Abstract
The follicular lymphoma (FL) T-cell microenvironment plays a critical role in the biology of this disease. We therefore determined the lineage, differentiation state, and functional potential of FL-infiltrating CD4+ T-helper cells (TH) compared with reactive and normal lymph node (NLN) TH cells. Relative to NLNs, FL cells have decreased proportions of naive and central memory but increased proportions of effector memory TH cells. We further show differences in the distribution and anatomical localization of CXCR5+ TH populations that, on the basis of transcription factor analysis, include both regulatory and follicular helper T cells. On Staphylococcus enterotoxin-B stimulation, which stimulates T cells through the T-cell receptor, requires no processing by APCs, and can overcome regulator T cell-mediated suppression, the proportion of uncommitted primed precursor cells, as well as TH2 and TH17 cells is higher in FL cells than in reactive lymph nodes or NLNs. However, the proportion of TH1 and polyfunctional TH cells (producing multiple cytokines simultaneously) is similar in FL cells and NLNs. These data suggest that, although TH-cell differentiation in FL is skewed compared with NLNs, FL TH cells should have the same intrinsic ability to elicit antitumor effector responses as NLN TH cells when tumor suppressive mechanisms are attenuated.
Introduction
The follicular lymphoma (FL) immune microenvironment plays a critical role in mediating the biologic and clinical activities of this disease. This is supported by recent data which show that the natural history of this disease for a given patient is predicted by the gene expression profile (GEP) of the nonmalignant immune-effector cells within the tumor.1 A GEP signature enriched in T cell–associated genes is associated with a relatively indolent natural history, whereas a GEP signature enriched in monocyte/macrophage genes is associated with a more aggressive course.1 In addition, IHC studies have shown a clear association between both the number and histologic pattern of infiltration within the FL node of helper CD4+ T cells (TH), regulatory T cells (Treg), and putative T follicular helper cells (TFH), as well as of other immune-effector cells such as macrophages, with clinical outcome.2-9
Many of the patients in these series received treatment with chemotherapy, immunotherapy with the anti-CD20 Ab rituximab, or combined chemo-immunotherapy. There are several possible reasons as to why the immune microenvironment might be predictive of outcome in such treated patients. The first is that these immune effector cells may directly or indirectly modulate FL viability and sensitivity to chemotherapy or immunotherapy or both. The second relates to emerging data suggesting that one potential mechanism of action of rituximab, as well as of certain chemotherapy agents, is the elicitation of a tumor-specific immune response. This is thought to be mediated through the induction of an “inflammatory” tumor cell death that results in the release of tumor Ags in a proimmunogenic environment.10-12 This environment favors the uptake and processing of such tumor Ags by APCs that elicit tumor-specific T cells that mediate, in part, the therapeutic benefit of such agents.10-12 Indeed, we have recently shown “proof of principal” of such a “vaccinal” effect of rituximab, showing the elicitation of lymphoma idiotype-specific T cells after rituximab treatment.12 It would thus be anticipated that an immune microenvironment that would optimize the elicitation of such tumor-specific T-cell responses would be associated with a more favorable survival.
Although the myriad of interactions between the immune effector and FL cells in the tumor microenvironment have not fully been defined, it is clear that TH cells play a pivotal role in FL biology. As such, a thorough understanding of the lineage, differentiation state, and functional potential of FL TH cells are essential. This can only be fully assessed, however, with the use of multiple parameter flow cytometric analyses of FL single-cell suspensions (SCSs) in concert with IHC and molecular analysis. In addition, such findings need to be compared not only with that of reactive lymph nodes (RLNs) and tonsils (TNs), as is done in many studies, but with that of noninflamed normal lymph nodes (NLNs) because FL cells, RLNs, and TNs may share similar inflammatory and immune activation signals. Such a detailed analysis of FL-infiltrating TH cells is the objective of this study.
Herein, we show that the pattern of FL TH differentiation differs from that of NLNs, the pattern of RLNs being intermediate to that of FL cells and NLNs. Despite such differences in differentiation, the ability of FL TH cells to differentiate into TH1 and polyfunctional T-cell populations that simultaneously secrete IL-2, TNF-α, and IFN-γ are similar to that seen in NLN TH cells on stimulation with Staphylococcus enterotoxin-B (SEB). Because SEB stimulates T cells through the TCR but requires no processing by APCs and has been reported to inhibit Treg-mediated effector T-cell suppression, such stimulation may bypass several potential immune-suppressive mechanisms in the microenvironment, thus allowing an assessment of the intrinsic functional potential of these cells.13-15 These studies therefore suggest that FL TH cells should be intrinsically competent to elicit effective cellular immune responses when tumor-associated immune suppression is attenuated.
Methods
Patient samples
Primary FL cells, RLNs, and TNs were obtained from patients undergoing routine biopsy. NLNs were obtained from patients undergoing vascular surgery when NLNs are removed to gain access to vascular structures. Whole blood was obtained from healthy donors by venipuncture. Viable SCSs were prepared as previously described.16-18 All tissues were obtained under protocols approved by the University of Rochester Institutional Review Board. The pathologic and clinical data from the patients with FL are shown in Table 1.
Functional assays
SCSs were thawed and incubated in RPMI plus 10% heat-inactivated FBS overnight at 37°C and 5% CO2. Cells were left unstimulated or were stimulated with SEB (1 μg/mL; Sigma-Aldrich) or soluble anti-CD3 (OKT3) and anti-CD28 (CD28.2) monoclonal Abs (1 μg/mL; eBioscience) for 2 hours, then stained for flow cytometric analysis.
Flow cytometry and cell sorting
The fluorochrome-conjugated Abs used are shown in Tables 2 through 4. For phenotyping and sorting, SCSs were thawed, washed in PBS containing 1% FBS, and surface stained, on ice, with specific Abs. For the intracellular cytokine and ICOS/PD-1 analysis, untreated, SEB, or anti-CD3/anti-CD28–stimulated cells were suspended in 1× PBS and stained with the live/dead aqua fluorescent reactive dye (Invitrogen). Cells were then washed with HBSS containing 1% BSA, blocked with 5% normal mouse serum, and surface stained with specific Abs. Cells were then fixed/permeabilized (BD PharMingen or eBiosciences; intracellular cytokine or ICOS/PD-1, respectively) followed by staining with intracellular-specific Abs. Stained cells were acquired on an LSR-II Flow Cytometer (BD Bioscience), and data were analyzed with FlowJo Version 8.8.4 software (TreeStar). Discrete cell populations were gated on the basis of fluorescence intensity of the indicated markers compared with the appropriate fluorescence minus one controls. Cell sorting of CXCR5+ subpopulations/ surface-stained cells were subjected to 4-way flow cytometric sorting with the use of a FACSAriaII cell sorter and FACSDiva Version 6.1.3 software (BD Bioscience).
RT-PCR analysis
RNA was isolated from cell pellets with the use of the Rneasy Micro Kit (QIAGEN). RNA concentration and integrity were determined with the Nanodrop spectrophotometer and Agilent 2100 Bioanalyzer (Agilent), respectively. cDNA was generated with the WT-Ovation PicoSL System (NuGEN Technologies). Quantitative PCR was performed on an Applied Biosystems 7900 HT instrument with the use of TaqMan Universal PCR Master Mix, No AmpErase UNG, and Taqman Gene Expression Assays.
CD57 and CD4 IHC analysis
Paraffin-embedded tissue sections were stained for CD57 or CD4 with the use of standard clinical diagnostic methods. CD57+ cell distribution was characterized by analyzing 10 RLN and 20 FL cases whereby random microscope fields were selected (100× magnification), and the location of 200 CD57+ cells was noted as being either follicular or otherwise. Images were acquired with an Olympus system (BX51 microscope, DP25 camera, Microsuite software). For images in Figure 4B, original magnification was 40× (objective 4×/0.13 NA) and calibrated scalebars are provided. For images in Figure 4C, original magnification was 400× (objective 40×/0.90 NA).
