The FDA in their revised approval of the warfarin prescribing information of 2007 noted the relevance of genetic testing for VKORC1 and CYP2C9 variants. Yet there is still a lack of prospective clinical trials using genetic testing, underscoring the difficulty of a pharmacogenetics-based initiation dosing strategy. In this issue of Blood, Gong and colleagues provide evidence for the clinical use of a newly established warfarin pharmacogenetics-initiation protocol (WRAPID).1
A vision to harness genomic research for effective therapy and improved health care has been recently discussed in a key article by Green and colleagues.2 They predict that, with few exceptions, translation of genomic information to clinical practice will be implemented only beyond 2020. One major barrier is the lack of research activities addressing the clinical use of genetic tests. The study by Gong and colleagues provides a good example of how a prospective evaluation of a novel pharmacogenetics-initiation protocol for warfarin therapy can improve safety and efficacy of drug therapy, using recently proposed translational research guidelines.3
Warfarin has been used for > 60 years as an oral anticoagulant and has been shown to impressively reduce the risk of stroke in patients with atrial fibrillation. Although other oral anticoagulants (eg, dabigatran,4 rivaroxaban, apixaban) are beginning to emerge as potential alternatives, warfarin will continue to be an important drug. Warfarin over-anticoagulation leads to a substantial increased risk for bleeding complications, as recently corroborated by a nationally representative public health surveillance of adverse drug reactions and a cross-sectional survey of outpatient medical visits. Here, warfarin was listed as 1 of the 10 most commonly implicated drugs in older adults for emergency department visits for adverse drug events.5
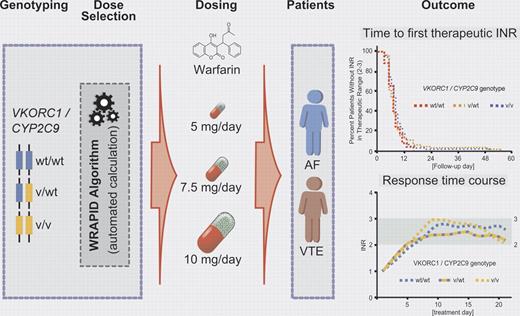
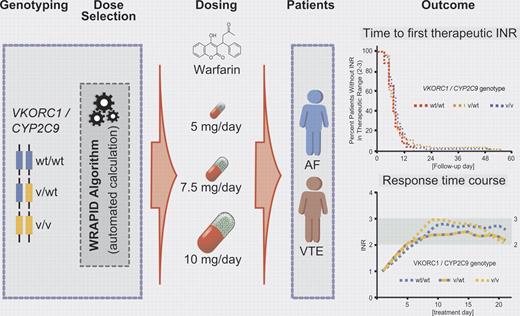
The impact of genetic variants in the vitamin K epoxide reductase complex subunit 1 (VKORC1) and the drug metabolizing enzyme cytochrome P450 2C9 (CYP2C9) has been well established by several retrospective and prospective studies. The dosing requirements of warfarin are unpredictable and highly variable (up to 20-fold).6 Variants in both VKORC1, which recycles the oxidized form to the reduced hydroquinone form of vitamin K1, and CYP2C9, which predominantly catalyzes (S)-warfarin to the 7-hydroxylated form as the major metabolite, determine the requirement of warfarin dosing. Although there have been previous attempts at pharmacogenetics-based initiation of warfarin dosing,6 the work by Gong et al for the first time employs VKORC1- and CYP2C9-dependent loading doses in patients treated for atrial fibrillation and venous thromboembolism. Because the dosing regimen includes loading and maintenance doses of warfarin, systematic pharmacokinetic and pharmacodynamic factors have been taken into consideration for the application of a novel standardized loading dose algorithm (WRAPID) in a prospective cohort study of outpatients (see figure).
Pharmacogenetics-guided initiation dosing of warfarin in patients with atrial fibrillation (AF) and venous thromboembolism (VT).
Pharmacogenetics-guided initiation dosing of warfarin in patients with atrial fibrillation (AF) and venous thromboembolism (VT).
In addition to VKORC1 and CYP2C9, other candidate genes such as CYP2C18, CYP2C19, CYP4F2, and apolipoprotein E, microsomal epoxide hydrolase, protein C, calumenin, γ-glutamyl carboxylase, and orosomucoid 1/2 have been targeted to optomize warfarin dosing, with inconsistent results. Although the contribution of CYP4F2 could not be excluded with certainty in the present study, it is important to note that, after warfarin initiation based on the WRAPID algorithm with the incorporation of VKORC1 and CYP2C9 genotypes only, no significant effects have been observed on the time required to reach the first international normalized ratios (INRs) within the therapeutic range of 2.0-3.0 and on the time spent in therapeutic range or above range during prestabilization or stabilization phases. In line with data from recent genome-wide association studies (GWAS) investigating warfarin dosing in large patient cohorts, again only the candidate genes VKORC1 and CYP2C9 and in some studies also CYP4F2 were confirmed.7,8 Thus, the clinical impact of variants in additional genes contributing to warfarin initiation dosing appears to be questionable and of minor importance. Whether or not rare variants in VKORC1 and possibly in CYP4F2 (for which systematic genetic work in various ethnic groups is still missing), need to be considered in the pharmacogenetics-guided warfarin dosing algorithms for distinct ethnic populations such as the Ashkenazi Jews9 should be explored in further studies.
All efforts toward a more individualized therapeutic approach will be judged by the success of translating promising, well-established pharmacogenetic strategies like the WRAPID algorithm into clinical practice.10 How can we convince treating physicians that the one-size-fits-all therapeutic concept is obsolete? Further studies are urgently needed to provide practical guidance for the implementation of complex pharmacogenetic strategies like WRAPID. The goal is not only to improve physicians' knowledge of pharmacogenetic fundamental principles but also to build health care teams to optimally implement complex, patient-dependent dosing regimens.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■