Abstract
Chromosomal instability is a defining feature of clonal myeloma plasma cells that results in the perpetual accumulation of genomic aberrations. In addition to its role in protein homeostasis, the ubiquitin-proteasome system is also involved in the regulation of DNA damage-repair proteins. In the present study, we show that proteasome inhibition induces a “BRCAness” state in myeloma cells (MM), with depletion of their nuclear pool of ubiquitin and abrogation of H2AX polyubiquitylation, an essential step for the recruitment of BRCA1 and RAD51 to the sites of DNA double-stranded breaks (DSBs) and the initiation of homologous recombination (HR)–mediated DNA repair. Inhibition of poly-ADP-ribose-polymerase 1 and 2 (PARP1/2) with ABT-888 induced transient DNA DSBs that were rapidly resolved and thus had no effect on viability of the MM cells. In contrast, cotreatment of MM cell lines and primary CD138+ cells with bortezomib and ABT-888 resulted in the sustained accumulation of unrepaired DNA DSBs with persistence of unubiquitylated γH2AX foci, lack of recruitment of BRCA1 and RAD51, and ensuing MM-cell death. The heightened cytotoxicity of ABT-888 in combination with bortezomib compared with either drug alone was also confirmed in MM xenografts in SCID mice. Our studies indicate that bortezomib impairs HR in MM and results in a contextual synthetic lethality when combined with PARP inhibitors.
Introduction
Genomic integrity is continuously challenged by both exogenous and endogenous stressors.1 To counteract DNA damage, cells have evolved repair mechanisms specific for many types of lesions.2-6 Single-strand DNA breaks (SSBs) are repaired through the nucleotide excision repair or the base excision repair machinery, which require the activation of poly-ADP-ribose polymerase (PARP). PARP1, and to a lesser extent PARP2, bind DNA SSBs and catalyze the synthesis and addition of large chains of poly-ADP-ribose (PAR) polymers on target proteins, including the histones H1 and H2B and PARP1 itself. These polymers serve to recruit variable proteins needed to activate DNA-damage repair (DDR).7-9 If persistent or left unrepaired, SSBs encountered by replication forks lead to the formation of potentially lethal double-strand DNA breaks (DSBs). These genomic DSBs encountered in the S/G2 phases are predominantly repaired by the homologous recombination (HR) pathway, in which the MRN (MRE11-RAD50-NBS1) complex senses the DSBs and initiates a dynamic protein recruitment to DNA-repair foci.10,11 MRN first recruits the ATM kinase to the vicinity of the lesions, with resulting ATM-mediated phosphorylation of the histone variant H2AX that leads to the accumulation of the MDC1 protein and its binding partners. These include the MRN complex and RNF8 and RNF168, 2 ubiquitin ligases that initiate histone H2AX Lys63 mono- and polyubiquitylation at sites of DNA damage. This histone ubiquitylation allows for a second wave of protein accumulation, including factors such as 53BP1 and the BRCA1 A complex that are critically important for DSB repair and for the maintenance of genomic integrity.12-14
Deregulation of the DDR machinery fuels the genomic instability needed to drive cancer-cell development and clonal evolution. Recognition of these deregulated DDR pathways has led to the discovery of novel therapeutics that result in synthetic lethality in transformed cells. Recent studies have demonstrated the efficacy of targeting PARP1 in tumors with impaired HR resulting from the homozygous loss of the BRCA1 or BRCA2 genes.15-17 Furthermore, genetic screens have identified a host of HR-related genes (including RAD51, ATR, and PCNA) that upon deletion or silencing render cells hypersensitive to PARP inhibitors.18 Therefore, tumor cells with any HR deficiency or “BRCAness” are likely to be particularly sensitive to PARP inhibitors because they are unable to cope effectively with the increase in lethal DSBs associated with replication fork collapse.
Multiple myeloma (MM) is a clonal malignancy of plasma cells characterized by a highly unstable genome with aneuploidy observed in nearly all patients.19-22 Whereas the exact mechanism for this karyotypic instability is largely unknown, recent observations have correlated these abnormalities with deranged DSB repair by nonhomologous end joining or elevated HR.23,24 Therefore, this impairment of the machinery involved in the maintenance of genomic stability in MM may allow the development of novel therapies that target the “residual” DNA-repair pathways upon which MM cells are now completely dependent. In addition, whereas the sensitivity of MM cells to proteasome inhibitors is thought to be related to their “high proteasome load and endoplasmic reticulum stress,”25,26 recent studies have suggested that proteasome inhibitors also impair the ability of MM cells to repair DNA interstrand crosslinks and DNA damage response.27-29 Furthermore, whereas the ubiquitin-proteasome system is best known for its role in targeting proteins for degradation, studies have significantly broadened the scope of the role of ubiquitylation to include nonproteolytic functions of ubiquitin. In particular, mono- or polyubiquitylation of several DNA-repair proteins (eg, H2AX, CtlP, BRCA1, FANCD2) is required for their regulatory functions in DDR.30-33
Based on these observations, we have postulated that inhibition of the 26S proteasome induces a BRCAness state in MM cells and sensitizes them to PARP inhibitors by blocking homology-mediated repair of DNA DSBs. In the present study, we have delineated the mechanisms of bortezomib-induced BRCAness and demonstrated a contextual synthetic lethality in MM cells treated with PARP and proteasome inhibitors.