Statistical analysis
The effect of tissue type on cell frequencies was first assessed with 1-way mixed ANOVAs, including tissue type as a factor and a random date-specific intercept to describe possible daily variations in flow cytometric measurements. To confirm the results obtained with these analyses we used 2-way ANOVA, including the date at which measurements were collected and tissue type as factors. The data were preliminary log-transformed in both of these analyses to correct for skewness. The influence of date on the flow cytometric measurements did not reach the 5% significance level in any of these analyses, and additional ANOVAs that assessed the effect of tissue type on cell frequencies, without controlling for date, were conducted with the distribution-free Kruskal-Wallis test. All statistical tests were 2- sided. Reported P values were not adjusted for multiple comparisons.
Unsupervised cluster analysis
To obtain a system-wide view of T-cell subset organization across nodal samples, data are displayed as clustered heat maps. Frequency data were log-transformed on the basis of the improvement of normality for most cell subsets because of Q-Q plots. Then, for each subset, the transformed frequency data were expressed as the number of SDs away from the mean across all samples. Samples and T-cell subsets were hierarchically clustered with the use of Euclidean distance and correlation, respectively, as distance measures. Samples were either clustered together or independently for each cell type. Clustering was accomplished with Matlab (The Mathworks).
Results
FL TH cells are skewed toward effector memory cells compared with NLN TH cells
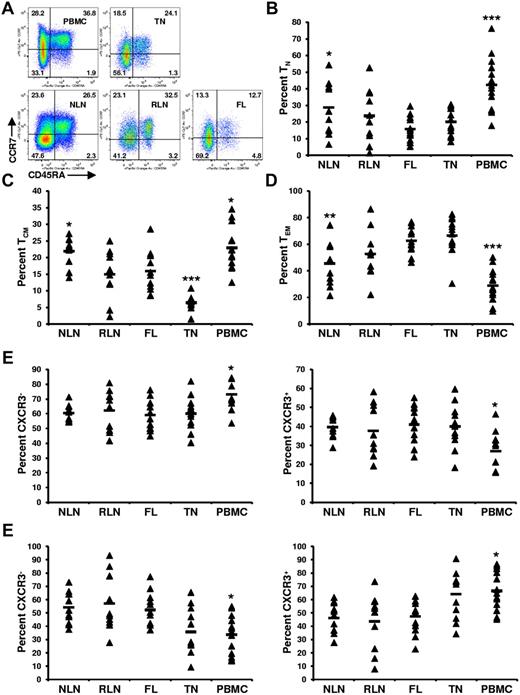
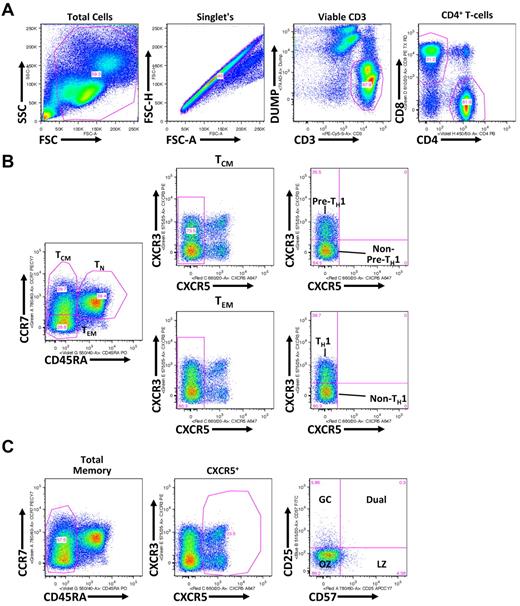
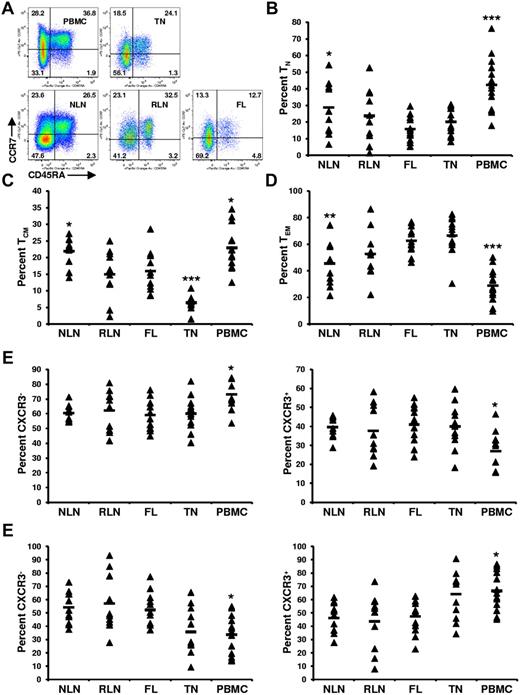
To determine whether FL TH differentiation is skewed compared with NLNs and RLNs, the proportion of TH cells that are naive (TN; CD45RA+), central memory (TCM; CD45RA−CCR7+), and effector memory (TEM; CD45RA−CCR7−) was determined.19 SCSs were analyzed with the gating strategy shown in Figure 1, with representative data shown in Figure 2A. The proportion of TH cells that are TN was significantly lower in FL cells than in NLNs (Figure 2B; P < .05) but was not statistically different from RLNs. In addition, the proportion of TN cells in FL was similar to that in TNs but significantly less than that in PBMCs (P < .001). Similarly, the proportion of TH cells that are TCM was significantly lower in FL cells than in NLNs (P < .05) but not statistically different from RLNs (Figure 2C). The proportion of TCM cells in FL was higher than that in TNs (P < .001) but less than that in PBMCs (P < .05). In contrast the proportion of TEM cells was significantly higher in FL than in NLNs (P < .01) but not significantly different from RLNs or TNs (Figure 2D). Finally, TCM and TEM populations can be subdivided on the basis of CXCR3 expression, with CXCR3+ cells being associated with a TH1-like phenotype.20,21 No significant difference was observed in the proportion of TCM or TEM cells that express CXCR3 (Figure 2E-F; P > .05). Taken together these data show that, compared with NLNs, FL TH cells are skewed away from TN and TCM differentiation and toward TEM differentiation, similar to what is seen in RLNs and TNs.
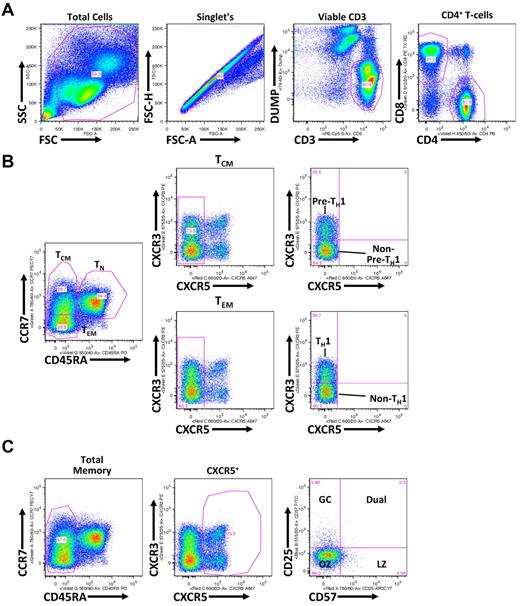
Flow cytometric gating strategy for discerning TH subsets. (A) Shown is a representative healthy donor PBMC analysis, whereby gated populations (from left to right) are indicated defining viable CD3+CD4+ T cells. (B) As indicated, viable CD3+CD4+ T cells, defined in panel A, are differentiated into TCM (CD45RA−CCR7+), TEM (CD45RA−CCR7−), and TN (CD45RA+) populations. TCM and TEM populations are further gated on the basis of CXCR3 expression with CXCR3+ TCM and TEM populations being associated with a pre-TH1 and TH1 population, respectively, and CXCR5+ TCM and TEM populations being associated with non–pre-TH1 and non-TH1 populations, respectively. (C) Total CD45RA− memory CD4+ T cells are gated based on CXCR5 expression. CXCR5+ cells are further divided into CD25 and CD57 subsets: germinal center (GC; CD57−CD25+), Dual (CD57+CD25+), light zone (LZ; CD57+CD25−), and outer zone (OZ; CD57−CD25−).
