Abstract
Next-generation sequencing of follicular lymphoma and diffuse-large B-cell lymphoma has revealed frequent somatic, heterozygous Y641 mutations in the histone methyltransferase EZH2. Heterozygosity and the presence of equal quantities of both mutant and wild-type mRNA and expressed protein suggest a dominant mode of action. Surprisingly, B-cell lymphoma cell lines and lymphoma samples harboring heterozygous EZH2Y641 mutations have increased levels of histone H3 Lys-27–specific trimethylation (H3K27me3). Expression of EZH2Y641F/N mutants in cells with EZH2WT resulted in an increase of H3K27me3 levels in vivo. Structural modeling of EZH2Y641 mutants suggests a “Tyr/Phe switch” model whereby structurally neutral, nontyrosine residues at position 641 would decrease affinity for unmethylated and monomethylated H3K27 substrates and potentially favor trimethylation. We demonstrate, using in vitro enzyme assays of reconstituted PRC2 complexes, that Y641 mutations result in a decrease in monomethylation and an increase in trimethylation activity of the enzyme relative to the wild-type enzyme. This represents the first example of a disease-associated gain-of-function mutation in a histone methyltransferase, whereby somatic EZH2 Y641 mutations in lymphoma act dominantly to increase, rather than decrease, histone methylation. The dominant mode of action suggests that allele-specific EZH2 inhibitors should be a future therapeutic strategy for this disease.
Introduction
Non-Hodgkin lymphomas represent a diverse spectrum of distinct entities with the 2 most common types represented by follicular lymphoma and diffuse large B-cell lymphoma (DLBCL). There are 2 molecular subtypes of DLBCL based on cell-of-origin distinctions: the activated B-cell type and the germinal center B-cell (GCB) type. Both follicular lymphoma and the GCB subtype of DLBCL derive from germinal center B cells. We have shown that, in 7% of follicular lymphomas and 22% of GCB-type DLBCL, a single point mutation in EZH2, which results in a single amino-acid change at position 641, is selected for; EZH2 (Tyr641 or WT) was mutated to phenylalanine (Y641F), asparagine (Y641N), histidine (Y641H), or serine (Y641S).1 EZH2 has been implicated as an oncoprotein often overexpressed in many solid tumors.2-4 Initial analysis of the activity of Y641 variants in cell-free reconstituted Polycomb Repressive Complex 2 (PRC2) complexes using unmethylated peptides suggested that the mutations behaved as a loss of function.1
EZH2 is the catalytic member of the PRC2; however, EZH2 alone has very weak histone-methylating activity. Other members of the PRC2 complex include EED, SUZ12, AEBP2, and RbAp48 and are required for full activity. The PRC2 complex has been shown to exhibit in vitro enzyme activity on histone peptide substrates and nucleosomes. EZH2 is a member of the Su(var)3,9, enhancer of zest, Trithorax (SET) domain containing family of histone methyltransferases (HMTases); all contain a conserved SET domain. Genetic and biochemical analysis of EZH2 SET domain has revealed their histone methyltransferase function associated with histone H3 Lys-27–specific trimethylation (H3K27me3) in vivo. Recently, a structural basis for the allosteric modulation of EZH2 activity by EED has been elucidated.5
Structural and biochemical data from other SET domain HMTases have shed light not only on the molecular mechanism of histone methylation but also the specific residues of the conserved SET domain responsible for these reactions. For example, all active HMTases contain a catalytic triad—the asparagine-histidine-serine (NHS) motif,6 and mutation of any one of these residues in the active site abolishes the activity of the enzyme. Studies have also implicated residues important for the binding of the S-adenosyl-methionine (SAM)7 and for recognition of the amino-acid sequence of the histone peptide tail.8 However, these studies have not shed light on the role of the highly evolutionarily conserved Tyr 641 of EZH2.
Through the use of molecular modeling and recently published molecular and biochemical data from an allelic series of G9a9 and by analogy to the “Tyr/Phe” switch alleles in other methyltransferases,10,11 we show that Tyr 641 of EZH2 is implicated in substrate and product specificity. Taken together with the finding that DLBCL cell lines heterozygous for EZH2 mutants exhibit higher steady-state levels of H3K27me3 and in vitro enzyme studies, a possible mechanism may be proposed. This report provides evidence that EZH2 Y641 mutant protein-containing PRC2 complexes exhibit increased activity on dimethylated peptides compared with wild-type containing PRC2 complexes, thereby shifting the steady state of H3K27 to favor trimethylation in vivo. As previously described, the EZH2 mutant-containing PRC2 complexes are inactive on unmethylated histone peptides,1 implying that the Y641F/N mutations can only act in the presence of a wild-type EZH2.
Methods
Reagents
Antibodies.
Rabbit polyclonal anti-histone H3 (ab1791), anti-histone H3K9me3 (ab8898), anti-histone H4K20me3 (ab9053), anti-EED (ab4469), and mouse monoclonal anti-histone H3K27me3 (ab6002) were purchased from Abcam. Rabbit polyclonal anti-histone H3K27me1 (07-448) and anti-histone H3K27me2 (05-821), anti-SUZ12 (ab12073), and mouse monoclonal anti-histone H3K27me3 (07-449) and anti-SUZ12 (04-046) were purchased from Millipore; the specificity of methyl-specific anti-histone H3K27 antibodies was tested using H3K27 methylated peptides (supplemental Figure 3, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Mouse monoclonal anti-EZH2 (612667) was purchased from BD Biosciences. Mouse monoclonal anti-Flag M2 (F1804) was purchased from Sigma-Aldrich, and mouse monoclonal anti-green fluorescent protein (GFP; MMS-118P) was from Covance Research Products. Antibiotin was from Cell Signaling Technology, and glyceraldehyde-3-phosphate dehydrogenase was from RDI Division of Fitzgerald Industries International.
