Abstract
To evaluate the prognostic value of genetic mutations for acute myeloid leukemia (AML) patients, we examined the gene status for both fusion products such as AML1 (CBFα)–ETO, CBFβ-MYH11, PML-RARα, and MLL rearrangement as a result of chromosomal translocations and mutations in genes including FLT3, C-KIT, N-RAS, NPM1, CEBPA, WT1, ASXL1, DNMT3A, MLL, IDH1, IDH2, and TET2 in 1185 AML patients. Clinical analysis was mainly carried out among 605 cases without recognizable karyotype abnormalities except for 11q23. Of these 605 patients, 452 (74.7%) were found to have at least 1 mutation, and the relationship of gene mutations with clinical outcome was investigated. We revealed a correlation pattern among NPM1, DNMT3A, FLT3, IDH1, IDH2, CEBPA, and TET2 mutations. Multivariate analysis identified DNMT3A and MLL mutations as independent factors predicting inferior overall survival (OS) and event-free survival (EFS), whereas biallelic CEBPA mutations or NPM1 mutations without DNMT3A mutations conferred a better OS and EFS in both the whole group and among younger patients < 60 years of age. The use of molecular markers allowed us to subdivide the series of 605 patients into distinct prognostic groups with potential clinical relevance.
Introduction
Acute myeloid leukemia (AML) is a group of heterogeneous diseases with considerable diversity in terms of clinical behavior and prognosis.1-5 Clinical and genetic prognostic markers are now crucial in the evaluation of AML patients and in guiding rational management. Among them, cytogenetic abnormalities are considered to be the most important prognostic factors of AML.5-11 For example, acute promyelocytic leukemia (APL, AML-M3) with t(15;17) translocation is associated with a favorable prognosis,12-14 and core-binding factor (CBF) leukemias with t(8;21) translocation in AML-M2 variant (M2v) or inv(16) rearrangement in AML-M4 with eosinophilia (M4eo) have also been reported to have relatively good outcome.5,7,15 Nevertheless, ∼ 1/2 of AML patients lack typical prognostic karyotypic changes. To improve clinical outcome, it is important to identify specific and accurate predictors in this group of patients using molecular approaches.
It has been proposed that, according to their roles in pathogenesis, genetic abnormalities in leukemia can be roughly grouped into 2 classes: (1) mutations involving signal transduction pathways and giving rise to proliferative advantages to leukemia clones (class I) and (2) those affecting transcription factors or cofactors and causing impaired differentiation (class II).16 Numerous gene mutations have been discovered in AML patients without cytogenetic markers, and these abnormalities have been considered to belong to either class I, as exemplified by internal tandem duplications (ITDs) or mutations of the tyrosine kinase domain (TKD) of both C-KIT17,18 and FMS-like tyrosine kinase 3 (FLT3) genes11,19,20 and point mutations of the neuroblastoma RAS viral oncogene homolog gene (NRAS)17,21-24 or to class II, as exemplified by mutations of nucleophosmin gene (NPM1),2,4,22,25 the CCAAT/enhancer binding protein α gene (CEBPA), the Wilm tumor (WT1) gene, and the additional sex combs like 1 (ASXL1) gene.26-30 Recently, a new category of gene mutations associated with epigenetic regulation have drawn much attention, including the mutations of isocitrate dehydrogenase1 (IDH1), isocitrate dehydrogenase2 (IDH2), and ten-eleven-2 (TET2), which result in a hypermethylation phenotype with impairment of hematopoietic differentiation.31-33 The mixed-lineage leukemia (MLL) gene, which can be affected either through chromosomal translocation or via an intragenic partial tandem duplications (PTDs) to form a fusion gene, actually encodes a histone methyltransferase.22,34-36 The discovery of these genetic events has raised the possibility of a new class of leukemogenic genes.
The above-mentioned molecular aberrations exert profound effects on individual response to the therapy and treatment outcome of the disease.14,16,37 It has been described that CEBPA mutations27,28 and NPM1 mutations without FLT3-ITD16,20,23,25 are associated with a favorable prognosis, whereas gene mutations associated with a poor prognosis include C-KIT involvement among CBF AMLs,17,18,23 FLT3-ITD without NPM1 mutations,16,20,23,25 and MLL-PTD mutations among AML with normal cytogenetics.22,34,35 However, the role of leukemic IDH1 and IDH2 mutations and WT1, TET2, and ASXL1 mutations in predicting the prognosis of AML are not clearly established.31-33,38 In addition, these markers may also provide potential molecular targets for tailored therapies, as recently reported by several groups demonstrating potential clinical values of sorafenib in the patients with FLT3 ITD and TKD, and azacitidine and decitabine in patients with MLL abnormalities.39
More recently, genomic sequencing research in AML and other malignancies has greatly facilitated the identification of new oncogenic mutations. We40 and others37 have discovered mutations in the DNA methyltransferase 3A (DNMT3A) gene in > 20% of acute monocytic leukemia patients using exome sequencing and subsequent Sanger sequencing. The enzyme encoded by the DNMT3A gene is responsible for de novo DNA cytosine methylation.37,41-43 Both studies suggested that DNMT3A mutations are associated with hyperleukocytosis at disease presentation, elderly age, and poor prognosis. With the accumulation of more new data, decision making for risk-stratified therapy will be possible and should be integrated into the individualized treatment of AML.
We performed this study to systemically investigate the frequencies and the prognostic relevance of previously known genetic events and newly established molecular markers in a large series of adult AML patients. In particular, our intent was to stratify the “ambiguous” AML patients who lack cytogenetic prognostic markers into appropriate prognostic groups using molecular markers.
Methods
Patients
BM and peripheral blood samples were collected from 1185 patients with de novo AML from 1998-2010 from the centers of Shanghai Institute of Hematology (SIH) and Zhejiang Institute of Hematology (ZIH). French-American-British (FAB) criteria were used to define the AML subtypes (M0 through M7, with a few cases not classifiable according to the FAB nomenclature). Patients with leukemia either transformed from myelodysplastic syndrome or secondary to other malignancies were excluded from this study.
