 genetic events caused by the t(15;17) translocation have now been studied in mouse models. These studies have shown that although PML-RARA initiates APL, the disease can also be shaped and influenced by the other 3 translocation-caused events. As whole genome sequencing begins to identify the specific breakpoints of translocations, future modeling studies will need to thoroughly consider all of the resultant elements and their potential influences on disease pathogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof:
In the Introduction of this paper, we stated that no fusion proteins involving RARB, RARG or any RXRA have been reported. However, a recent paper (Such et al. Blood. 2011;117(1):242-245) described the first case of an RARG fusion transcript associated with t(11;12)(p15;q13) and a classic hypergranular acute promyelocyte leukemia. The authors identified a resulting NUP98-RARG fusion transcript, but the reciprocal RARG-NUP98 transcript was not detected. To the best of our knowledge, the report of Such et al is the first published case of an oncogenic RARG fusion transcript.
Acknowledgments
We thank Mieke Hoock for excellent mouse colony management, Callie Corsa for expert technical assistance, Pierre Chambon for the generous gift of the Rara L− mice, and the Alvin J. Siteman Cancer Center for the use of the High-Speed Cell Sorter Core Laboratory, which provided stem cell analysis.
This work was supported by the National Institutes of Health (grants CA83962 and CA101937), by the Barnes-Jewish Hospital Foundation (grant 00335-0505-02 to T.J.L.), and by a Leukemia & Lymphoma Society Fellows award (no. 5340-10 to J.S.W.).
National Institutes of Health
Authorship
Contribution: J.S.W. designed and performed research, contributed vital new reagents and analytical tools, analyzed data, and wrote the paper; J.M.K. designed and performed research; N.V. and R.N. analyzed data; and T.J.L. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: R.N. is on the scientific advisory board of Persistent Systems. Persistent Systems does not fund R.N.'s research. The remaining authors declare no competing financial interests.
Correspondence: Timothy J. Ley, MD, 660 South Euclid Ave, Box 8007, Washington University School of Medicine, St. Louis, MO 63110; e-mail: timley@wustl.edu.