Methods
Cell culture and treatments
The human MM cell lines NCI-H929, RPMI-8226 (ATCC), KMS11, MM1S, and OPM2 (provided by Dr Lawrence Boise, Emory University, Atlanta, GA) were maintained in RPMI 1640 medium (Sigma-Aldrich) with l-glutamine and NaHCO3 containing 10% FBS (Sigma-Aldrich), 100 U/mL of penicillin, and 100 μg/mL of streptomycin (Sigma-Aldrich). INA-6 cells were provided by Dr Renate Burger34 (University of Erlangen-Nuernberg, Erlangen, Germany) and cultured in the presence of 2.5 ng/mL of recombinant human IL-6 (Sigma-Aldrich).
ABT-888 (2-[(R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide) was obtained from Abbott Laboratories. Bortezomib was purchased from Millennium Pharmaceuticals at the concentration of 1 mg/mL and diluted in culture medium as indicated. The NF-κB inhibitor SN50 was obtained from Enzo Life Sciences.
MM tissue microarray
Myeloma tissue microarrays were generated from formalin-fixed paraffin sections of pretreatment diagnostic BM biopsies (n = 83). Detailed methods for the tissue microarray PARP1 immunostaining and scoring are provided in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Cell viability and apoptosis assays
Cell viability of MM cells after ABT-888 and/or bortezomib treatment was assessed with MTT (Chemicon), apoptotic and necrotic cell death was evaluated by FITC-conjugated annexin V (BioVision) and propidium iodide (PI) staining using flow cytometric analysis as described previously.35,36
For primary MM-cell studies, mononuclear cells that had been fraction separated by Ficoll gradients from the BM aspirates of consenting MM patients (in accordance with the Declaration of Helsinki) were plated at a cell density of 5 × 105 cells/mL in RPMI medium with 20% FBS plus the indicated concentrations of ABT-888 and bortezomib. After 24 hours in culture, cells were double stained with anti–CD138-PE (Pharmingen) and FITC-conjugated annexin V (BioVision) and cell viability was assessed by flow cytometry, as described previously.37
CFU assay
CD34+ peripheral blood stem cells were obtained from discarded peripheral blood apheresis products. A hematopoietic colony-forming cell assay was performed using the CytoSelect 96-well Hematopoietic Colony Forming Cell Assay kit according to the manufacturer's protocol (Cell Biolabs). Briefly, 1-5 × 105 cells/mL were plated in CytoSelect methylcellulose medium in 96-well culture plates in the presence of G-CSF. At 10 days, colonies were quantified using a fluorescence plate reader (485/520 nm filter set).
Western blot analysis
MM cells were exposed to the indicated doses of ABT-888 and/or bortezomib, harvested, lysed, and immunoblotted with specific Abs as described in supplemental Methods.
Immunostaining
Immunofluorescence techniques were used to analyze ubiquitin and polyubiquitin cellular localization, as well as γ-H2AX, BRCA1, and RAD-51 foci formation induced by irradiation with 4 Gy or ABT-888 and/or bortezomib treatment. Cells cultured on glass coverslips were fixed in 4% paraformaldehyde in PBS for 10 minutes and permeabilized with PBS containing 0.5% Triton X-100 for 15 minutes. After treatment with blocking solution (PBS containing 3% BSA), cells were incubated with the appropriate primary Ab: anti–γ-H2AX (Millipore) and anti-ubiquitylated proteins (clone FK2, Millipore); anti-ubiquitin (Cell Signaling Technology); and anti-RAD51 and anti-BRCA1 (Santa Cruz Biotechnology). Cells were then washed in PBS containing 0.1% Tween 20 for 15 minutes, followed by visualization with fluorescence-labeled secondary Abs: Alexa 555–conjugated goat anti–mouse, Cy3-conjugated goat anti–rabbit IgG, and Cy5-conjugated goat anti–rabbit IgG (Invitrogen). Slides were then mounted with ProLong Gold antifade mounting medium containing DAPI (Invitrogen). Image acquisition was performed with an epifluorescence microscope (BX51; Olympus) and multispectral color camera (Nuance FX; CRi) with either a 60× or 100× magnification lens and oil immersion.