All the samples were assessed for overview of pattern and distribution of gene mutations, and they were further divided into 3 groups. Group I, containing 605 patients without prognostic cytogenetic markers except for 11q23, represented the focus of this study for clinical relevance of gene abnormalities and prognostic analysis. Because 11q23 rearrangements are mostly associated with MLL fusion genes and have been considered to bear similar clinical impact as MLL-PTD mutations,34-36,44 which are not recognizable at karyotypic level, patients with these chromosomal changes were included into the series of 605 patients for prognostic analysis. Groups II and III consisted of, respectively, the 2 most common leukemia subtypes with translocation, namely CBF leukemias with AML1 (CBFα)–ETO (158 cases) or CBFβ-MYH11 fusion (18 cases) and APL with PML-RARα fusion (387 cases) or the rare variant NPM1-RARα (1 case).6,14,15 Although the prognostic value of these 2 groups has been established, these patients were included to overview the frequencies, distributions, and correlations of molecular mutations in the whole AML setting. In addition, there was a small group of 16 patients with relatively rare prognostic cytogenetic markers,45 including t(3;3), t(9;22), −7, del(5q), del(7q), and complex translocation (for details see supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
This study was approved by the ethics committees of all participating centers. All patients were given informed consent for both treatment and cryopreservation of BM and peripheral blood according to the Declaration of Helsinki.
Treatment protocols
For APL (AML-M3) patients with t (15;17), all-trans retinoic acid and arsenic trioxide–based treatment was given for the induction and consolidation therapy.14
Other AML patients received standard first-line treatment in a DA-like regimen, which consisted of daunorubicin 45 mg/m2 on days 1-3 and cytarabine 100-150 mg/m2 on days 1-7. In the consolidation therapy, young patients were treated with high-dose cytarabine-based chemotherapy. Because of small number of the patients (n = 47) who received allogeneic stem cell transplantation, these patients were not separated out for further analysis. The chemotherapy consolidation for elderly patients was decided by the physicians in an individualized manner, as described previously.46
Cytogenetic and molecular genetic analysis
Cytogenetic and molecular studies were performed centrally at SIH and ZIH. The BM samples of de novo AML patients were studied mostly by R- and/or G-banding analysis, and were confirmed in most cases with relevant molecular markers. Chromosomal abnormalities were described according to the International System for Human Cytogenetic Nomenclature.9
Genomic DNA and total RNA were extracted as described previously.40 Initially, we screened mutations in several genes, including MDR1, BCL2, P53, XPA, ATM, SULT1C2, KIAA1244, COL7A1, N-RAS, NPM1, and IDH1 in some patients. Because sequence variations in the first 8 genes proved to be single nucleotide polymorphisms in 384 control samples from unrelated healthy individuals (data not shown), and because they were not included in current guidelines or practices of prognostic predicting,8,45 no further analysis were performed on these genes. The remaining 3 genes revealed a certain percentage of mutations, and therefore our efforts were focused on these genes and on other previously known mutations such as FLT3-ITD and FLT3-TKD and C-KIT, CEPBA, WT1, ASXL1, DNMT3A, MLL, IDH2, and TET2 gene mutations. Because the mutations of FLT3-TKD, N-RAS, NPM1, IDH1, and IDH2 were clearly concentrated,24,25,33 we used a chip-based matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis system (iPLEX; Sequenom) to perform high-throughput genotyping assays for analysis of the mutational status of these genes. For mutations of FLT3-ITD (in ITD region) and those in the C-KIT, CEPBA, WT1, ASXL1, DNMT3A, and TET2 genes, samples were analyzed by whole-gene sequencing. In addition, a multiplex RT-PCR strategy was used to detect 6 MLL-related common fusion genes, including MLL-AF9, MLL-AF10, MLL-AF6, MLL-ELL, MLL-ENL, and MLL-AF17. Briefly, all samples were screened with 2 parallel multiplex RT-PCR reactions. If there were positive PCR fragments in the samples, split-out PCR was performed to determine the fusion gene type.47 The mutational status of MLL-PTD and fusion genes such as AML1 (CBFα)–ETO, CBFβ-MYH11, and PML-RARα were determined by RT-PCR.22
Quantitative real-time RT-PCR
After Turbo DNase (Ambion) treatment, 1 μg of total RNA was used for cDNA synthesis using the M-MLV First Strand Kit (Invitrogen). Real-time PCR was performed in ABI PRISM 7900HT using SYBR Premix Ex Taq (TaKaRa). The -fold change was calculated based on the 2−ΔCt × 1000 method after normalization to the transcript level of the housekeeping gene GAPDH. Primer sequences used in the real-time RT-PCR are listed in supplemental Table 2.
Microarray expression profiling and methylation analysis
Affymetrix Human Genome U133 Plus 2.0 Array GeneChip microarrays were used to assess the total RNA samples (Affymetrix). DNA samples were extracted for the HG18 Methylation 2.1M Deluxe Promoter Array (NimbleGen) to identify the methylated DNA regions. The procedure and statistical analysis were performed as described previously.40 All microarray data are available in the NCBI Gene Expression Omnibus (GEO) under accession number GSE27244.
Statistical analyses
For clinical analysis, complete remission (CR) was defined according to the criteria of the International Working Group.48 The Fisher exact P test was used to compare differences in the CR rates. A 1-way ANOVA was used to compare the age, WBC count, and BM blasts at diagnosis in different groups. The relationship between different gene mutations was analyzed by Kendall's τ-b correlation coefficients. Overall survival (OS) was measured as the time from the date of disease diagnosis to death (failure) or alive at last follow-up (censored).48 Event-free survival (EFS) was defined as the time from disease diagnosis to treatment failure such as relapse, refractory disease, death, or alive in CR at last follow-up (censored). Kaplan-Meier analysis was used to calculate the distribution of OS and EFS.48 A log-rank comparison was performed to compare the differences in survival times. Binary logistic regression and the Cox model was used for the multivariate analysis of associations between mutational status and the achievement of CR and OS and of EFS, respectively.49 A limited backward selection procedure was used to exclude redundant variates.49 All of the above statistical procedures were performed with the SPSS Version 16.0 statistical software package.