Plasmids.
pDONR223 human EZH2 was from the human ORFeome collection.12 Point mutations were made to generate EZH2Y641F and EZH2Y641N. These ORFs were then transferred into pcDNA6.2 GFP-V5 (Invitrogen) using the LR cloning kit (Invitrogen). Plasmids containing mouse Ezh2, pCMV FLAG-Ezh2WT, and FLAG-Ezh2H689A were gifts from D. Reinberg (New York University, New York, NY).13 Plasmid inserts were fully sequenced to confirm their identity. For viral vectors, mutant constructs were generated using site-directed mutagenesis of the RefSeq EZH2 (NM_004456) as previously described.14 For transfer into a retroviral vector system, EZH2WT and EZH2Y641F were amplified by polymerase chain reaction using Phusion polymerase (New England Biolabs) according to the manufacturer's recommendations. The polymerase chain reaction product was purified, A-tailed using Taq polymerase, and cloned into pCR8/GW/TOPO (Invitrogen) from where it was transferred into a Gateway-adapted version of pSF9114 upstream of the internal ribosomal entry site and the enhanced GFP gene. As a control, the pSF91 vector carrying only the internal ribosomal entry site-enhanced GFP cassette was used. Constructs were confirmed by sequencing. Generation of recombinant ecotropic retrovirus-producing GP + E86 cells was performed as previously described.15
Primers
Primers include the following: EZH2-HA fw N-terminal MluI, gagagaacgcgtaaccatgtacccatacgacgtcccagactacgctatgggccagactgggaag; EZH2-HA fw N-terminal MfeI, gagagacaattgaaccatgtacccatacgacgtcccagactacgctatgggccagactgggaag; and EZH2-HA rev, gagagagtcgactcaagggatttccatttctcttt.
Peptides.
Biotinylated peptides, H3 (1-21) (#12-403), H3 (21-44) (#12-404), H3(21-44)K27(me1) (#12-567), H3(21-44)K27(me2) (#12-566), and H3(21-44)K27(me3) (#12-565), were purchased from Upstate Biotechnology. The purity for each peptide was assessed using mass spectrometry to be more than 95% (data not shown) and identity confirmed by dot blotting (supplemental Figure 3).
Clinical samples.
Snap-frozen initial diagnostic lymph node biopsies with a confirmed diagnosis of DLBCL were studied. In total, 10 cases were analyzed by Western blotting, including 5 cases with confirmed wild-type EZH2 and 5 mutant cases. All cases were identified by whole-transcriptome shotgun sequencing, and the DNA status then confirmed by Sanger sequencing.9 Informed consent for these studies was obtained for all patients together with research ethics approval in accordance with the British Columbia Cancer Agency policy and the Declaration of Helsinki.
Cell lines and transfection
DLBCL cell lines harboring EZH2WT or heterozygous for EZH2Y641F or EZH2Y641N (imported from the DSMZ; www.dsmz.de) were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (Invitrogen). HEK293T cells (150 000 cells per well of a 6-well plate) were transfected using Lipofectamine 2000 (Invitrogen) or TransIT−LT1 (Mirus) with 1 to 5 μg of plasmids. Stable cell lines were made by transfection of HEK293T cells with the respective plasmids and then selection with blasticidine (Sigma-Aldrich) for 1 week. Pools of stable lines expressing GFP-alone or GFP-tagged proteins were used for the experiments.
Western blotting and immunoprecipitation
Standard methods of Western blotting and immunoprecipitation were used; details are in the supplemental data. Densitometry measurements of bands were used to calculate H3K27me3/totalH3 by integrating each peak in ImageJ (1.44n, http://imagej.nih.gov/ij) software.
Large-scale immunoprecipitation and mass spectrometry
A total of 3 × 108 cells from the WSU-DLCL2 and DOHH2 cell lines were harvested, and nuclear lysates were prepared as described in the supplemental data. For large-scale immunoprecipitations, 160 mg of nuclear lysates was diluted with binding buffer and precleared with Sepharose 4B for 1 hour at 4°C. EZH2 complexes were immunoprecipitated with 30 μg of anti-EZH2 antibody (#612667, BD Biosciences) by rotating overnight at 4°C. Antibody-bound protein complexes were then incubated with 120 μL of Protein G Dynabeads (Invitrogen) for 1 hour at 4°C. Bound EZH2 complexes were washed 5 times with 1 mL of phosphate-buffered saline and resuspended in 30 μL of sodium dodecyl sulfate–polyacrylamide gel electrophoresis loading dye. Samples were resolved using a 4% to 12% gradient gel (NuPAGE, Invitrogen) and stained with colloidal Coomassie stain. After destaining, the gel was cut into 16 slices per lane corresponding to a specific molecular weight range. In-gel trypsin digestion was performed, and peptides were extracted from the gel using standard protocols.16
High-performance liquid chromatography-electrospray-mass spectrometry (MS) was performed on a 4000 QTrap mass spectrometer (Applied Biosystems/Sciex) coupled to an Agilent 1100 Nano-HPLC (Agilent Technologies) using a nano-electrospray interface, as described in supplemental data.
Histone methyltransferase assay
Similar batches of recombinant reconstituted active PRC2 complexes as used previously1 containing EZH2WT(51 004), EZH2Y641N, or EZH2Y641F were purchased from BPS Biosciences. Methyltransferase assays were performed using a kit (#17-330, Millipore) as per the manufacturer's instructions, except that biotinylated peptides were used as substrates; 250 ng of active PRC2 complex was incubated with 0.67μM 3H-SAM (PerkinElmer Life and Analytical Sciences), 1μM biotinylated peptide in 50mM Tris-HCl, pH 9.0, and 0.5mM dithiothreitol for 30 minutes at 30°C in a 10-μL volume. A total of 5 μL was spotted on a P81 square paper (Millipore), washed (twice with 10% trichloroacetic acid and once with 100% ethanol) to remove unincorporated SAM, air-dried, and placed in a glass scintillation vial with 5 mL of scintillation fluid (ScintiSafe Econo1, Fisher Chemical) and counted on a 1900TR Liquid Scintillation Analyzer (PerkinElmer Life and Analytical Sciences).