qRT-PCR
The mRNA levels of FANCD2 (forward: GTTCGCCAGTTGGTGATGGAT and reverse GGGAAGCCTGTAACCGTGAT primers), RAD-51 (forward CGAGCGTTCAACACAGACCA; and reverse GTGGCACTGTCTACAATAAGCA primers), BRCA1 (forward CTGAAGACTGCTCAGGGCTATC and reverse AGGGTAGCTGTTAGAAGGCTGG primers), BRCA2 (forward TGCCTGAAAACCAGATGACTATC and reverse AGGCCAGCAAACTTCCGTTTA) and GAPDH (forward AACAGCGACACCCATCCTC and reverse CATACCAGGAAATGAGCTTGACAA primers) were detected by real-time quantitative PCR (qRT-PCR) in MM cell lines after treatment with bortezomib for the indicated times. RT-PCR amplification was carried out with 250 ng of c-DNA in a 25-μL reaction mixture containing 12.5 μL of SYBR Green (Bio-Rad) and 1 μL of RT2 qRT-PCR primers (SA Biosciences). Data quantification was carried out by the 2−ΔΔCt method.
Promoter constructs and luciferase assay
A pSGG_prom vector containing an approximately 3-kb fragment of the human promoter for BRAC1, BRAC2, FANCD2, RAD51, or GAPDH (control) cloned into the MCS and fused to a luciferase reporter gene (SwitchGear genomics) was transfected into MM cells using Lipofectamine LTX (Invitrogen). After bortezomib treatment, the activity of promoters was evaluated with the Steady-Glo Luciferase Assay System (Promega). The levels of firefly luciferase activity were normalized to GAPDH luciferase activity.
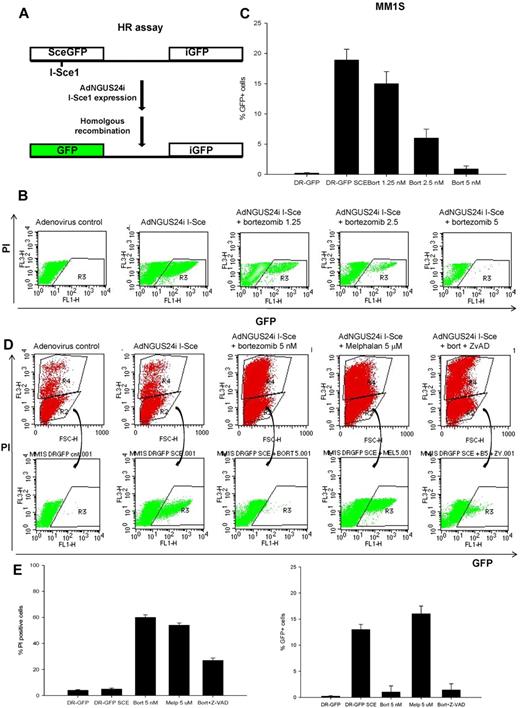
HR assay
To measure HR-mediated repair of DNA DSBs, we used the DR-GFP/SceI assay as described previously,38,39 except instead of transfecting cells with an I-SceI yeast endonuclease–expressing plasmid, we used an adenovirus AdNGUS24i–expressing I-SceI (kindly provided by Dr Silvia Bacchetti and Frank Graham, McMaster University, Hamilton, ON, and Dr Alain Nepveu, McGill University, Montreal, QC).40 The DR-GFP plasmid (kindly provided by Dr Maria Jasin, Memorial Sloan-Kettering Hospital, New York, NY) contains a modified GFP reporter in which GFP is modified to contain an I-SceI site (Sce-GFP) in tandem with a 5′ and 3′-truncated GFP gene (iGFP). In this assay, cell lines stably expressing (puromycin-selected) the DR-GFP HR reporter substrate are infected with AdNGUS24i to produce a DSB in the mutant Sce-GFP gene. Intragenic HR then repairs these DSBs, resulting in a functional GFP gene. In our studies, the myeloma MM1S cells stably expressing the DR-GFP plasmid were cultured for 12 hours in regular medium or medium supplemented with AdNGUS24i particles with or without bortezomib (1.25-5nM) or melphalan (5μM) or bortezomib (5nM) with ZVAD (20μM). The percentage of live (PI−) and GFP+ cells was measured 12 hours later by flow cytometry as an indicator for their ability to conduct homology-directed DNA repair.