Results
Patient characteristics
The characteristics of the 1185 de novo AML patients are summarized in Table 1. Particular attention for clinical analysis was given to 605 patients who lacked cytogenetic prognostic markers other than 11q23 abnormalities (group I, 51.1%). The relatively high number of patients with group III (APL, AML-M3) could have resulted from a preference for SIH by patients because of the successful all-trans retinoic acid and arsenic trioxide–based therapy. In addition, our data showed a relatively low incidence of M4eo after a careful examination of CBFβ-MYH11 was performed in all morphologically M4 patients, which might reflect the difference in genetic backgrounds between the Chinese and white populations, in agreement with a previous large Chinese AML series.50
Frequencies and distribution of gene mutations
Among the 605 group I patients, FLT3 mutations were found in 61 (10.8%), C-KIT in 30 (5.4%), N-RAS in 34 (5.9%), NPM1 in 122 (20.9%), CEBPA in 123 (22.0%), WT1 in 20 (3.7%), ASXL1 in 27 (5.2%), DNMT3A in 73 (12.3%), MLL in 83 (14.0%), IDH1 in 52 (9.3%), IDH2 in 53 (9.8%), and TET2 in 65 (12.7%); 452 (74.7%) patients were found to have at least one mutation. In group II patients with CBF leukemias, the most frequent mutations in addition to those caused by chromosomal translocations were C-KIT (25.6%) and N-RAS (9.7%), which differed from the situation in group III (APL, AML-M3), in whom mutations other than PML-RARα were mainly FLT3 (13.4%) and N-RAS (5.4%) mutations (supplemental Figure 1).
As we recently reported,40 a mutated DNMT3A gene was mostly associated with a myelomonocytic or monocytic morphology in the FAB classification (P < .001), with a frequency of 10.0% (22 of 219) and 18.2% (38 of 209) in M4 and M5, respectively. We identified 13 potential new types of DNMT3A mutations, which are shown in Figure 1A. All of these sequence variations were checked in a series of 384 control samples from unrelated healthy individuals and none was observed. We found that 3 M1, 1 M2 with AML1 (CBFα)–ETO fusion, 1 M3 with PML-RARα fusion, 2 M6, and 2 M2 without CBF fusion patients also carried previously reported DNMT3A mutations.37,40 The 2 patients with M6 presented monocytic features in nonerythroid lineages, whereas each of the M2 CBF and M3 patients experienced CNS involvement in spite of a standard intrathecal prophylaxis.
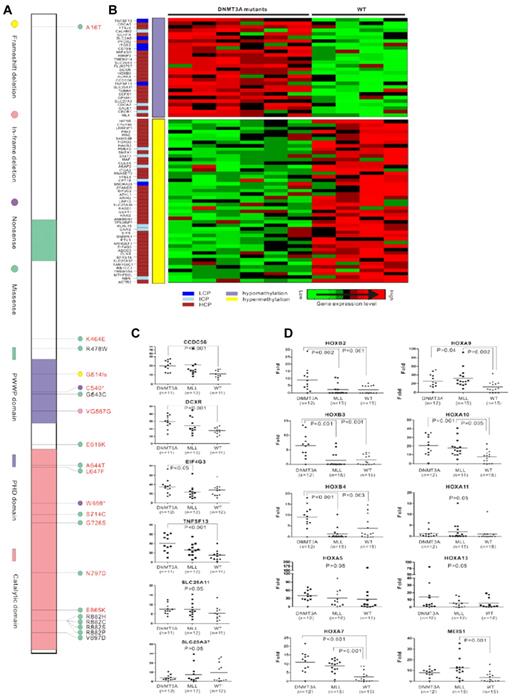
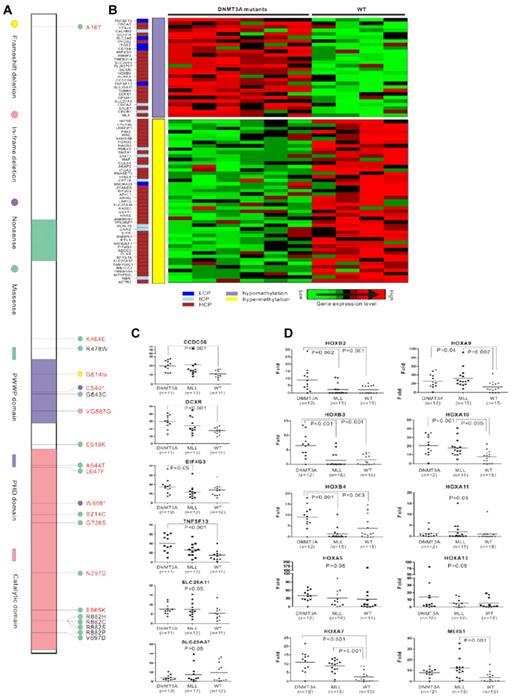
DNMT3A mutations in AML. (A) Three conserved domains in DNMT3A are shown: the PWWP domain, which targets the enzyme to nucleic acids; the cysteine-rich PHD zinc-finger domain, which interacts with unmodified histone H3; and the highly conserved catalytic domain in the C-terminal region. The mutations in AML previously reported by us are marked in black, and the newly detected mutations in the present study are in red. The most common missense mutations are predicted to affect amino acid R882. A total of 37 AML patients had the R882H mutation, 24 with R882C, 1 with R882S, and 1 with R882P in our 1178 samples. (B) Correlation analysis of gene expression and DNA methylation. The CpG content of the promoter sequences of the genes is color coded in a separate column (left lane), including low CpG content (LCP), intermediate CpG content (ICP), and high CpG content (HCP). Hypomethylation or hypermethylation in the middle lane indicates the CpG methylated level of genes in DNMT3A-mutated samples compared with samples without DNMT3A mutations. Cluster of differently expressed genes are shown on the right. Raw microarray data of gene expression and DNA methylation were published previously.40 (C) Quantitative RT-PCR analysis of genes associated with hematopoiesis and epigenetics regulation that were up- or down-regulated and accompanied by DNA methylation changes in microarray analysis in patients with DNMT3A mutations (DNMT3A), MLL abnormalities (MLL), or without these 2 types of aberrations (WT). (D) Quantitative RT-PCR analysis of genes in distinct HOX families in patients with DNMT3A mutations (DNMT3A), MLL abnormalities (MLL), or without these 2 types of aberrations (WT).