Modeling
A model of the EZH2 core catalytic domain lining the substrate lysine binding channel was built by homology to the G9a/EHMT2 structure (PDB code 2o8j), based on the alignment shown in supplemental Figure 6 and supplemental Figure 8. The EZH2 sequence was threaded onto the G9a backbone, and when homologous, G9a side chains were used as a template to position EZH2 residues, according to the “minimize tethers” command using ICM, Version 3.6-1g (Molsoft LLC17 ).
Retroviral infection and B-cell in vitro differentiation assay
Parental strain mice were bred and maintained as approved by the University of British Columbia Animal Care Committee. Primary mouse bone marrow cells were transduced as previously described with minor modifications.18 Details are in the supplemental data.
Results
EZH2Y641F/N proteins are expressed approximately 1:1 with wild-type EZH2 in DLBCL cell lines
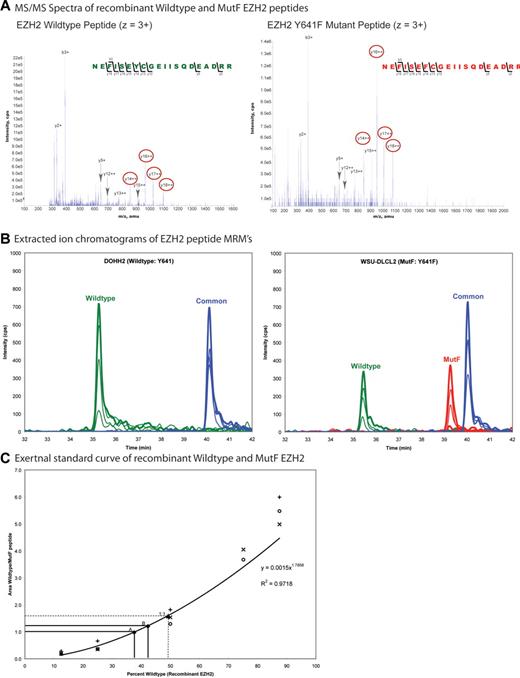
Conspicuously, the EZH2 mutations identified in GCB lymphomas are all located in exon 15 of EZH2, and all are missense mutations that change a single highly evolutionarily conserved tyrosine residue in the catalytic SET domain (Tyr641 or WT) to phenylalanine (Y641F), asparagine (Y641N), histidine (Y641H), or serine (Y641S).1 This recurrent pattern of a heterozygous somatic mutation in a functionally conserved region, affecting a single residue, is highly suggestive of a gain of dominant mode of action, either dominant negative or a gain-of-function/neomorph. Both wild-type (EZH2WT) and EZH2Y641 variant transcripts could be observed in lymphoma-derived cell lines (Y641N: DB, Karpas 422, SU-DHL-6, and Y641F: WSU-DLCL-2) bearing the mutations (supplemental Figure 1). Potentially, the mutations at EZH2Y641 could alter the stability of the protein. To investigate this possibility, we developed a multiple reaction monitoring (MRM) mode MS signature for tryptic peptides covering the mutated region of EZH2 (wild-type, MutF) and a common region from tandem MS spectra of recombinant EZH2WT and EZH2Y641F (Figure 1A; supplemental Figure 2A-B) and used this to estimate the abundance of mutant EZH2 in malignant cells. To determine the amount of mutant EZH2 in a DLCBL cell line, we immunoprecipitated total endogenous EZH2 from nuclear lysates of the WSU-DLCL2 cell line (EZH2Y641F) and the DOHH2 cell line (EZH2WT) from 2 independent experiments. Gel electrophoresis fractions of the immunoprecipitates encompassing the size of EZH2 were analyzed for the peptides by MRM-MS. Signals for the wild-type, MutF, and common peptides were observed in the WSU-DLCL2 cell line, whereas only wild-type and common signals were observed in the DOHH2 cell line (Figure 1B; supplemental Figure 2C; and data not shown), demonstrating that both EZH2WT and EZH2Y641F proteins were present in the WSU-DLCL2 cell line. The wild-type/MutF peak area ratios were compared with the wild-type/MutF peak area ratios of an external standard curve of mixtures of recombinant EZH2WT and EZH2Y641F (Figure 1C). The percentage EZH2WT was calculated from an equation fit to the standard curve. The comparison indicated that there was a 40:60 EZH2WT/EZH2Y641F protein ratio in the WSU-DLCL2 sample (Figure 1C; supplemental Figure 2C); thus, approximately equal quantities of mutant EZH2 protein are present, showing that the mutations do not adversely affect the stability of the protein in vivo and therefore that any differences in activity should not be explained by a highly skewed ratio of wild-type and mutant proteins alone.
Quantitation of EZH2 wild-type and EZH2 Y641F (MutF) protein expression by mass spectrometry. (A) Tandem MS spectra of recombinant EZH2 wild-type and MutF peptides (residues 635-654) used in the MRM assay. Peptide fragments (y and b ions) identified are labeled, and transitions used in the MRM assay are circled in red (supplemental Figure 2). (B) Extracted ion chromatograms of MRM signals for EZH2 from the gel fractions containing total EZH2 from the DOHH2 and WSU-DLCL2 samples. Traces correspond to the wild-type (green), MutF (red), and common (blue, residues 442-456) peptides. For MRM quantification, peak areas for each coeluting transition were summed for each peptide. (C) External standard curve of mixtures of recombinant EZH2WT and EZH2Y641F proteins, in triplicate. The wild-type and MutF peptides show different inherent ionization ability as illustrated by the difference in heights and areas of their specific peaks, thus necessitating the use of an external calibration curve to determine the corresponding percentage of EZH2WT and EZH2Y641F proteins in the sample. The equation fit to the curve used for the calculations is shown. Points A and B on the graph represent 2 biologic replicates of total endogenous EZH2 immunoprecipitated from the WSU-DLCL2 cell line. An independent 50:50 mixture of EZH2WT/EZH2Y641F recombinant proteins was analyzed (labeled as point “1:1”) to validate the accuracy of the procedure.