Flow cytometric gating strategy for discerning TH subsets. (A) Shown is a representative healthy donor PBMC analysis, whereby gated populations (from left to right) are indicated defining viable CD3+CD4+ T cells. (B) As indicated, viable CD3+CD4+ T cells, defined in panel A, are differentiated into TCM (CD45RA−CCR7+), TEM (CD45RA−CCR7−), and TN (CD45RA+) populations. TCM and TEM populations are further gated on the basis of CXCR3 expression with CXCR3+ TCM and TEM populations being associated with a pre-TH1 and TH1 population, respectively, and CXCR5+ TCM and TEM populations being associated with non–pre-TH1 and non-TH1 populations, respectively. (C) Total CD45RA− memory CD4+ T cells are gated based on CXCR5 expression. CXCR5+ cells are further divided into CD25 and CD57 subsets: germinal center (GC; CD57−CD25+), Dual (CD57+CD25+), light zone (LZ; CD57+CD25−), and outer zone (OZ; CD57−CD25−).
Skewed proportions of CD4+ TN, TCM, and TEM cells in FL. (A) Shown are representative plots of healthy donor PBMCs, reactive TN, NLN, RLN, and FL defining CD4+ TN (CD45RA+), TCM (CD45RA−CCR7+), and TEM (CD45RA−CCR7−) populations (numbers above each gate are representative of the percentage of total CD4+ T cells). Proportions of TN, (B), TCM (C), and TEM (D) are expressed as the percentage of the total CD4+ T cells in the indicated tissues. Also shown are the proportions of TCM that are either CXCR3− or CXCR3+ (E) or the proportions of TEM that are either CXCR3− or CXCR3+ (F). Triangles represent values for individual samples, and the rectangles represent averages for each tissue type expressed as a percentage of the total CD4+ T cells (B-D); TCM cells (E-F), or TEM cells (G-H). *P < .05, **P < .01, and ***P < .001 compared with FL.
Skewed proportions of CD4+ TN, TCM, and TEM cells in FL. (A) Shown are representative plots of healthy donor PBMCs, reactive TN, NLN, RLN, and FL defining CD4+ TN (CD45RA+), TCM (CD45RA−CCR7+), and TEM (CD45RA−CCR7−) populations (numbers above each gate are representative of the percentage of total CD4+ T cells). Proportions of TN, (B), TCM (C), and TEM (D) are expressed as the percentage of the total CD4+ T cells in the indicated tissues. Also shown are the proportions of TCM that are either CXCR3− or CXCR3+ (E) or the proportions of TEM that are either CXCR3− or CXCR3+ (F). Triangles represent values for individual samples, and the rectangles represent averages for each tissue type expressed as a percentage of the total CD4+ T cells (B-D); TCM cells (E-F), or TEM cells (G-H). *P < .05, **P < .01, and ***P < .001 compared with FL.
Proportion of memory T cells expressing CXCR5 is greater in FL cells than NLNs
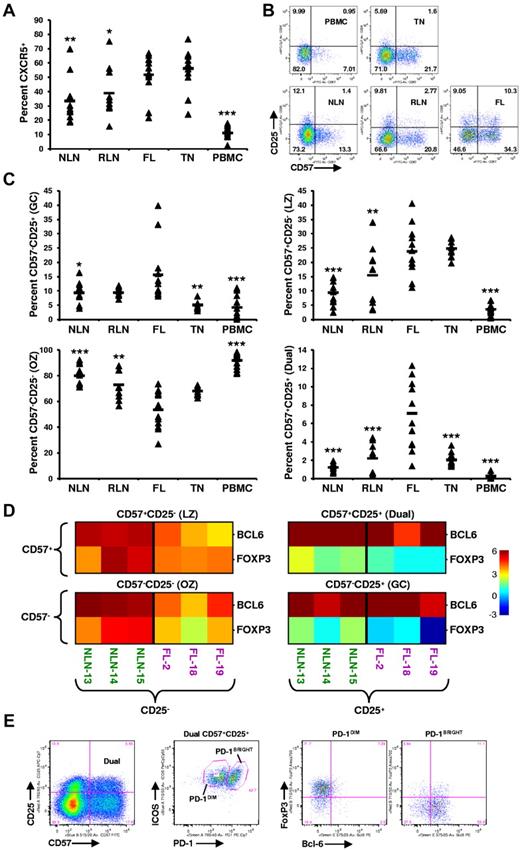
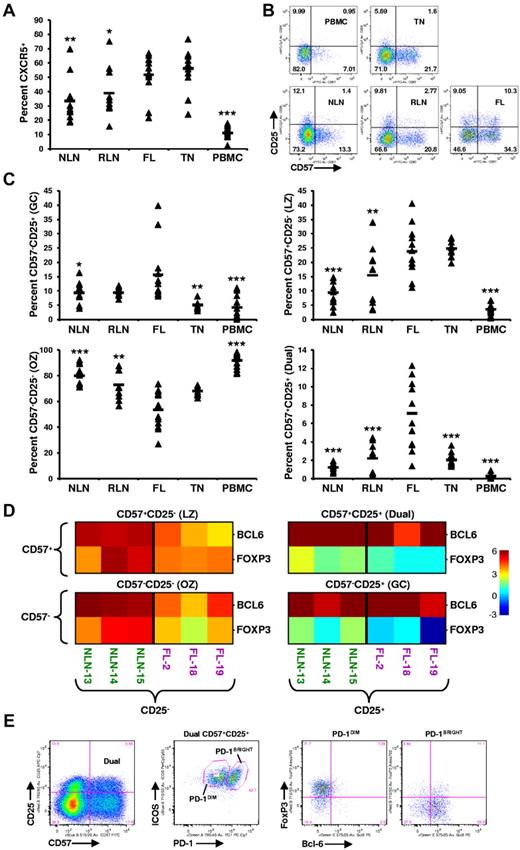
Memory T cells (TM) expressing CXCR5, the receptor for the CXCL13 chemokine, include a population of TH cells known as TFH cells, which play a critical role in facilitating germinal center (GC) B-cell differentiation into high-affinity plasma and memory cells.22 As shown in Figure 3A, the proportion of TM cells expressing CXCR5 in FL (51.8% ± 14.7%) is greater than that of NLNs (33.5% ± 15.8%; P < .01), RLNs (39.0% ± 17.5%; P < .05), and PBMCs (11.1% ± 5.1%; P < .0001) but similar to that in TNs (56.29% ± 14.63%; P > .05).