DNMT3A mutations in AML. (A) Three conserved domains in DNMT3A are shown: the PWWP domain, which targets the enzyme to nucleic acids; the cysteine-rich PHD zinc-finger domain, which interacts with unmodified histone H3; and the highly conserved catalytic domain in the C-terminal region. The mutations in AML previously reported by us are marked in black, and the newly detected mutations in the present study are in red. The most common missense mutations are predicted to affect amino acid R882. A total of 37 AML patients had the R882H mutation, 24 with R882C, 1 with R882S, and 1 with R882P in our 1178 samples. (B) Correlation analysis of gene expression and DNA methylation. The CpG content of the promoter sequences of the genes is color coded in a separate column (left lane), including low CpG content (LCP), intermediate CpG content (ICP), and high CpG content (HCP). Hypomethylation or hypermethylation in the middle lane indicates the CpG methylated level of genes in DNMT3A-mutated samples compared with samples without DNMT3A mutations. Cluster of differently expressed genes are shown on the right. Raw microarray data of gene expression and DNA methylation were published previously.40 (C) Quantitative RT-PCR analysis of genes associated with hematopoiesis and epigenetics regulation that were up- or down-regulated and accompanied by DNA methylation changes in microarray analysis in patients with DNMT3A mutations (DNMT3A), MLL abnormalities (MLL), or without these 2 types of aberrations (WT). (D) Quantitative RT-PCR analysis of genes in distinct HOX families in patients with DNMT3A mutations (DNMT3A), MLL abnormalities (MLL), or without these 2 types of aberrations (WT).
With regard to the class I gene mutations, FLT3-ITD and/or FLT3-TKD mutations exhibited an extremely lower incidence in CBF leukemias (2 of 176, 1.1%) than in other subtypes of AML (P < .001), whereas these were frequently present in addition to the PML-RARα fusion in the M3 subtype. Conversely, C-KIT mutations were most commonly seen in CBF leukemias (45 of 176, 25.6%, P < .001). In contrast, N-RAS mutations were distributed evenly in the different AML subtypes. Interestingly, among 259 patients bearing class I mutations, only 8 (3.1%) had overlapping of these markers, whereas all others carried mutation of only one gene.
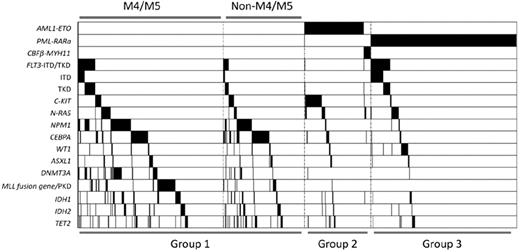
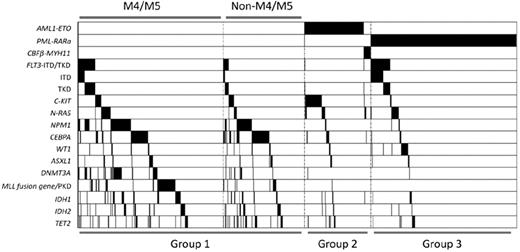
Class II mutations involving NPM1 and CEBPA, as well as mutants of genes related to epigenetic regulation such as DNMT3A, MLL, IDH1, IDH2, and TET2, tended to occur in AML patients without chromosome translocations (all P < .001). However, ASXL1 (P = .116) and WT1 (P = .296) mutations seemed to be distributed equally in cytogenetically normal or abnormal AML groups. Interestingly, in contrast to DNMT3A (P = .002) and MLL (P = .004) mutations, which were closely related to a M4 or M5 phenotype, CEBPA mutations favored a non-M4 and non-M5 phenotype (P < .001). Mutation of another epigenetic regulator, TET2, also showed this tendency (P = .056). The phenomenon of overlapping mutations in the same patient was quite frequent. Some mutations were even statistically correlated. DNMT3A (43 of 129, P < .001), FLT3 (23 of129, P < .001), IDH1 (17 of 129, P = .046), IDH2 (26 of 129, P < .001), and TET2 (16 of 129, P = .030) were significantly associated with NPM1 mutations. Associations between DNMT3A and FLT3, IDH1, and IDH2 were also observed (P = .003, .010, and .001, respectively). Among group I patients, frequent associations between the mutations of NPM1 and FLT3 (P < .001), DNMT3A (P < .001), IDH1 (P = .056), and IDH2 (P < .001), but not TET2 (P = .710), were observed. Whereas mutations of CEBPA and TET2 were highly correlated in all the patients (P < .001), only marginal correlation was identified in group I patients (P = .053). Notably, MLL abnormalities seldom coexisted with other mutations. Figure 2 shows the gene mutational status, including distributions and frequencies among distinct AML subtypes.
Mutation status of AML1-ETO, PML-RARα, CBFβ-MYH11, FLT3, C-KIT, N-RAS, NPM1, CEBPA, WT1, ASXL1, DNMT3A, MLL, IDH1, IDH2, and TET2. Black shadow indicates mutation cases.
Mutation status of AML1-ETO, PML-RARα, CBFβ-MYH11, FLT3, C-KIT, N-RAS, NPM1, CEBPA, WT1, ASXL1, DNMT3A, MLL, IDH1, IDH2, and TET2. Black shadow indicates mutation cases.