In vivo animal studies
CB-17 SCID mice were purchased from Charles River Laboratories and maintained and monitored in our animal research facility. All animal studies were conducted according to protocols approved by the University of Calgary institutional animal welfare committee. CB-17 SCID mice were irradiated with 150 cGy and 24 hours later were subcutaneously (SC) inoculated in the interscapular area with 2 × 106 MM1S cells.41 After the detection of measurable tumors, animal cohorts were treated with bortezomib (0.4 mg/kg SC every Monday and Thursday) or ABT-888 (50 mg/kg by oral gavage twice a day 5 days/wk) or ABT-888 in combination with bortezomib or vehicle alone until the animal was killed. Tumor sizes were measured every 3 days in 2 dimensions using an electronic caliper, and the tumor volume was calculated using the formula: V = 0.5a × b2, where a and b are the long and short diameter of the tumor, respectively. Survival was evaluated by Kaplan-Meier from the first day of tumor appearance until death. Animals were killed when their tumors reached 2 cm in the largest diameter or when paralysis or other major distress occurred.
Statistical analysis
Statistical significances of differences were determined using the Student t test. The minimal level of significance was P < .05.
Results
High PARP1 expression is correlated with poor survival in MM
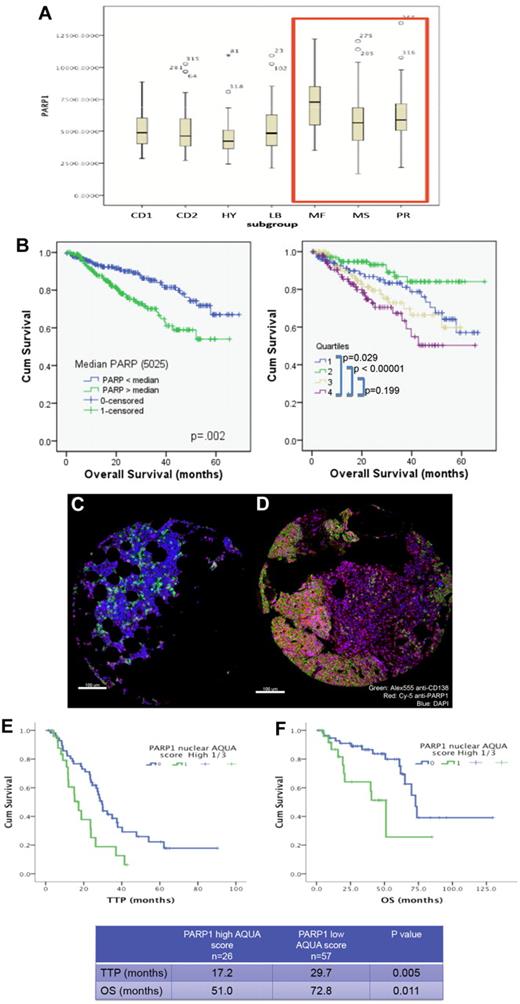
We have first examined whether PARP1 expression in primary MM cells were correlated with or predicted for worse survival. Interrogation of the dataset GSE4581 (deposited by Dr J. D. Shaughnessy Jr, University of Arkansas for Medical Sciences) revealed that high-risk MM molecular subgroups (MF, MS, and PR) did have a higher PARP1 mRNA expression compared with the remaining subgroups (CD1, CD2, HY, and LB; Figure 1A). In addition, higher expression of PARP1 (top quartile Q4 or higher than median Affymetrix probe 208644_at) was correlated with shortened survival (Figure 1B). We also examined PARP1 protein expression using immunofluorescence staining and automated quantitation with the HistoRx platform and AQUA analysis software in a cohort of MM patients (n = 83) uniformly treated with high-dose melphalan and autologous stem cell transplantation (Figure 1C-D and supplemental Figure 1). Higher PARP1 expression (top tertile) in this cohort was also correlated with shortened time to progression and overall survival (Figure 1E-F). These results suggest that PARP1 may be implicated in MM-cell survival or drug resistance, and led us to further investigate its role in this disease.