Skewed proportions of CD4+ CXCR5+ memory T-cell subsets in FL. (A) Proportions of CXCR5+ memory (CD45RA−) CD4+ T cells in the indicated tissues are expressed as the percentage of the total CD45RA− memory T-cell population. (B) Shown are representative plots of healthy donor PBMCs, reactive TN, NLN, RLN, and FL defining CD4+CD45RA−CXCR5+ subsets defined by CD57 and CD25 expression. (C) Proportions of GC (CD57−CD25+), LZ (CD57+CD25−), OZ (CD57−CD25−), and the dual (dual, CD57+CD25+) CD4+CXCR5+ memory T-cell in the indicated tissues expressed as the percentage of the total CXCR5+CD45RA− memory T-cell population. Triangles represent values for individual samples, and rectangles represent averages for each tissue type. *P < .05, **P < .01, and ***P < .001 compared with FL. (D) Quantitative RT-PCR analysis of sorted CXCR5+ TFH subsets defined in panel B, from FL or NLN (BCL-6 or FOXP3 expression normalized to GAPDH). Color indicates the number of cycles more (red) or less (blue) than the GAPDH control. (E) Further characterizing the FL CD57+CD25+ dual population (first plot) into PD-1 and ICOS populations (second plot) defining a ICOS+PD-1DIM and a ICOS+PD-1BRIGHT population that are predominately FoxP3+ (third plot) and Bcl-6+ (forth plot), respectively. Shown is a representative analysis of one FL specimen, with similar results obtained when analyzing an additional 3 FL patient specimens (total n = 4).
Skewed proportions of CD4+ CXCR5+ memory T-cell subsets in FL. (A) Proportions of CXCR5+ memory (CD45RA−) CD4+ T cells in the indicated tissues are expressed as the percentage of the total CD45RA− memory T-cell population. (B) Shown are representative plots of healthy donor PBMCs, reactive TN, NLN, RLN, and FL defining CD4+CD45RA−CXCR5+ subsets defined by CD57 and CD25 expression. (C) Proportions of GC (CD57−CD25+), LZ (CD57+CD25−), OZ (CD57−CD25−), and the dual (dual, CD57+CD25+) CD4+CXCR5+ memory T-cell in the indicated tissues expressed as the percentage of the total CXCR5+CD45RA− memory T-cell population. Triangles represent values for individual samples, and rectangles represent averages for each tissue type. *P < .05, **P < .01, and ***P < .001 compared with FL. (D) Quantitative RT-PCR analysis of sorted CXCR5+ TFH subsets defined in panel B, from FL or NLN (BCL-6 or FOXP3 expression normalized to GAPDH). Color indicates the number of cycles more (red) or less (blue) than the GAPDH control. (E) Further characterizing the FL CD57+CD25+ dual population (first plot) into PD-1 and ICOS populations (second plot) defining a ICOS+PD-1DIM and a ICOS+PD-1BRIGHT population that are predominately FoxP3+ (third plot) and Bcl-6+ (forth plot), respectively. Shown is a representative analysis of one FL specimen, with similar results obtained when analyzing an additional 3 FL patient specimens (total n = 4).
Distribution of CXCR5+ T-cell subsets differs in FL compared with NLN
Distinct subsets of CXCR5+ TM cells, defined by their coexpression of CD57 and CD25 (Figure 3B), have been shown to have unique functions as well as distinct anatomical niches within human TNs.23-25 The proportion of CXCR5+ T cells (Figure 3C) that are CD57−CD25+ (GC) is higher in FL (15.6% ± 10.3%) than in NLNs (9.3% ± 3.9%; P = .013), TNs (5.1% ± 1.3%; P = .0013), or PBMCs (4.2% ± 4.2%; P = .0006) but not statistically different from RLNs (9.3% ± 1.5%; P > .05). The proportion of CD57+CD25− (LZ) cells in FL (23.8% ± 8.6%) is greater than that in NLNs (9.4% ± 4.3%; P < .0001), RLNs (15.5% ± 10.2%; P = .021), and PBMCs (3.6% ± 1.7%; P < .0001) but similar to that of TNs (24.8% ± 2.9%; P > .05). In contrast, significantly fewer CD57−CD25− (OZ) cells were seen infiltrating FL (53.4% ± 14.9%) than NLNs (80.1% ± 8.2%; P < .0001), RLNs (72.9% ± 11.3%; P = .0013), or PBMCs (91.9% ± 5.7%; P < .0001) but similar to that in TNs (P > .05). Finally, we identify a distinct dual CD57+CD25+ population (Dual) that is overrepresented in FL compared with that of the other tissues examined; 7.11% ± 3.71% of the FL CXCR5+ TM cells are CD57+CD25+, whereas only 1.24% ± 0.42%, 2.21% ± 1.58%, 2.03% ± 0.69%, and 0.27% ± 0.29% of such cells are in NLNs, RLNs, TNs, and PBMCs, respectively (P < .0001). Taken together, in addition to there being an increased proportion of TM cells expressing CXCR5 in FL, the distribution of CXCR5+ TM subsets are different in FL, and a novel population of CXCR5+CD57+CD25+ cells exist in FL.
CXCR5+ T-cell subsets include cells that express the transcription factors BCL-6 or FOXP3 or both
To better characterize these CXCR5+ cells, we sorted nodal CD4+CD45RA−CXCR5+ T cells on the basis of their coexpression of CD57 and CD25 and performed quantitative PCR analysis for BCL-6 and FOXP3, transcription factors associated with TFH and Treg cells, respectively.26,27 As shown in Figure 3D, relative to GAPDH, each CXCR5+ subset, from either NLNs or FL, expressed detectable amounts of both BCL-6 and FOXP3. However, the CD25+ subsets expressed greater levels of FOXP3 than that of the CD25− subsets, regardless of their level of CD57 expression. Specifically, there are roughly equivalent cycles worth of FOXP3 in the FL CD25+ subsets compared with GAPDH, 20.76 ± 0.40 compared with 20.86 ± 1.25, respectively, whereas there are only 23.91 ± 0.55 cycles of FOXP3 in the FL CD25− subsets compared with 20.65 ± 0.36 cycles of GAPDH. Given that the GAPDH cycle time (CT) was consistently within 20.5 cycles regardless of the subset (average of all GAPDH CT was 20.49 ± 0.76), there is roughly 7.8-fold more FOXP3 message in the FL CD25+ subsets than in the FL CD25− subsets (each cycle corresponds to a 2.5-fold increase in message). In contrast, the FL CD25− subsets expressed greater levels of BCL-6 than the FL CD25+ subsets regardless of their CD57 expression level. Taken together, the CXCR5+ populations defined by their coexpression of CD57 and CD25 probably include both Treg and TFH cells and possibly cells at different stages of inter-conversion between these populations.
A more detailed analysis of the FL CXCR5+ dual CD25+CD57+ population was next conducted (Figure 3E). This population was shown to consist of 2 distinct ICOS+ populations, one that is PD-1DIM, the other PD-1BRIGHT. The PD-1DIM cells express FoxP3, whereas the PD-1BRIGHT cells express Bcl-6, consistent with Treg and TFH cells, respectively. Although we show this novel CXCR5+CD57+CD25+ dual population to include both Treg and TFH cells, we cannot rule out the possibility that these cells transition back and forth between these 2 populations, with the relatively small proportions (∼ 10%) of dual Bcl-6+ and FoxP3+ cells representing those in a transitional state.