Correlation between gene-expression levels and DNA methylation status associated with DNMT3A mutations
We previously reported that aberrant DNA methyltransferase activity due to DNMT3A mutations could change DNA methylation and alter gene expression.40 Interestingly, through in-depth exploration of the possible consequences of the DNMT3A mutations using microarray data from gene-expression profiling and whole-genomic DNA methylation, we identified a correlation between gene-expression levels and the DNA methylation status in some genes; 28 up-regulated genes with DNA hypomethylation and 47 down-regulated genes accompanied with DNA hypermethylation were revealed (Figure 1B). Among these, some genes were associated with hematopoiesis and epigenetics regulation, including CCDC56, DCXR, TNFSF13, and SLC25A11 (up-regulated) and SCL25A37 and EIF4G3 (down-regulated). A correlation was revealed between the gene-expression levels of CCDC56, DCXR, and TNFSF13 and the mutation of DNMT3A, whereas no up-regulation of these 3 genes was observed in patients with MLL rearrangement (Figure 1C).
It has been reported that increased expression of multiple homeobox (HOX) genes such as HOXA7, HOXA9, HOXA10, and MEIS1 due to MLL abnormalities play an important role in leukemogenesis.40 Recently, we demonstrated an up-regulation of HOXB family genes and the IDH1 gene among patients with DNMT3A mutations.40 We also observed from this study that although MLL and DNMT3A mutations seldom overlapped, they shared a common feature of poor prognosis in distinct patient populations. Therefore, we tried to address possible association of expression levels of different HOX genes with MLL or DNMT3A abnormalities. Interestingly, members of the HOXB family were found to be overexpressed only in a group with DNMT3A mutations, whereas up-regulation of the MEIS1 gene could be observed only in patients with MLL abnormalities. Overexpression of HOXA7, HOXA9, and HOXA10 genes was observed in both the DNMT3A and MLL mutation groups, contrarily to the situation of HOXA5 and HOXA13, for which expression was not affected by DNMT3A and MLL abnormalities (Figure 1D). Although we previously found hypomethylation in genomic sequences of some HOXB family members, which might contribute to the overexpression status of these genes,40 no obvious changes were found in HOXA family genes in the present study, so the exact molecular mechanisms for their up-regulation need further investigation.
Molecular markers and clinical aspects
Regarding possible association with clinical features, we found that mutations of NPM1, CEBPA, and nearly all of the genes regulating epigenetics (DNMT3A, IDH1, IDH2, and TET2) except for MLL rearrangements were associated with elderly age at diagnosis; FLT3, NPM1, CEBPA, DNMT3A, MLL, and IDH2 mutations were associated with high WBC count at presentation; and NPM1, FLT3, DNMT3A, and MLL mutations were associated with a higher percentage of blasts in the BM (Table 2).
Response to induction therapy
In univariate analysis of the 605 patients in group I, DNMT3A mutations, MLL abnormalities, and N-RAS mutations were associated with a statistically significant lower CR rate (45.8%, 48.2%, and 41.2%, respectively; P = .014, .022, and .030, respectively) in contrast to CEBPA mutations (67.5%), which conferred a higher CR rate (P = .030). However, only biallelic (75.3%), but not monoallelic (56.0%), CEBPA mutations were associated with favorable response to the treatment (P = .003 and 1.000, respectively). Because NPM1 mutations were frequently associated with abnormalities of DNMT3A, FLT3, IDH1, and IDH2, we performed further analysis using the combination of NPM1 mutations with each of the above-mentioned mutations to investigate theirpotential prognostic impact. Obviously, DNMT3A mutations could separate the patients with NPM1 mutations into 2 distinct prognostic groups: a subpopulation of NPM1 mutations without DNMT3A mutations (NPM1m+/DNMT3Am−) was related to a significantly higher CR rate (72.3%, P = .017); however, mutations of IDH1 (P = .265) and IDH2 (P = .218), and FLT3-ITD (P = .164) or FLT3-TKD (P = .318) did not add more prognostic value among patients with the NPM1 mutation (supplemental Table 3). In M4 and M5 patients, similar results were achieved (P = .056 for DNMT3A mutations and P = .742, .736, .661, and .737 for IDH1, IDH2, FLT3-ITD, and FLT3-TKD mutations, respectively).When 89 cases without gene mutations were grouped, they seemed to have an intermediate prognosis in terms of CR (64.0%).
A complete list of covariates that entered the multivariate model is provided in Table 3. Multivariate analysis of group I patients indicated that NPM1m+/DNMT3Am− and biallelic CEBPA mutations (CEBPAm+) were independent factors associated with favorable CR rate, and DNMT3A mutations (DNMT3Am+) were associated with a lower CR rate. Two other independent clinical factors, WBC count and age, were also unfavorable for CR rate. In the age-adjusted population (n = 481) who were younger than 60 years and treated intensively, MLL abnormalities, TET2 mutations, and BM blasts proportion predicted unfavorable CR rate independently (Table 4).
Survival analysis
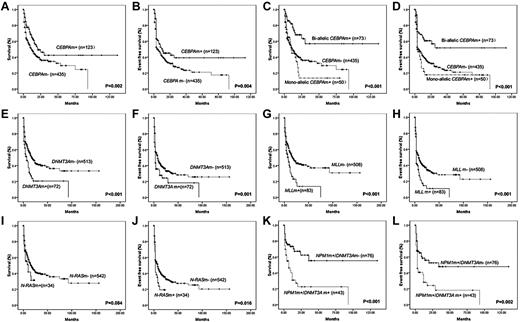
Among 605 group I patients, the median OS and EFS were 15.0 ± 1.9 and 8.0 ± 1.2 months, respectively. In univariate analysis, DNMT3A mutations and MLL abnormalities suggested a poor prognosis (both P < .001 for OS; and P = .001 and P < .001 for EFS, respectively). Although N-RAS mutation cases showed an inferior EFS (P = .016), OS was only marginally affected (P = .084). Favorable OS and EFS were achieved in the CEBPAm+ patients (P = .002 and .005 for OS and EFS, respectively). Further analysis showed that only biallelic CEBPAm+ status was associated with better treatment outcome (both P < .001 for OS and EFS, respectively). There was no statistical significance in OS and EFS in FLT3, C-KIT, WT1, ASXL1, IDH1, IDH2, and TET2 mutations (P = .169 and .371, .317 and .165, .815 and .587, .765 and .717, .257 and .731, .339 and .770, .148 and .074, respectively). NPM1 mutation did not predict OS (P = .409) and EFS (P = .274). However, in patients with NPM1 mutations, it was the mutation of DNMT3A (P < .001 and P = .002 for OS and EFS, respectively), but not of FLT3 (ITD: P = .231 and .156, for OS and EFS, respectively; TKD: P = .314 and .204, for OS and EFS, respectively), IDH1 (P = .457 and .552 for OS and EFS, respectively) or IDH2 (P = .863 and .843 for OS and EFS, respectively) that helped to discriminate the 2 prognostic groups. Figure 3 shows the Kaplan-Meier curves for OS and EFS according to genotypes with statistical significance in univariate analysis. In M4 and M5 patients, only DNMT3A mutations (P = .001 and .006 for OS and EFS, respectively) could subdivide the patients with NPM1 mutations. The existence of the mutation of IDH1 (P = .585 and .941 for OS and EFS, respectively), IDH2 (P = .275 and .118 for OS and EFS, respectively), FLT3-ITD (P = .807 and .507 for OS and EFS, respectively), and TKD (P = .988 and .621, respectively) could not help to further stratify the NPM1m+ patients.