Quantitation of EZH2 wild-type and EZH2 Y641F (MutF) protein expression by mass spectrometry. (A) Tandem MS spectra of recombinant EZH2 wild-type and MutF peptides (residues 635-654) used in the MRM assay. Peptide fragments (y and b ions) identified are labeled, and transitions used in the MRM assay are circled in red (supplemental Figure 2). (B) Extracted ion chromatograms of MRM signals for EZH2 from the gel fractions containing total EZH2 from the DOHH2 and WSU-DLCL2 samples. Traces correspond to the wild-type (green), MutF (red), and common (blue, residues 442-456) peptides. For MRM quantification, peak areas for each coeluting transition were summed for each peptide. (C) External standard curve of mixtures of recombinant EZH2WT and EZH2Y641F proteins, in triplicate. The wild-type and MutF peptides show different inherent ionization ability as illustrated by the difference in heights and areas of their specific peaks, thus necessitating the use of an external calibration curve to determine the corresponding percentage of EZH2WT and EZH2Y641F proteins in the sample. The equation fit to the curve used for the calculations is shown. Points A and B on the graph represent 2 biologic replicates of total endogenous EZH2 immunoprecipitated from the WSU-DLCL2 cell line. An independent 50:50 mixture of EZH2WT/EZH2Y641F recombinant proteins was analyzed (labeled as point “1:1”) to validate the accuracy of the procedure.
EZH2Y641 mutation bearing B-cell lymphoma cell lines and GCB lymphoma containing tissues exhibit higher levels of H3K27me3 than wild-type
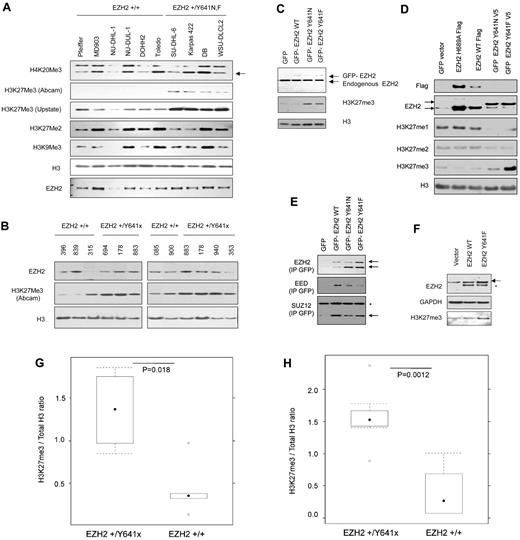
Initial studies1 of EZH2Y641F/N cell-free, reconstituted PRC2 complexes showed that the mutant EZH2-containing PRC2 complexes had 7-fold less activity in converting unmethylated H3K27 target peptides, compared with wild-type. This would suggest that mutation-bearing cell lines and tumors should have globally reduced H3K27me3 levels. A direct comparison is complicated by the nonisogenic status of the cell lines and, in the case of primary tumor material, by variable tumor cellularity. Nevertheless, we measured H3K27me3 (with 2 different H3K27me3 antibodies, verified for monospecificity; see “Western blotting and immunoprecipitation”; supplemental Figure 3) with respect to total H3, in a series of GCB-derived lymphoma cell lines, sequenced to determine the presence or absence of Y641 mutations. Unexpectedly, we found that EZH2Y641F/N bearing cell lines (Figure 2A,G) and tumors (Figure 2B,H) generally exhibited greater levels of H3K27me3 than wild-type cases, suggesting hyperactivity of the PRC2 complexes.
Y641 mutations in B-cell lines and tumors increase steady state of H3K27me3. (A) Steady-state H3K27me3 levels in DLBCL. Nuclear lysates from DLCBCLs with either wild-type EZH2 (Pfeiffer, MD903, NU-DHL-1, NU-DUL-1, DOHH2, and Toledo) or heterozygous for EZH2 (SU-DHL-6 (+/Y641N), Karpas 422 (+/Y641N), DB (+/Y641N), WSU-DLCL-2 (+/Y641F)) were probed with the respective antibodies to the following epitopes: H4K20me3 (arrowed lower band), H3K27me3, H3K27me2, H3K9me3, or EZH2. Levels of H3 were used as a loading control for histones. (B) Whole cell lysates from frozen tumor sections (each 5 × 20-μm slices) of 10 patients with either wild-type EZH2 (IDs 396, 839, 315, 085, or 900) or heterozygous for EZH2 (IDs 694 (+/Y641F), 178 (+/Y641H), 883 (+/Y641N), 940 (+/Y641F), or 353 (+/Y641S)) were probed with the respective antibodies. This a composite figure assembled to reflect similar levels of H3 from the 2 blots. (C) Nuclear lysates from HEK293T cells stably expressing GFP-tagged EZH2 and Y641 mutants were probed with anti-Ezh2 to assess ectopically expressed levels (top band) or endogenous (lower band) Ezh2 levels and anti-H3K27me3. Anti-H3 was used to assess histones as a loading control. (D) Respective plasmids encoding GFP, EZH2, or mutants (as indicated) were transfected in HEK293T cells; the lysates were probed with the antibodies (anti-FLAG M2, anti-GFP, anti-EZH2 (top band shows GFP-tagged EZH2, lower band shows endogenous EZH2 and FLAG-tagged EZH2), monomethyl, dimethyl, or trimethyl specific H3K27. These antibodies are specific for the respective methylation states (supplemental Figure 3). (E) GFP-tagged proteins from nuclear lysates of HEK293T lines stably expressing GFP-EZH2 wild-type and mutants were immunoprecipitated with GFP-trap (“Western blotting and immunoprecipitation”) and probed with anti-EZH2 to show precipitation of GFP-tagged EZH2 (top band) with copurification of endogenous EZH2 (lower band) and EED and SUZ12 (lower band, arrowed). *Nonspecific band. (F) Mouse bone marrow stem cells were infected with the respective retroviruses expressing wild-type or Y641F EZH2 and differentiated in vitro into B cells before whole cell lysates were probed with the respective antibodies. The top band (arrowed) in the EZH2 blot represents ectopic EZH2-HA and endogenous EZH2. *The lower band indicates degradation products. (G) Densitometry measurements of H3K27me3 to total H3 ratio in wild-type and mutant cell lines. The boxplots represent the distribution of ratios in mutant and wild-type. Significance testing by unpaired Student t test. (H) Densitometry measurements of H3K27me3 to total H3 ratio in wild-type and lymphoma samples. The boxplots represent the distribution of ratios in mutant and wild-type. Significance testing by unpaired Student t test.