Proportions of T cells expressing CD57 is higher in FL and have variable anatomical localizations
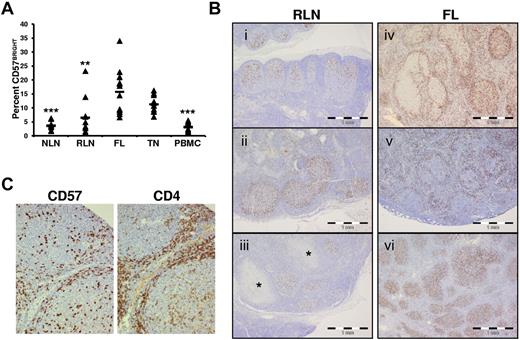
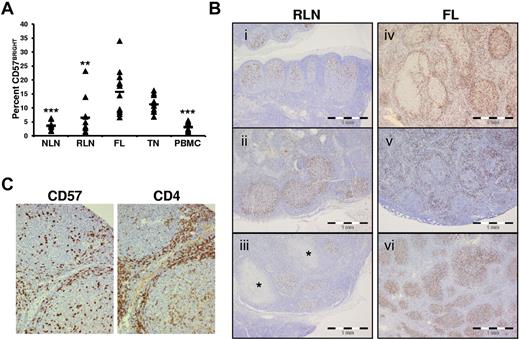
CD4+CD57+ T cells are one of the main TFH populations in the GC.22,28 In TNs, these cells have a dual function, providing classic B-cell help and suppressing conventional CD4+ T-cell activation.29 We show in Figure 4A that the proportion of FL CD57BRIGHT TH cells, which are associated with TFH cells,24 is higher than in NLNs. In addition, IHC analysis of nodal tissue sections show that the distribution of CD57+ cells in RLNs (Figure 4Bi-iii) is consistently seen in the follicle in 10 of 10 specimens evaluated. In contrast, the distribution of CD57+ cells varied widely among FL specimens and even within FL specimens, (Figure 4Biv-vi). In 20 cases of FL, 3 had more than three-quarters of the CD57+ cells within follicles (Figure 4Bvi), 11 showed more than three-quarters of CD57+ cells outside follicles (Figure 4Bv), and 6 showed a mixed distribution of cells (Figure 4Biv). In all cases, CD57+ cells colocalized with CD4+ cells (Figure 4C).
Increased proportions and altered anatomical localization of CD57+ T cells in FL. (A) Proportions of CD57BRIGHT CD4+ T cells in the indicated tissue are expressed as the percentage of the total CD4+ T cells. Triangles represent values for individual samples, and rectangles represent averages for each tissue type expressed as a percentage of the total CD4+ T cells. *P < .05, **P < .01, and ***P < .001 compared with FL. (B) ICH analysis of CD57+ cells within RLNs and FL. In RLNs, CD57+ cells are highly restricted to the GC and show polarization, with a relative abundance in the LZ (Bi-iii). This stereotypic distribution was seen in 10 of 10 RLN specimens, even when the node showed other features of inflammation such as necrotizing granulomata (* in Biii). In contrast, the distribution of CD57+ cells varied widely among FL specimens. Random microscope fields were selected (original magnification, ×100) from 20 cases of FL, and the location of 200 CD57+ cells was noted as either follicular or otherwise. Three of 20 cases had more than three-quarters of the CD57+ cells within follicles (Bvi), 11 of 20 showed more than three-quarters of CD57+ cells outside follicles (Bv, in which the CD57+ cells appear associated with a mantle zone), and 6 of 20 showed a mixed distribution of cells (Biv). (C) IHC analysis of CD57 and CD4 shows CD57+ cells colocalized to areas of CD4+ cells in a FL nodal section.
Increased proportions and altered anatomical localization of CD57+ T cells in FL. (A) Proportions of CD57BRIGHT CD4+ T cells in the indicated tissue are expressed as the percentage of the total CD4+ T cells. Triangles represent values for individual samples, and rectangles represent averages for each tissue type expressed as a percentage of the total CD4+ T cells. *P < .05, **P < .01, and ***P < .001 compared with FL. (B) ICH analysis of CD57+ cells within RLNs and FL. In RLNs, CD57+ cells are highly restricted to the GC and show polarization, with a relative abundance in the LZ (Bi-iii). This stereotypic distribution was seen in 10 of 10 RLN specimens, even when the node showed other features of inflammation such as necrotizing granulomata (* in Biii). In contrast, the distribution of CD57+ cells varied widely among FL specimens. Random microscope fields were selected (original magnification, ×100) from 20 cases of FL, and the location of 200 CD57+ cells was noted as either follicular or otherwise. Three of 20 cases had more than three-quarters of the CD57+ cells within follicles (Bvi), 11 of 20 showed more than three-quarters of CD57+ cells outside follicles (Bv, in which the CD57+ cells appear associated with a mantle zone), and 6 of 20 showed a mixed distribution of cells (Biv). (C) IHC analysis of CD57 and CD4 shows CD57+ cells colocalized to areas of CD4+ cells in a FL nodal section.
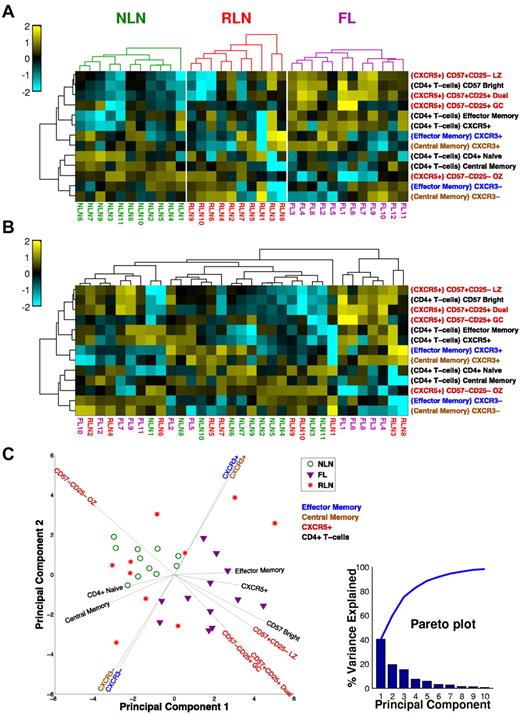
Unsupervised cluster analysis segregates FL from NLNs
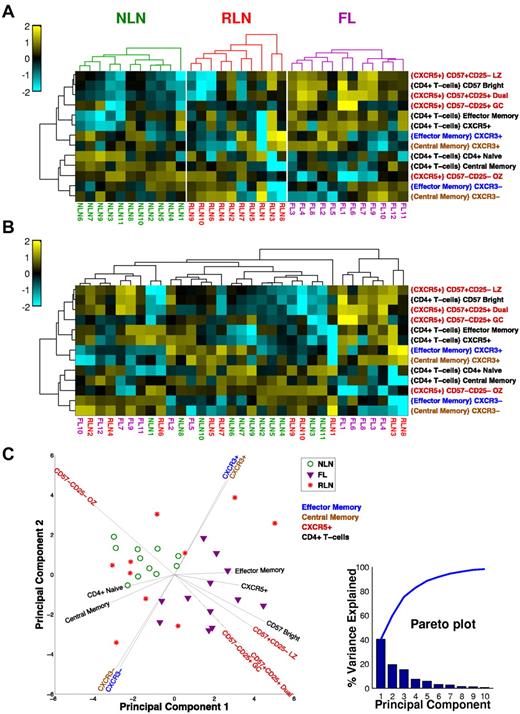
When all of the TH cell subsets are considered together as an immune profile, the FL tissue segregates from NLNs. Figure 5 illustrates this as a clustered heat map in which color relates to the deviation from the mean of all the samples for a particular subset. In Figure 5A, samples are clustered separately for each tissue type. Clearly, there are several subsets that segregate FL and NLNs, notably the CXCR5+ subsets that tend to be relatively more abundant in FL (except for the OZ subtype, which is more abundant in NLNs). In Figure 5B, all of the samples are clustered together so that their inherent organization is shown and can be related to tissue type. Note that there appears to be a bimodal distribution of FL populations which are distinct from the NLN cluster. In contrast, the RLN samples are distributed across both NLN and FL samples. This shows a clear phenotypic difference between NLNs and FL, whereas the RLNs are not well defined, some looking phenotypically like NLNs and others like FL. The differences observed between the NLN and FL samples are even more evident with the use of a principal component (PC) analysis of the TH subset data (Figure 5C) whereby the FL and NLN samples clearly segregate from one another (with the PCs driving this distinction being the CXCR5+ subsets), whereas the RLN samples are interspersed throughout. In addition, the TH1-like CXCR3 memory populations appear to drive the bimodal distribution of the FL samples.