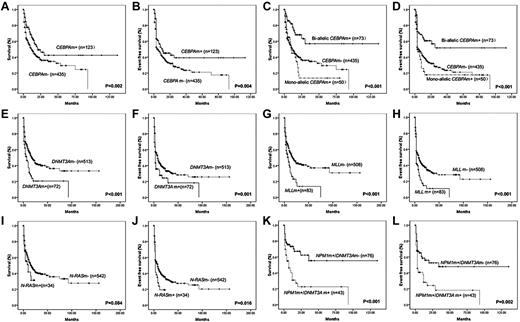
Kaplan-Meier curves for OS and EFS according to genotypes with statistical significance in univariate analysis. (A-B) The median OS and EFS of patients with or without CEBPA mutations (CEBPAm+ or CEBPAm−) were 21.0 ± 5.8 months (mo) and 12.0 ± 1.5 mo (P = .002), 11.0 ± 5.1 mo, and 5.0 ± 0.5 mo (P = .004), respectively. (C-D) The median OS and EFS of patients with biallelic or monoallelic CEBPA mutations were not reached (NR) and 10.0 ± 1.6 mo (P < .001), and NR and 3.0 ± 0.7 mo (P = .001), compared with wild-type CEBPA patients. (E-F) The median OS and EFS of patients with or without DNMT3A mutations (DNMT3Am+ or DNMT3Am−) were 7.0 ± 2.1 mo and 18.0 ± 2.3 mo (P < .001), 3.0 ± 0.3 mo and 8.0 ± 1.2 mo (P = .001), respectively. (G-H) The median OS and EFS of patients with or without MLL abnormalities (MLLm+ or MLLm−) were 8.0 ± 2.2 mo and 17.0 ± 2.4 mo (P < .001), 3.0 ± 0.2 mo and 8.0 ± 1.4 mo (P < .001), respectively. (I-J) The median OS and EFS of patients with or without N-RAS mutations (N-RASm+ or N-RASm−) were 10.0 ± 4.2 mo and 17.0 ± 2.1 mo (P = .084), 3.0 ± 0.3 mo and 8.0 ± 1.3 mo (P = .006), respectively. (K-L) The median OS and EFS of patients with NPM1 mutation but no DNMT3A mutation (NPM1m+/DNMT3Am−) were NR and 34.0 mo, whereas NPM1 mutation cases with DNMT3A mutations (NPM1m+/DNMT3Am+) had inferior OS (7.0 ± 3.4 mo, P < .001) and EFS (3.0 ± 0.6 mo, P = .002).
Kaplan-Meier curves for OS and EFS according to genotypes with statistical significance in univariate analysis. (A-B) The median OS and EFS of patients with or without CEBPA mutations (CEBPAm+ or CEBPAm−) were 21.0 ± 5.8 months (mo) and 12.0 ± 1.5 mo (P = .002), 11.0 ± 5.1 mo, and 5.0 ± 0.5 mo (P = .004), respectively. (C-D) The median OS and EFS of patients with biallelic or monoallelic CEBPA mutations were not reached (NR) and 10.0 ± 1.6 mo (P < .001), and NR and 3.0 ± 0.7 mo (P = .001), compared with wild-type CEBPA patients. (E-F) The median OS and EFS of patients with or without DNMT3A mutations (DNMT3Am+ or DNMT3Am−) were 7.0 ± 2.1 mo and 18.0 ± 2.3 mo (P < .001), 3.0 ± 0.3 mo and 8.0 ± 1.2 mo (P = .001), respectively. (G-H) The median OS and EFS of patients with or without MLL abnormalities (MLLm+ or MLLm−) were 8.0 ± 2.2 mo and 17.0 ± 2.4 mo (P < .001), 3.0 ± 0.2 mo and 8.0 ± 1.4 mo (P < .001), respectively. (I-J) The median OS and EFS of patients with or without N-RAS mutations (N-RASm+ or N-RASm−) were 10.0 ± 4.2 mo and 17.0 ± 2.1 mo (P = .084), 3.0 ± 0.3 mo and 8.0 ± 1.3 mo (P = .006), respectively. (K-L) The median OS and EFS of patients with NPM1 mutation but no DNMT3A mutation (NPM1m+/DNMT3Am−) were NR and 34.0 mo, whereas NPM1 mutation cases with DNMT3A mutations (NPM1m+/DNMT3Am+) had inferior OS (7.0 ± 3.4 mo, P < .001) and EFS (3.0 ± 0.6 mo, P = .002).
In multivariate analysis, DNMT3Am+ and MLL rearrangements (MLLm+) were independent factors predicting poor prognosis; biallelic CEBPAm+ and NPM1m+/DNMT3Am− conferred a better OS and EFS (Table 3). Age and WBC count were also independent factors related to prognosis. Among younger patients who received standard induction and consolidation, the results were similar to the entire group, and the mutational status of the above-mentioned 4 genes still bore prognostic significance, whereas WBC count no longer predicted OS and EFS.