Y641 mutations in B-cell lines and tumors increase steady state of H3K27me3. (A) Steady-state H3K27me3 levels in DLBCL. Nuclear lysates from DLCBCLs with either wild-type EZH2 (Pfeiffer, MD903, NU-DHL-1, NU-DUL-1, DOHH2, and Toledo) or heterozygous for EZH2 (SU-DHL-6 (+/Y641N), Karpas 422 (+/Y641N), DB (+/Y641N), WSU-DLCL-2 (+/Y641F)) were probed with the respective antibodies to the following epitopes: H4K20me3 (arrowed lower band), H3K27me3, H3K27me2, H3K9me3, or EZH2. Levels of H3 were used as a loading control for histones. (B) Whole cell lysates from frozen tumor sections (each 5 × 20-μm slices) of 10 patients with either wild-type EZH2 (IDs 396, 839, 315, 085, or 900) or heterozygous for EZH2 (IDs 694 (+/Y641F), 178 (+/Y641H), 883 (+/Y641N), 940 (+/Y641F), or 353 (+/Y641S)) were probed with the respective antibodies. This a composite figure assembled to reflect similar levels of H3 from the 2 blots. (C) Nuclear lysates from HEK293T cells stably expressing GFP-tagged EZH2 and Y641 mutants were probed with anti-Ezh2 to assess ectopically expressed levels (top band) or endogenous (lower band) Ezh2 levels and anti-H3K27me3. Anti-H3 was used to assess histones as a loading control. (D) Respective plasmids encoding GFP, EZH2, or mutants (as indicated) were transfected in HEK293T cells; the lysates were probed with the antibodies (anti-FLAG M2, anti-GFP, anti-EZH2 (top band shows GFP-tagged EZH2, lower band shows endogenous EZH2 and FLAG-tagged EZH2), monomethyl, dimethyl, or trimethyl specific H3K27. These antibodies are specific for the respective methylation states (supplemental Figure 3). (E) GFP-tagged proteins from nuclear lysates of HEK293T lines stably expressing GFP-EZH2 wild-type and mutants were immunoprecipitated with GFP-trap (“Western blotting and immunoprecipitation”) and probed with anti-EZH2 to show precipitation of GFP-tagged EZH2 (top band) with copurification of endogenous EZH2 (lower band) and EED and SUZ12 (lower band, arrowed). *Nonspecific band. (F) Mouse bone marrow stem cells were infected with the respective retroviruses expressing wild-type or Y641F EZH2 and differentiated in vitro into B cells before whole cell lysates were probed with the respective antibodies. The top band (arrowed) in the EZH2 blot represents ectopic EZH2-HA and endogenous EZH2. *The lower band indicates degradation products. (G) Densitometry measurements of H3K27me3 to total H3 ratio in wild-type and mutant cell lines. The boxplots represent the distribution of ratios in mutant and wild-type. Significance testing by unpaired Student t test. (H) Densitometry measurements of H3K27me3 to total H3 ratio in wild-type and lymphoma samples. The boxplots represent the distribution of ratios in mutant and wild-type. Significance testing by unpaired Student t test.
EZH2Y641F/N mutants act dominantly in HEK293T and in vitro differentiated B cells to increase H3K27me3 levels
The first 2 sets of observations strongly suggested that EZH2Y641F/N mutants could act dominantly in cells to increase H3K27me3. To test this idea, we expressed wild-type and EZH2Y641F/N proteins in HEK293T cells (wild-type for EZH2) and measured the effect on total H3K27me3 levels. Both stable and transient expression of EZH2Y641F/N mutant proteins in a wild-type context (HEK293T cells) results in a significant increase in H3K27me3 (Figure 2C; supplemental Figure 4), whereas wild-type EZH2 or the methyltransferase dead EZH2H689A,19 expressed in the same cell types results in a barely detectable increase in H3K27me3 (Figure 2D). Notably, the increase in the steady-state levels of H3K27me3 occurred, even though stable expression of GFP-EZH2Y641 protein was lower compared with endogenous EZH2 (Figure 2C upper band). To show that GFP-EZH2 was expressed as part of the PRC2,13 we immunoprecipitated GFP-tagged EZH2 and probed with EZH2, EED, or SUZ12. As shown in Figure 2E, mutant EZH2 was able to coprecipitate with other members of the PRC2 complex. EZH2Y641 mutants thus shifted the in vivo equilibrium levels of H3K27 methylation from monomethylation and dimethylation to predominantly trimethylation (Figure 2D). Interestingly, the cell lines bearing EZH2Y641F/N mutations are also uniformly and markedly more resistant (supplemental Figure 5) to 3-deazaneplanocin A, a global small-molecule inhibitor of single-carbon transfer methyltransferases, which has been reported as a nonspecific inhibitor of EZH2 activity20,21 (specific inhibitors have yet to be described). Finally, we asked whether Y641 variants were capable of conferring a gain of H3K27me3 on B-cell lineages in the mouse. Primary murine primary bone marrow cells were transduced with retroviruses expressing GFP and either wild-type EZH2 or EZH2Y641F and differentiated into the B-cell lineage in the presence of IL7 and FLT3-L. Measurement of total H3K27me3 again showed increased H3K27me3 (Figure 2F) in cells bearing EZH2Y641F, whereas EZH2WT constructs did not confer this phenotype (Figure 2F).