Unsupervised cluster analysis of CD4+ TH populations. (A) Clustered heat map showing samples (columns) and T-cell subsets (rows). For each subset, the frequency data (parent population is in braces) is expressed as the number of SDs above (yellow) and below (cyan) the mean across all samples, after log-transformation. Samples are clustered (within each tissue type) on the basis of Euclidean distance; subsets clustered by correlation coefficient. Dendrograms were constructed with complete linkage. (B) Same as panel A, but all samples are clustered together to show how the samples group independent of the tissue type. (C) PC analysis of dataset. The first 2 PCs of the 21 original variables (normalized cell subset frequencies) are shown. Each symbol represents a NLN (green circle), FL (purple triangle), or RLN (red star) sample. Most of the variance in the data is explained by the first 3 PCs (see pareto plot, inset, which shows that the first 3 PC account for ∼ 80% of the overall variance), indicating that several of the cell subsets are correlated with each other. Vectors emanating out from the center show how the original variables (cell subsets) are related to the first 2 PCs. The horizontal and vertical projections of a vector indicate the relative contributions of that variable to the first and second PC, respectively. The color of the cell subset label for each vector indicates the parent population used to compute the frequencies (see the key, top right corner of plot).
Unsupervised cluster analysis of CD4+ TH populations. (A) Clustered heat map showing samples (columns) and T-cell subsets (rows). For each subset, the frequency data (parent population is in braces) is expressed as the number of SDs above (yellow) and below (cyan) the mean across all samples, after log-transformation. Samples are clustered (within each tissue type) on the basis of Euclidean distance; subsets clustered by correlation coefficient. Dendrograms were constructed with complete linkage. (B) Same as panel A, but all samples are clustered together to show how the samples group independent of the tissue type. (C) PC analysis of dataset. The first 2 PCs of the 21 original variables (normalized cell subset frequencies) are shown. Each symbol represents a NLN (green circle), FL (purple triangle), or RLN (red star) sample. Most of the variance in the data is explained by the first 3 PCs (see pareto plot, inset, which shows that the first 3 PC account for ∼ 80% of the overall variance), indicating that several of the cell subsets are correlated with each other. Vectors emanating out from the center show how the original variables (cell subsets) are related to the first 2 PCs. The horizontal and vertical projections of a vector indicate the relative contributions of that variable to the first and second PC, respectively. The color of the cell subset label for each vector indicates the parent population used to compute the frequencies (see the key, top right corner of plot).
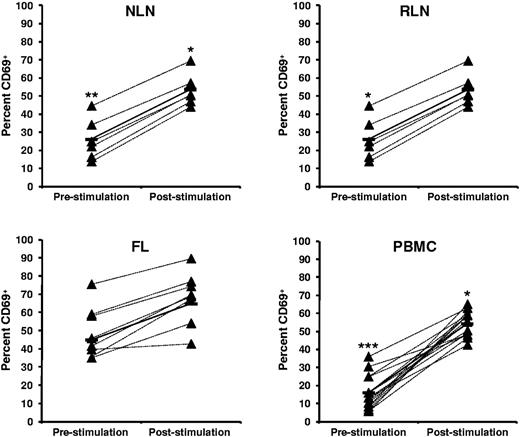
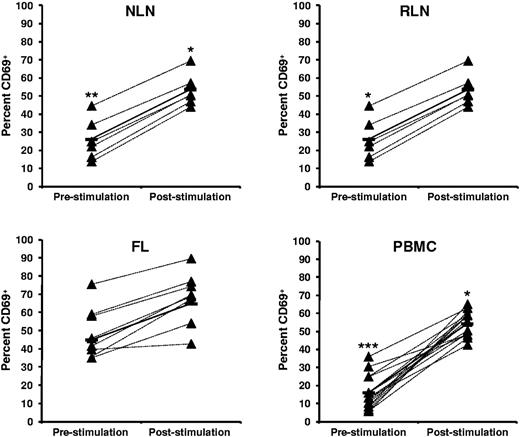
FL TH cells display a more activated phenotype before SEB stimulation than NLNs and RLNs
The activation status of TH cells within FL, NLNs, RLNs, and PBMCs, both before and after SEB stimulation, was assessed by analysis of CD69 expression (Figure 6). Before SEB stimulation, the FL TH cells have a more activated phenotype than either NLNs (P < .01) or RLNs (P < .05). However, on SEB stimulation the FL TH cells can be further activated, to an extent similar to that observed for the NLN or RLN cells (P < .05).
Higher proportions of CD69+ CD4+ T cells in FL before SEB stimulation. Activation status, as measured by CD69, before and after SEB stimulation of CD4+ T cells from (A) NLN, (B) RLN, (C) FL, and (D) healthy donor PBMCs are shown expressed as the percentage of the total CD4+ T cells. Triangles represent values for individual samples, and rectangles represent averages for each tissue type expressed as a percentage of the total CD4+ T cells. *P < .05, **P < .01, and ***P < .001 compared with FL, either before or after SEB stimulation.
Higher proportions of CD69+ CD4+ T cells in FL before SEB stimulation. Activation status, as measured by CD69, before and after SEB stimulation of CD4+ T cells from (A) NLN, (B) RLN, (C) FL, and (D) healthy donor PBMCs are shown expressed as the percentage of the total CD4+ T cells. Triangles represent values for individual samples, and rectangles represent averages for each tissue type expressed as a percentage of the total CD4+ T cells. *P < .05, **P < .01, and ***P < .001 compared with FL, either before or after SEB stimulation.
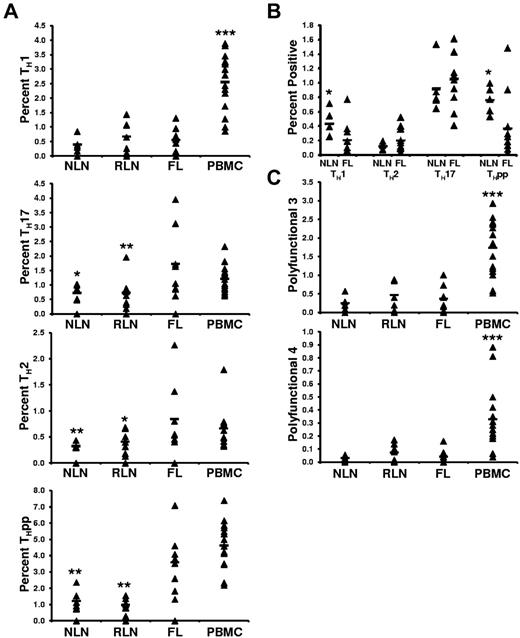
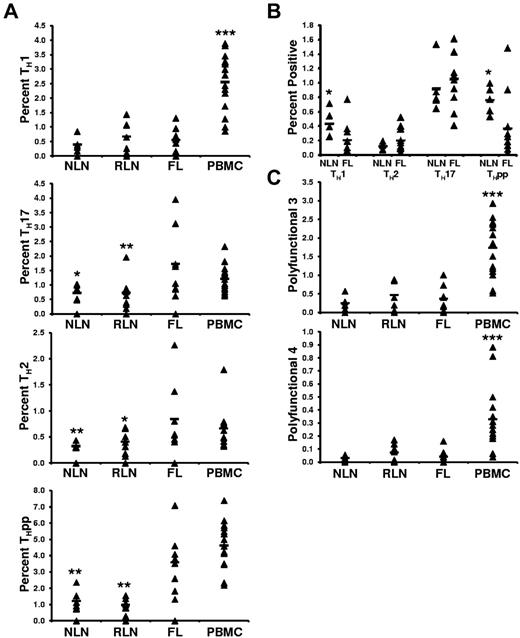
FL TH cells are skewed toward TH2, TH17, and THpp cells but not TH1 cells after SEB stimulation compared with NLN TH cells
Although we show the FL TH microenvironment to differ from that of NLNs, a fundamentally important question is whether the intrinsic effector function of these cells differs from NLN TH cells. To address this, unseparated SCSs were stimulated with SEB, and the proportions of TH cells producing the prototypical TH1 (IFN-γ+IL-2+), TH2 (IL-4−IFN-γ+), TH17 (IL-17+IFN-γ−IL-4−), and THpp (IL-2+IFN-γ−IL-4−) cytokines were determined (Figure 7A).30,31 On SEB stimulation, we observed no significant differences in the proportions of TH1 cells in FL compared with NLNs or RLNs (0.55% ± 0.42% compared with 0.40% ± 0.23% and 0.66% ± 0.54%, for the FL, NLNs, and RLNs, respectively; P > .05). In contrast, the proportions of FL TH cells that are TH2 or TH17 cells are greater than in NLNs or RLNs (0.85% ± 0.66%, 0.32% ± 0.05%, and 0.41% ± 0.22% TH2 cells, P < .02; whereas 1.73% ± 1.20%, 0.72% ± 0.26%, and 0.73% ± 0.59% are TH17 cells, P < .05; in FL, NLNs, and RLNs, respectively). In addition, the proportion of FL TH that are THpp cells (3.59% ± 1.81% of the FL TH) is greater than that seen in NLNs and RLNs (1.22% ± 0.62% and 0.97% ± 0.41%, respectively; P < .002).