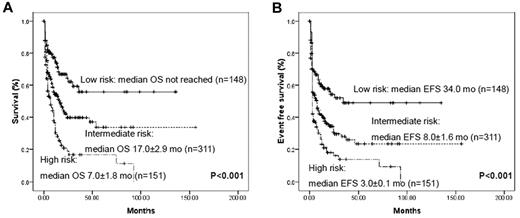
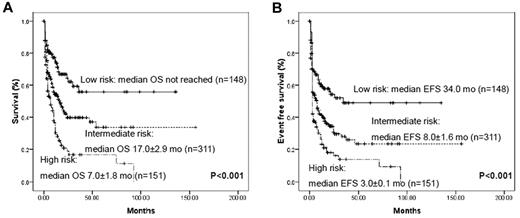
Using molecular markers that had proved to be significantly related to prognosis in multivariate analysis, we could stratify AML patients without cytogenetic markers into 3 prognostic groups: (1) a favorable-risk group with biallelic CEBPAm+ or NPM1m+/DNMT3Am− status; (2) a poor-risk group with DNMT3Am+ or MLLm+ status; and (3) an intermediate group with all of the other remaining cases (Figure 4). The prognosis for the 89 patients without detectable gene mutations also corresponded to an intermediate status in terms of both OS and EFS (supplemental Figure 3). Therefore, according to the gene mutational status, the 605 group I patients could be clearly classified into distinct prognostic subgroups.
Kaplan-Meier curves for OS and EFS according to genotypes with statistical significance in multivariate analysis. All AML patients without cytogenetic prognostic markers could be divided into 3 prognostic groups using 4 marker combinations: low- risk, biallelic CEBPAm+ and/or NPM1m+/DNMT3Am−; high-risk, DNMT3Am+ and/or MLLm+; and intermediate, all remaining cases. Very few patients were repeatedly calculated in each group because of the concurrence of different mutations.
Kaplan-Meier curves for OS and EFS according to genotypes with statistical significance in multivariate analysis. All AML patients without cytogenetic prognostic markers could be divided into 3 prognostic groups using 4 marker combinations: low- risk, biallelic CEBPAm+ and/or NPM1m+/DNMT3Am−; high-risk, DNMT3Am+ and/or MLLm+; and intermediate, all remaining cases. Very few patients were repeatedly calculated in each group because of the concurrence of different mutations.
Discussion
It has long been appreciated that cytogenetic factors are independent predictors for the prognosis of AML patients.7,8,10,15,45 However, for > 50% of AML patients, no cytogenetic markers can be found. Genetic mutations that escape cytogenetic detection have increasingly been discovered, and these mutations may serve as potential markers to extend the prognostic parameters in AML. Enormous efforts have been made to clarify the correlation between molecular changes and the clinical outcome of AML patients, allowing further dissection of AML into molecular subtypes with distinctive prognosis and therapy responses.16,37,45 Nevertheless, the clinical value of some genetic mutations remain controversial, and the frequency and prognostic impact of some newly discovered mutations have not yet been well documented. A systematic investigation of genetic mutations in large series of patients is essential to determine their clinical relevance.
In this study, we attempted to clarify the value of a cluster of molecular markers other than cytogenetic factors in the stratification of AML patients into different prognostic groups. One important finding was that the recently reported DNMT3A mutations indeed had a higher incidence in M4 and M5 subtypes (10.0% and 18.2%, respectively, P < .001) in a much larger patient cohort in this study.40 The fact that DNMT3A mutations were also identified in 2 cases of M6 with erythromonocytic leukemia provides further evidence that DNMT3A mutations are restricted to the monocytic lineage involvement in AML. We also detected, for the first time, DNMT3A mutations in one M2 patient carrying AML1(CBFα)-ETO and in one M3 patient with PML-RARα. These 2 patients quickly developed CNS leukemia even under intrathecal prophylaxis, which is reminiscent of the characteristic extramedullary involvement in monocytic leukemia. In addition, in support of the report by Ley37 and extending our previous results,40 DNMT3A mutations were highly associated with a very poor prognosis in AML patients.
Combining the mutations discovered by Ley's study and the potential sequence variations identified in this series, an interesting situation emerged: the sequence changes occurring in the PHD domain of the DNMT3A protein were microdeletions or nonsense mutations resulting in loss-of-function of the protein (except for a homozygous G543C mutation with abnormal interaction to histone H3 that we reported previously40 ). In contrast, the sequence changes found in the catalytic domain consisted mostly of missense alterations, leading to reduction of enzymatic activity according to biochemistry assay.40 DNA methylation is a crucial epigenetic modification of the genome that is involved in many cellular processes, including the regulation of gene expression and structural remodeling of chromatin.30-32 In the present study, we further analyzed the aberrant DNA methylation status due to DNMT3A mutations in relationship to gene-expression patterns in AML patients. Directly or indirectly due to abnormal DNA methyltransferase activity, these deregulated genes might further contribute to the pathogenesis of leukemia. We found that whereas overexpression of HOXB genes could be detected only in the DNMT3A-mutated group and MEIS1 was found only in MLL abnormal patients, up-regulation of HOXA7, HOXA9, and HOXA10 genes was found in both DNMT3A mutated and MLL abnormal patients. The leukemogenesis of DNMT3A mutations and MLL aberration might share some common pathways, albeit with different characteristics. Some other important genes associated with hematopoiesis and epigenetics regulation, such as CCDC56, DCXR, and TNFSF13, were also affected by DNMT3A mutation, which deserves mechanistic study with regard to leukemogenesis. Considering the poor prognosis of patients with DNMT3A mutations, together with the biologic data,40 we propose DNMT3A mutation as a “driver” mutation that plays an essential role in the pathogenesis of leukemia involving the monocytic lineage.