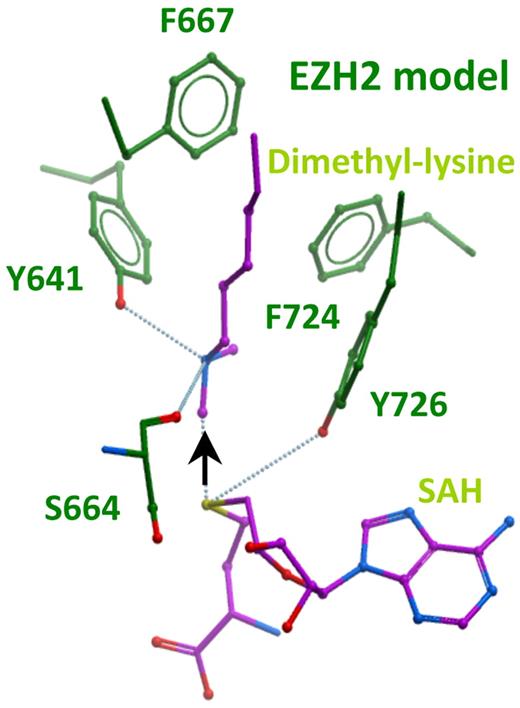
Structural modeling suggests that Tyr 641 residue may be involved in product equilibrium
We sought to understand why EZH2Y641F/N variants act dominantly through a structural analysis of this region in EZH2. Scanning mutagenesis and solved structures have provided insights into regions governing substrate preferences and specificity of several methytransferases; however, a fully solved structure of the EZH2 protein is not currently available. We therefore undertook structural homology modeling of the catalytic core region of EZH2 on the high resolution crystal structures of the lysine dimethyltransferase, G9a, and the lysine monomethyltransferase, SETD7 for which there are also detailed enzymatic data9,10 (Figure 3; supplemental Figures 6-8). Y641 of EZH2 aligns with Y1067 in G9a, which when mutated to Phe was recently shown to convert G9a into a trimethyltransferase with a concomitant decrease in monomethylation and dimethylation activity.9 Similarly, the analogous residue in SETD7 (Y245) can also confer trimethyltransferase activity on SETD7 when mutated to Ala.10 The structures reveal that Tyr in this position forms a hydrogen bond with the substrate lysine amino nitrogen moiety and keeps one of its lone pair electrons oriented away from the SAM methyl donor (Figure 3 black arrow). Thus, this Tyr residue “ties up” the face of the recipient lysyl nitrogen to which the third methyl would otherwise be transferred. This Tyr-substrate hydrogen bond also contributes to the preferred binding affinity of the enzyme for monomethylated and unmethylated substrates as evidenced by the weaker Km values of the Y1067F G9a mutants for unmethylated and monomethylated substrates.9 Furthermore, the Km of G9a Y1067F for dimethylated substrate reflects a stronger affinity for H3K27me2 than for monomethylated or unmethylated substrates.9 In the trimethyltransferases, such as EZH2, this tyrosine residue is also conserved, probably facilitating the first 2 methyl transfer reactions. Because the active sites of EZH2WT can also accommodate a third methyl group, it has probably evolved a compromise between monomethylation, dimethylation, and trimethylation (relative to G9a, for example), perhaps via small structural adjustments that allow it to carry out trimethylation, even in the presence of the Y641 residue. Thus, mutation of Y641 in EZH2 to residues, such as Phe (F) or Asn (N), which do not disturb the structure, can be predicted to abrogate the affinity for unmethylated or monomethylated lysine substrates while at the same time reducing steric crowding in the active site, thereby potentially facilitating the catalytic efficiency for trimethylation.
Homology model of the active site of EZH2. The model is based on sequence and structure alignment with histone dimethyltransferase G9a/EHMT2 (PDBID 2O8J).9 This was also structurally aligned with GLP/EHMT1 bound to an H3K9me2 peptide substrate (PDBID 2RFI), and SETD7 (PDBID 1O9S) using ICM software, Version 3.6-1g (Molsoft LLC).17 See supplemental Figures 6-8 for sequence and structural alignments. The model shows the reaction products dimethyllysine and S-adenosylhomocysteine (SAH) in purple. The black arrow indicates the path of methyl transfer from the sulfur atom (yellow) of the cofactor to the nitrogen atom (blue) of substrate lysine. Hydrogen bonds are shown as dotted blue lines.
Homology model of the active site of EZH2. The model is based on sequence and structure alignment with histone dimethyltransferase G9a/EHMT2 (PDBID 2O8J).9 This was also structurally aligned with GLP/EHMT1 bound to an H3K9me2 peptide substrate (PDBID 2RFI), and SETD7 (PDBID 1O9S) using ICM software, Version 3.6-1g (Molsoft LLC).17 See supplemental Figures 6-8 for sequence and structural alignments. The model shows the reaction products dimethyllysine and S-adenosylhomocysteine (SAH) in purple. The black arrow indicates the path of methyl transfer from the sulfur atom (yellow) of the cofactor to the nitrogen atom (blue) of substrate lysine. Hydrogen bonds are shown as dotted blue lines.
Mutations of these tyrosine residues (analogous to Y641 in EZH2) result in a shift of product equilibrium to favor the trimethylated state. The structural and functional basis of the substitution of Tyr to smaller residues, particularly Phe, is known as the Tyr/Phe switch.11,22 Although the Tyr/Phe switch has been described at other residues from scanning mutagenesis in vitro, this is the first example of a disease-associated Tyr/Phe switch.