Proportions of cytokine producing CD4+ T cells after SEB stimulation. (A) The TH subsets defined by their cytokine secretion profile after SEB stimulation are expressed as the percentage of the total CD4+ T cells; TH1 (IL-2+IFN−γ+IL-4−), TH2 (IL-2+IFN-γ−IL-4+), TH17 (IL-17+), or THpp (IL-2+IFN-γ−IL-4−). (B) The TH subsets defined by their cytokine profile as above, TH1, TH2, TH17, and THpp, after stimulation with soluble anti-CD3 and anti-CD28 mAbs. (C) Proportions of CD4+ T cells producing either 3 or 4 cytokines simultaneously after stimulation with SEB, polyfunctional T cells 3 (PFT 3; IL-2+IFN-γ+TNF-α+) or PFT 4 (IL-2+IFN-γ+TNF-α+MIP-1β+), respectively. For all plots, triangles represent values for individual samples, and the rectangles represent averages for each tissue type expressed as a percentage of total CD4+ T cells. *P < .05, **P < .01, and ***P < .001 compared with FL.
Proportions of cytokine producing CD4+ T cells after SEB stimulation. (A) The TH subsets defined by their cytokine secretion profile after SEB stimulation are expressed as the percentage of the total CD4+ T cells; TH1 (IL-2+IFN−γ+IL-4−), TH2 (IL-2+IFN-γ−IL-4+), TH17 (IL-17+), or THpp (IL-2+IFN-γ−IL-4−). (B) The TH subsets defined by their cytokine profile as above, TH1, TH2, TH17, and THpp, after stimulation with soluble anti-CD3 and anti-CD28 mAbs. (C) Proportions of CD4+ T cells producing either 3 or 4 cytokines simultaneously after stimulation with SEB, polyfunctional T cells 3 (PFT 3; IL-2+IFN-γ+TNF-α+) or PFT 4 (IL-2+IFN-γ+TNF-α+MIP-1β+), respectively. For all plots, triangles represent values for individual samples, and the rectangles represent averages for each tissue type expressed as a percentage of total CD4+ T cells. *P < .05, **P < .01, and ***P < .001 compared with FL.
Given that we hypothesize that the cytokine profile of SEB-stimulated T-cell models that of T cells stimulated under conditions whereby some of the immunosuppressive networks in FL, such as Treg suppression, are bypassed, we next determined the cytokine profile of T cells stimulated with soluble anti-CD3 and anti-CD28 Abs, stimulation conditions whereby such immunosuppressive networks may not be bypassed. In this regard, we have previously shown that anti-CD3/CD28 stimulated FL T cells are suppressed by FL Treg cells.17 Under these conditions, the proportion of T cells having a TH1 and THpp profile were significantly lower in FL than in NLNs (Figure 7B; P < .05). This was not surprising, given our previous findings that FL Treg cells inhibit FL T-cell production of both IFN-γ and IL-2 to a greater extent than NLN Treg cells inhibit NLN effector T-cell production of such cytokines. Finally, in contrast to the differences seen in the proportion of FL TH17 and TH2 cells compared with that seen in NLNs after SEB stimulation, we observed no significant differences in these populations after anti-CD3/anti-CD28 stimulation (Figure 3B; P > .05).
FL TH cells have the same polyfunctional response on SEB stimulation as NLN TH cells
The proportions of TH cells that simultaneously produce either 3 (IFN-γ+IL-2+TNF-α+) or 4 (IFN-γ+IL-2+TNF-α+MIB-1β+) cytokines simultaneously, called polyfunctional T cells,32 was next determined (Figure 7C). We show that the proportion of FL TH cells that are polyfunctional on SEB stimulation was similar to that of NLNs and RLNs. Specifically, 0.37% ± 0.35%, 0.25% ± 0.17%, and 0.47% ± 0.39% of the FL, NLN, and RLN TH cells, respectively, produced 3 cytokines (P > .05). Similarly, 0.041% ± 0.054%, 0.030% ± 0.018%, and 0.073% ± 0.067% of the FL, NLN, and RLN TH cells, respectively, simultaneously produced 4 cytokines (P > .05).
Discussion
Numerous studies have shown that the T-cell compartment of the tumor microenvironment plays a pivotal role in FL biology.1-9,33 Given differences in the cytokine milieu, nature of antigenic stimulation, and immune-suppressive networks in FL, compared with that of NLNs, it is not surprising that differences exist in both the distribution and function of their TH populations. Indeed, we have previously shown that significant differences exist in both the number and function of Treg cells in FL compared with NLNs.17 A better understanding of these differences should provide insight into the immunobiology of FL as well as into the potential of augmenting tumor immunity elicited by immunotherapy with or without chemotherapy.
We first show there to be skewing of FL TH cell differentiation, with the proportion of TN and TCM cells being decreased and the proportion of TEM cells being increased compared with that of NLNs. One possible explanation for the decrease in FL TN cells is that the FL microenvironment is altered in such way that it cannot provide the necessary signals for TN cell homeostasis and survival. An alternative explanation is that chronic antigenic stimulation drives TN cells into TEM and TCM pools.34,35 In this regard, sustained T-cell activation drives TN into TEM cells, whereas transient stimulation drives TN into TCM cells.34,35 Because FL may be driven, in part, by a yet undefined auto-Ag, the possible sustained Ag stimulation in FL may favor the generation of TEM. Our results differ from those described by Anicihine et al36 who noted TH skewing toward TN and TCM in indolent lymphomas. In contrast to our series, they included tumor cells from the blood, LNs, and marrow from patients with several histologic subtypes of lymphoma which were compared with that of healthy PBMCs and tumor-free LNs from patients with breast cancer.36 Such differences in the tissues studied may account for these different findings. Finally, the findings that FL TH cell differentiation is skewed in a similar direction as that of RLNs and TNs suggests that the inflammatory and immune activation signals common to all these tissues play a role in shaping the microenvironment. However, it is probable that tumor-specific signals also play a critical role in modulating the immune microenvironment as shown by the fact that the FL TH cell effector function after SEB stimulation differs from that seen with RLNs and as shown by our prior work that showed that FL Treg-mediated T-cell anergy was significantly greater than that seen in the RLNs.17
TH cells that provide the necessary B-cell help for the generation of high-affinity Ab responses are a distinct TH lineage, TFH cells, whose differentiation program is under the control of the Bcl-6 transcription factor.22 Whereas GEP studies suggest that FL TFH cells play a critical role in FL biology,37 a detailed analysis of the discrete TFH populations within FL is lacking. TFH cells express CXCR5 that allows homing to LN follicles.22 Although CXCR5 expression has indeed been associated with TFH cells, it is not specific for these cells. For example, activated T cells express CXCR5 (intermediate staining) because the follicle provides a critical environment for the survival and expansion of such cells.38,39 In addition, a subset of Treg cells also express CXCR5, as well as ICOS and PD-1, costimulatory molecules also associated with TFH.25,40 Our data in total, however, suggest that there are clear differences in the distribution of CXCR5+ TH populations in FL and NLNs on the basis of on their coexpression of CD57 and CD25. The CD25+ populations are probably more enriched in Treg cells (because they express more FOXP3), and the CD25− populations are probably more enriched in TFH cells (because they express more BCL-6). Because there is known functional interplay between Treg and TFH cells,25,41 it is probable that it is the balance of CXCR5+ GC Treg and TFH cells that affects FL biology.