We found that leukemia samples from 452 of 605 cases (74.7%) in group I contained at least one of the mutations. It has been generally accepted that 2 classes of gene mutations cooperate in AML pathogenesis. Class I mutations such as those of C-KIT, FLT3, and N-RAS are associated with activated signal transduction and provide a proliferative and survival advantage to the hematopoietic progenitors.16 However, they often show subtype-restricted distribution in AML, as evidenced by a rather specific C-KIT mutation as the second hit in the pathogenesis of CBF leukemias18 and the unique high incidence of FLT3 mutations in APL (AML-M3),14 although N-RAS mutations displayed an even distribution across all major subtypes of AML. The class II gene mutations affecting transcription regulation and causing impaired differentiation often overlapped with other molecular defects, and coexistence patterns of some mutations were recognizable. Nevertheless, MLL mutations seemed to be mutually exclusive events, seldom overlapping with other mutations. Interestingly, in our preliminary morphology analysis, CEBPA, MLL, and DNMT3A seemed to be correlated with distinct morphologic phenotypes within the AML-M4 subtype: patients with DNMT3A mutations and MLL variants tended to have a major monocytic lineage involvement in BM, in contrast to those with CEBPA mutations, who had more blasts of granulocytic lineage. This might be explained by the respective roles of CEBPA in myeloid differentiation and of MLL and DNMT3A in the control of differentiation/growth of monocytic cells. Our results suggest a necessity of cooperation between distinct genetic events in leukemogenesis, although further investigation of the underlying mechanism is warranted.
It is worth pointing out that abnormalities of epigenetic regulation seem to play an essential role in the pathogenesis of AML, as evidenced by the fact that DNMT3A is a DNA methyltransferase whereas MLL is a histone methyltransferase. In addition, IDH1, IDH2 and TET2 mutations were found to result in DNA hypermethylation,31-33,51 and a group of histone methyltransferase genes (eg, MLL2, UTX, and SETD2) have recently been found to be mutated in hematologic malignancies. Mutations of EVI1, the protein product of which is assumed to interact with DNMT3A and DNMT3B, was recently reported to be associated with poor prognosis in AML.1 Conversely, mutations of EZH2, encoding a histone methyltransferase that could interact with DNMT3A, were described in lymphomas and myelodysplastic syndromes.52,53 Taking these data into consideration, we propose that gene mutations involved in epigenetic regulation may be considered as a third class, apart from class I and II mutations, because they not only belong to a distinct regulatory network, but also might share common features of aggressive disease, poor prognosis, and older age onset (with the exception of MLL abnormalities).
In the present study, logistic-regression analyses showed that DNMT3A mutations represented independent, unfavorable prognostic factors for remission induction with conventional daunorubicin and cytarabine-based chemotherapy. In contrast, biallelic CEBPA or NPM1 mutations without DNMT3A mutations were associated with a favorable response. Age and WBC count also proved to be independent factors of adverse outcome. However, in a relatively young patient population, only MLL and TET2 abnormalities were independently associated with unfavorable induction results. Cox regression analysis revealed that DNMT3A mutations and MLL rearrangements were associated with an inferior OS and EFS, whereas biallelic CEBPA mutations or NPM1 mutations without DNMT3A changes independently predicted favorable OS and EFS. Again, these 4 genes were proved to be the only independent prognostic factors in relatively young patients in predicting OS and EFS. NPM1 mutations with wild-type FLT3-ITD was shown to be an important favorable genotype in AML patients without cytogenetic changes in previous studies.20-22 Our series failed to show the difference between NPM1m+/FLT3-ITDm+ and NPM1m+/FLT3-ITDm− groups, possibly due to the lower frequency of FLT3-ITD mutations in our series in cytogenetically normal patients (23 of 605, 3.8%), which might hamper further stratification of NPM1m+ patients and decrease the sensitivity of the statistics.
In summary, we have established a new stratification system with which to subclassify the prognosis of AML without cytogenetic prognostic factors according to the mutation patterns of 4 genes. Evaluation of molecular markers in AML, especially through detection of DNMT3A, MLL, NPM1, and CEBPA mutations, can therefore be recommended. Further studies will be focused on using different treatment strategies according to AML genotypes. For example, enhanced therapies, such as high-dose anthracyclines and DNA-methylation–regulatory agents in elderly patients with DNMT3A mutations, might be evaluated for their potential to improve clinical outcome in AML.
An Inside Blood analysis of this article can be found at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Wei-Li Zhao for assistance in clinical analysis of the AML samples and to Shu-Min Xiong for performing the hematologic morphological analysis.
This work was supported in part by the National High Tech Program for Biotechnology (863:2006AA02A405), the Chinese National Key Basic Research Project (973: 2010CB529200), the Mega-projects of Science Research for the 11th Five-Year Plan (2008ZX09312-026), the National Natural Science Foundation of China (30772744, 30830119, 30821063), the Shanghai Municipal Commission for Science and Technology (10411965600), the Shanghai Rising Star Program (11QA1404300), and the Samuel Waxman Cancer Research Foundation Co-PI Program.
Authorship
Contribution: S.-J.C, J.J., and Z.C. were the principal investigators who conceived the study; S.-J.C., J.J., Z.C., and Y.S. coordinated and oversaw the study; Y.S., Y.-M.Z., X.F., Q.-R.W., and J.-Y.S. performed most of the experiments; Z.-H.G. and X.-J.Y. were responsible for bioinformatics investigation; Q.-R.W. and J.-Y.S. participated in the validation experiments; C.-L.J and H.Y. contributed in sample treating and PCR amplification; Y.S., X.F., F.-F.C., Y.-Y.W, B.C., and H.-M.C. gathered detailed clinical information for the study and helped to perform the clinical analysis; Y.-M.Z. and J.-Y.S. participated in the PCR assay and Sequenom analysis; and Z.C., S.-J.C., and Y.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sai-Juan Chen and Zhu Chen, State Key Laboratory of Medical Genomics, Shanghai Institute of Hematology, Rui Jin Hospital Affilated to Shanghai Jiao Tong University School of Medicine, 197 Rui Jin Road II, Shanghai 200025, China; or Jie Jin, Department of Hematology, Institute of Hematology, the First Affiliated Hospital, Zhejiang School of Medicine, 79 Qing Chun Road, Hangzhou 310003, China; e-mail: sjehen@stn.sh.cn, jie.j@hzenc.com, or zchen@stn.sh.cn.
References
Author notes
Y.S., Y.-M.Z., X.F., J.-Y.S. and Q.-R.W. contributed equally to this work.