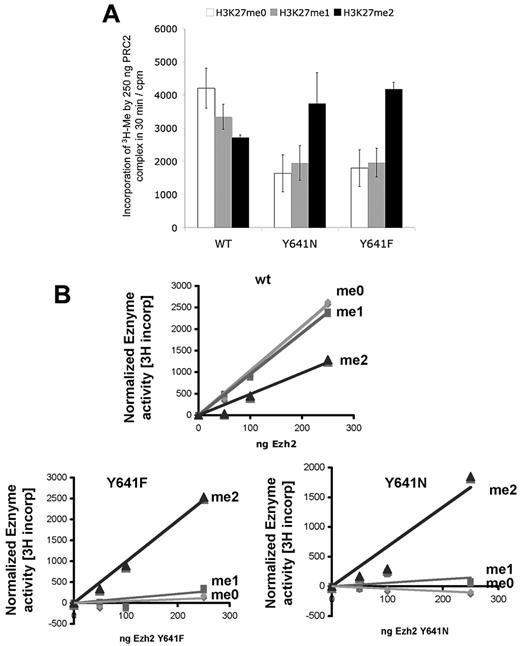
EZH2Y641-containing PRC2 complexes show increased substrate preference for dimethylated peptide in vitro
To test the structural hypothesis that EZH2Y641 mutations have a reduced ability to methylate H3K27me0/1 substrates but increased ability to methylate H3K27me2 substrates, we next studied the function of the enzyme in vitro. Because the relative catalytic efficiency of the EZH2/PRC2 complex for each methyl addition step is poorly understood, we carried out a 3H methyl incorporation assay using recombinant PRC2 complexes,23 containing EED, SUZ12, AEBP2, RbAp48, and wild-type or EZH2Y641 mutants, as previously described.1 We assayed the activity of the variant proteins by measuring 3H-methyl incorporation, using unmodified, chemically synthesized monomethylated and dimethylated K27 substrate peptides, in preference to nucleosomes because the latter contain endogenous histone modifications (supplemental Figure 3) that make nucleosome assays difficult to interpret (supplemental Figure 9). Strikingly, EZH2Y641 mutants showed (Figure 4) strongly reduced activity on unmethylated or monomethylated peptides but markedly enhanced activity on dimethylated peptides compared with EZH2 wild-type containing PRC2 complexes. We estimated the specific activity of the protein complexes (Table 1) and observed that the EZH2Y641F/N containing PRC2 complexes are approximately twice as active on dimethylated peptides compared with wild-type. These data indicate that EZH2Y641F/N mutations alter the substrate specificity, as suggested by the structural model, in favor of dimethylated peptides compared with PRC2 containing complexes with only wild-type EZH2.
In vitro PRC2 activity of EZH2. Histone methyltransferase reaction was performed using 1.2μM biotinylated peptide substrate (ie, peptide mimicking the H3 tail, H3(21-44)), which has been unmethylated (H3K27me0, open bar or me0), monomethylated (H3K27me1, gray bar or me1) or dimethylated (H3K27me2, black bar or me2) at Lys-27. The reactions were incubated with purified 250 ng (A) or various amounts (B) of PRC-2 complex containing wild-type EZH2 (WT), EZH2 mutants (Y641N or Y641F). (A) Error bars represent SD of replicates from 2 independent experiments.
In vitro PRC2 activity of EZH2. Histone methyltransferase reaction was performed using 1.2μM biotinylated peptide substrate (ie, peptide mimicking the H3 tail, H3(21-44)), which has been unmethylated (H3K27me0, open bar or me0), monomethylated (H3K27me1, gray bar or me1) or dimethylated (H3K27me2, black bar or me2) at Lys-27. The reactions were incubated with purified 250 ng (A) or various amounts (B) of PRC-2 complex containing wild-type EZH2 (WT), EZH2 mutants (Y641N or Y641F). (A) Error bars represent SD of replicates from 2 independent experiments.
Specific activity of PRC2 complexes
| PRC2 complex containing . | Enzyme activity* on various histone H3 (21-44) K27 substrates with indicated methylation states . | ||
|---|---|---|---|
| Unmethylated . | Monomethylated . | Dimethylated . | |
| EZH2WT | 10.28 (r2 = 0.99) | 9.55 (r2 = 1.00) | 4.90 (r2 = 0.95) |
| EZH2Y641N | −0.40 (r2 = 0.98) | 0.59 (r2 = 0.04) | 9.01 (r2 = 0.92) |
| EZH2Y641F | 0.45 (r2 = 0.37) | 1.09 (r2 = 0.61) | 9.86 (r2 = 0.99) |
| PRC2 complex containing . | Enzyme activity* on various histone H3 (21-44) K27 substrates with indicated methylation states . | ||
|---|---|---|---|
| Unmethylated . | Monomethylated . | Dimethylated . | |
| EZH2WT | 10.28 (r2 = 0.99) | 9.55 (r2 = 1.00) | 4.90 (r2 = 0.95) |
| EZH2Y641N | −0.40 (r2 = 0.98) | 0.59 (r2 = 0.04) | 9.01 (r2 = 0.92) |
| EZH2Y641F | 0.45 (r2 = 0.37) | 1.09 (r2 = 0.61) | 9.86 (r2 = 0.99) |
The specific activity (units/ng per minute) of PRC2 complexes containing EZH2WT, EZH2Y641N, or EZH2Y641F was calculated by linear regression from the graphs in Figure 4B.
Discussion
The relevance of somatic mutations in chromatin-modifying enzymes in cancer has recently been emphasized by discoveries of high-frequency somatic mutations in UTX, JMJD demethylases, ARID1A, and in EZH2.1,24-27 However, the mechanisms by which these mutations promote or enable carcinogenesis are largely unknown, as are the relevant modes of allelism (dominance, recessive). We show here, for the first time, a dominant (neomorphic) allele of a histone methyltransferase associated with a specific cancer.
Taken together, our data strongly suggest that EZH2Y641F/N variants in lymphoma act dominantly, through a mechanism that involves altered substrate catalytic specificity of the mutant EZH2 enzyme for its substrates. Our data show that, despite equimolar quantities of mutant and wild-type proteins in malignant cells, there is greater total H3K27me3 in mutant-bearing cell lines and primary tumors. Moreover, the EZH2Y641F/N variants have a strong dominant effect on H3K27me3 levels in cell lines that are wild-type for EZH2. They act in trans to provide a strong gain of function.