A bifunctional CD4+CD57+ population has been described in the TN that provides B-cell help as well as suppression of TH1-cell activation.29 We find an increase in CD4+CD57+ cells (particularly CD57BRIGHT cells that are more associated with TFH)24 in FL than in NLNs and show that a CD25+ subpopulation of these cells consists of both Bcl-6+ TFH cells and FoxP3+ Treg cells. Indeed, one would hypothesize that, if such populations provided both FL growth support and inhibition of antilymphoma T-cell responses, it would be associated with FL having a more aggressive clinical course. Indeed, an increased number of FL CD57+ T cells have been associated with a more aggressive clinical course.7 The variability of the distribution of FL CD57+ T cells compared with the uniform distribution seen within the follicle of RLNs (as also reported by others7,41 ) suggests that differences in the chemokine/cytokine milieu of these nodal tissues may modulate the follicular homing or differentiation or both of this population. Finally, CD57 expression has been associated with decreased telomere length; therefore, it is possible that the increase in FL CD57+ TH cells represents an increase in the proportion of senescent TH cells in FL compared with that in NLNs.42
It is ultimately the ability of TH cells to mediate their effector functions in the context of their microenvironment that dictates their biologic function(s). As previously discussed, we and others have shown that FL Treg cells mediate their effector function within its microenvironment, suppressing CD25− TH function.17,43,44 To determine the intrinsic effector functions of other FL TH subsets in the context of its microenvironment (modeled with the use of unseparated FL SCSs) and in a manner whereby immunosuppressive mechanisms often associated with cancer, such as defects in Ag presentation and Treg suppression, may be partially overcome, we stimulated TH cells with SEB. Before such ex vivo stimulation, FL TH cells have a more activated phenotype than NLNs or RLNs, as determined by CD69 expression, as has previously been reported.36 However, such cells can be activated further, because after stimulation they show a similar degree of activation as do NLN or RLN TH cells. After SEB stimulation, the FL TH cells show a functional skewing toward TH2 polarization compared with that of NLNs. Because IL-4 polarizes TH-cell differentiation toward TH2, the presence of IL-4 in the FL microenvironment, the main source of which has been suggested to be TFH cells, would in part explain this polarization.37
We also show that FL TH differentiation is skewed toward TH17 differentiation, compared with NLNs, after SEB stimulation. This is in contrast to the findings of Yang et al45 who showed the frequency of FL TH17 cells to be less than that seen in benign nodes. Recent studies have shown that CD39+ Tregs, a TH subset that we have previously shown to be increased in FL compared with NLNs, inhibits TH17 cells.46 It is probable that their use of phorbol 12-myristate 13-acetate and ionomycin to stimulate the unseparated CD4+ T cells may not have been sufficient to overcome potential Treg suppression, resulting in decreased numbers of TH17 cytokine-secreting cells being detected, thereby reflecting what may be seen in the FL microenvironment when Treg suppression is not abrogated.45 In the current study, however, such Treg attenuation was probably overcome by stimulation with SEB. This has potential clinical significance because TH17 cells have been shown to enhance tumor immunity, and, as such, an immunotherapeutic approach that includes abrogation of Tregs should result in the elicitation of such TH17 cells.47
Finally, FL TH differentiation is skewed toward THpp differentiation compared with that of NLNs. This population was first identified in the mouse as a proliferating but uncommitted population of primed precursor cells that can subsequently differentiate into either TH1 or TH2 cells, dependent on the cytokine microenvironment.31 Whereas such cells make few cytokines, they produce several chemokines and as such probably play a role in modeling the FL microenvironment and enhancing but not polarizing an immune response. To our knowledge this population has not previously been described in a human tumor, and its function needs to be further defined.
Despite the skewing described earlier, the proportion of TH1 cells was similar in FL and NLNs after SEB stimulation, and indeed it is this population that is critical in mediating tumor-specific cellular immunity. In addition, recent studies support the notion that polyfunctional T cells, those that simultaneously produce ≥ 2 different cytokines, particularly IL-2, TNF-α, and IFN-γ, elicit better control of certain pathogens than those mediated by monofunctional single-positive IFN-γ–producing T cells.32 Such polyfunctional T cells appear optimized for effector function both by their ability to elicit multiple cytokine-mediated effector functions simultaneously, as well as their ability to produce ≤ 10-fold more cytokine on a cell-to-cell basis as monofunctional T cells.32 In this study, FL had the same proportion of polyfunctional T cells as NLNs after SEB stimulation. Taken together with the TH1 data above, these data suggest that FL TH cells should have the same intrinsic ability to elicit antitumor effector responses as NLN TH cells when tumor-suppressive mechanisms are attenuated. Therefore, the elicitation of a tumor-specific immune response by tumor vaccines, rituximab, and chemotherapy should be successful if done in concert with strategies to abrogate tumor-mediated immune suppression.
Limitations to this study exist. This was not a homogenous population of patient samples, with samples being derived from both previously untreated and treated patients. In addition, several FL grade 3 cases were included. This was due, in part, to the difficulty in obtaining the excess biopsy material needed to perform these detailed studies. Despite these differences, the FL cases essentially clustered together phenotypically with the use of unsupervised cluster analysis (see supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In addition, the FL samples do not tend to cluster by diagnosis or prior treatment when examining cytokine production (supplemental Figure 2), suggesting that our data are not biased on the basis of either prior treatment or histology. However, because the overall sample number is relatively small, we cannot completely rule out an effect of either diagnosis or prior treatment on cytokine production. Despite these limitations, our data suggest that, although the FL TH microenvironment is clearly distinct from that of NLNs, FL TH cells have the intrinsic ability to be activated in such a fashion that would provide antitumor immunity.
Presented in part at the 52nd annual meeting of the American Society of Hematology, Orlando, FL, December 4-7, 2010.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Andrea Berry, MS, for her assistance in running some of the statistical analysis.
This work was supported in part by a University of Rochester Human Immunology Center Pilot Project (grant 5-29712), US Public Health Service (grant RO1-CA122645), and a SPORE in Lymphoma (grant P50-CA130805).
National Institutes of Health
Authorship
Contribution: S.P.H. and W.R.B. designed and performed research, analyzed data, and wrote the paper; A.F.R., O.H., and I.S. analyzed data and wrote the paper; S.S-S., M.R.C., M.T.B., and J.-C.E.W. performed research, analyzed data, and wrote the paper; and S.A.Q. and S.H.B. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven H. Bernstein, James P. Wilmot Cancer Center, University of Rochester School of Medicine and Dentistry, 601 Elmwood Ave, Box 704, Rochester, NY 14642; e-mail: steven_bernstein@urmc.rochester.edu.