The probable mechanism is suggested by structural and enzymatic analysis of the EZH2Y641F/N variants. As shown in the cell-free assays of reconstituted PRC2 complexes, EZH2Y641F/N cannot carry out trimethylation of unmodified or monomethylated substrates (Figure 4)1 but still are able to form PRC2 complexes (Figure 2E) and increase the steady state of H3K27 trimethylation in vivo (Figure 2C-D). This means that it is probable that the gain of function of EZH2Y641 mutants would be to shift the monomethylated and/or dimethylated state of H3K27 to that of trimethylation (Figure 2D) on some bona fide EZH2 targets. It is clear that in chromatin immunoprecipitation experiments using anti-histone H3K27me1 and anti-histone H3K27me3,28 that expression of EZH2Y641 mutants could shift the steady-state levels from H3K27me1 (which is correlated with gene activation) and H3K27me2 to H3K27me3, which is correlated with gene repression.29 Whether or not these EZH2Y641 mutants are able to catalyze higher order methylation on non-EZH2 (such as EZH130 ) targets remains an area of active investigation. The mechanism and the in vitro assays show that EZH2Y641F/N cannot catalyze monomethylation and dimethylation efficiently; therefore, the mutant protein is completely dependent on the presence of wild-type proteins for the gain-of-function effect. This requirement for a wild-type copy of EZH2 is highly analogous to the behavior of the Drosophila E(z)1 allele31,32 ; E(z)1 (homologous to EZH2Y641N) leads to a phenotypic gain of function and has been shown to act as a dominant repressor of E(z) target genes. However, the unique phenotype of the E(z)1 allele depends on the presence of the wild-type allele, and the E(z)1 enzyme appears to be inactive when assayed in vitro.32
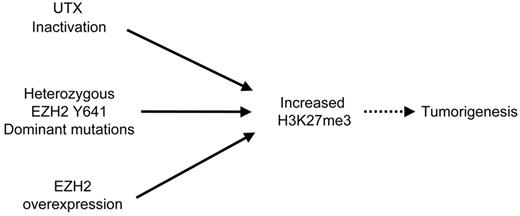
It has been shown that overexpression of EZH2 is a feature of some cancers,2-4 whereas histone H3K27 demethylase UTX was found to be inactivated in other tumors.24,25 This would suggest that the increase in H3K27me3 levels may be achieved in cancer cells by redundant pathways induced by overexpression of EZH2 or inactivation of UTX. Our findings are consistent with this idea, as dominant EZH2Y641 mutations also increase the levels of H3K27me3 (Figure 5). It is currently unknown whether the gain of H3K27me3 in germinal center-derived B-cell lymphomas reflects an increase of methylation at normal PRC2 targets, or possibly a gain of methylation at noncanonical PRC2 targets. The significance of H3K27me3 may well be lineage/cell type dependent, and this will require future conditional alleles and lineage studies to dissect as well as improved efficiency of ChIP-seq methods in small cell populations. However, recent studies have emphasized that GCB lymphomas are dependent on EZH2 and that knockdown of EZH2 in GCB cell lines bearing Y641 mutations results in cell cycle arrest in germinal center B cells.33 Knockdown of EZH2 in this context results in increased expression of PRC2-regulated tumor suppressors. This is consistent with the physiologic roles of EZH2 in B-cell development and differentiation,34-36 although it should be noted that a full understanding of the PRC2 targets in B cells remains to be elucidated. Taken together with the present study, these support the notion that EZH2 is causally implicated.
Model depicting pathways to increase H3K27me3 levels that result in cancer. In this schematic diagram: solid arrow represents a direct effect; dotted arrow, multiple steps that lead to an effect.
Model depicting pathways to increase H3K27me3 levels that result in cancer. In this schematic diagram: solid arrow represents a direct effect; dotted arrow, multiple steps that lead to an effect.
The observation of a dominant, gain-of-function mutation through altered catalytic preference in histone methyltransferases is unprecedented in cancer and carries important implications for drug development against EZH2. It is interesting to note that the substrate pockets of the mutant enzymes probably have a new, larger shape (smaller side chains of His, Asn, or Phe relative to wild-type Tyr), which potentially could be selectively targeted therapeutically in a strategy similar to that used to develop selective inhibitors of glycine gatekeeper mutants of protein kinases.37
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank D. Reinberg for the kind gifts of pCMV Flag-Ezh2 and Flag-Ezh2H689A, C. Ennis for expert project management, and T. Wee for technical assistance. DLBCL cell lines were imported from the DSMZ (www.dsmz.de).
S.A.J.R.A. was supported by a Canada Research Chair in Molecular Oncology. R.K.H. was supported by the Leukemia Lymphoma Society of Canada and the Terry Fox Foundation. T.B. was supported by the Deutsche Forschungsgemeinschaft (Bonn, Germany; grant no. BE4198/1–1). A.J.M. was supported by the Leukemia and Lymphoma Society (Career Development Program Fellowship). This work was supported by the BC Cancer Foundation and Pfizer Worldwide Research and Development (San Diego, CA). The Structural Genomics Consortium is a registered charity (no. 1097737) that receives funds from the Canadian Institutes of Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co, Inc, the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.
Authorship
Contribution: S.A.J.R.A., M.A.M., G.B.M., D.B.Y., R.K.H., and T.B. conceived of the study and led the design of the experiments; S.W.G.C., A.M., and G.B.M. produced Figure 1; D.B.Y., T.B., and J.C. performed the experimental work and produced Figures 2, 4, and 5 and the supplemental figures; T.B., M.S., and C.H.A. produced Figure 3; S.A.J.R.A., G.B.M., D.B.Y., C.H.A., and T.B. wrote the manuscript; R.D.M. and A.J.M. contributed to Figure 3 and supplemental Figure 1; V.E.M. provided the 3-deazaneplanocin A and contributed to supplemental Figure 5; and R.D.G., B.M., and M.B. provided patient samples and contributed to Figure 2.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Samuel A. J. R. Aparicio, Department of Molecular Oncology, BC Cancer Agency, 675 West 10th, Vancouver V5Z 1L3, BC, Canada; e-mail: saparicio@bccrc.ca.