High PARP1 expression is correlated with poor survival in MM. (A) Box plot displaying PARP1 mRNA range of expression (Affymetrix probe 208644_at) according to the molecular subgroup in a cohort of MM patients from the Arkansas dataset (GSE4581). Note the higher PARP1 expression in the MR, MS, and PR subgroups. (B) Kaplan-Meier overall survival curves of MM patients treated in total therapy 1 and 3 according to PARP1 mRNA expression above or below the median value of 5025 (left panel) or according to the quartiles distribution (right panel) of PARP1 mRNA expression based on National Institutes of Health GEO dataset deposited by Dr J. D. Shaughnessy Jr under accession number GSE4581. Quartile 4 represents the subgroup with the highest PARP1 expression. The 60-month OS for patients with PARP1 below and above the median are 67% and 54%, respectively (P = .02). (C-D) Myeloma tissue microarray constructed from the BM biopsies of 83 newly diagnosed MM patients was used to evaluate the expression of PARP1 by immunofluorescence staining and its impact on prognosis. Shown are representative histospots with low and high AQUA score for PARP1 expression, respectively, in panels C and D. AQUA scores are calculated based on the average PARP1 signal intensity per compartment area (CD138+ cells) of 3 representative histospots per patient (detailed methodology for AQUA score calculation is provided in supplemental Methods). Staining for DAPI, CD138, and PARP1 is merged together in the figure. (E) Kaplan-Meier survival curves indicate the shorter time to progression (TTP) and overall survival (OS) for patients with MM cells expressing high protein levels of PARP1.
High PARP1 expression is correlated with poor survival in MM. (A) Box plot displaying PARP1 mRNA range of expression (Affymetrix probe 208644_at) according to the molecular subgroup in a cohort of MM patients from the Arkansas dataset (GSE4581). Note the higher PARP1 expression in the MR, MS, and PR subgroups. (B) Kaplan-Meier overall survival curves of MM patients treated in total therapy 1 and 3 according to PARP1 mRNA expression above or below the median value of 5025 (left panel) or according to the quartiles distribution (right panel) of PARP1 mRNA expression based on National Institutes of Health GEO dataset deposited by Dr J. D. Shaughnessy Jr under accession number GSE4581. Quartile 4 represents the subgroup with the highest PARP1 expression. The 60-month OS for patients with PARP1 below and above the median are 67% and 54%, respectively (P = .02). (C-D) Myeloma tissue microarray constructed from the BM biopsies of 83 newly diagnosed MM patients was used to evaluate the expression of PARP1 by immunofluorescence staining and its impact on prognosis. Shown are representative histospots with low and high AQUA score for PARP1 expression, respectively, in panels C and D. AQUA scores are calculated based on the average PARP1 signal intensity per compartment area (CD138+ cells) of 3 representative histospots per patient (detailed methodology for AQUA score calculation is provided in supplemental Methods). Staining for DAPI, CD138, and PARP1 is merged together in the figure. (E) Kaplan-Meier survival curves indicate the shorter time to progression (TTP) and overall survival (OS) for patients with MM cells expressing high protein levels of PARP1.
PARP1/2 inhibition results in transient DNA DSBs but does not affect MM-cell survival
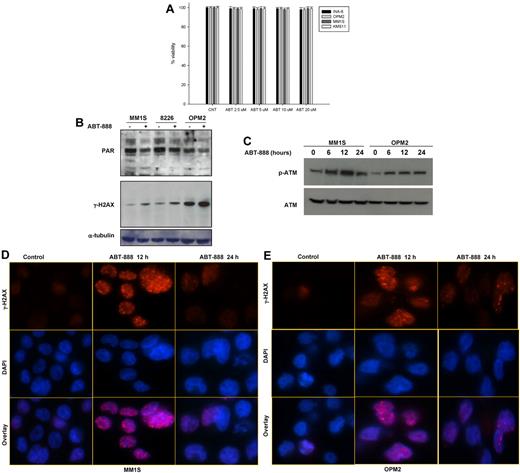
Using a panel of human MM cell lines, we first evaluated the effects of a potent and selective PARP1 and PARP2 inhibitor, ABT-888,42,43 on MM-cell survival. As shown in Figure 2A, exposure to increased doses of ABT-888 did not affect MM-cell viability despite effective PARP inhibition, as evidenced by the reduced PAR polymer levels measured by Western blot analysis (Figure 2B). To explore whether, as postulated, PARP inhibition does result in DNA damage and DNA DSBs, we measured the level of γH2AX (phospho-Ser139-H2AX) and p-ATM in MM cells treated with ABT-888. As shown in Figure 2B-C, ABT-888 did induce an increase in the level of γH2AX and p-ATM in treated cells, confirming that PARP1/2 inhibition does induce DNA damage and DSBs with effective ATM activation. Increased γH2AX foci formation was also confirmed by immunofluorescence within 12 hours of ABT-888 treatment, and these foci nearly fully resolved by 24 hours, which is consistent with the ability of the cells to repair these DNA DSBs induced by ABT-888 (Figure 2D-E).
Effects of ABT-888 on MM-cell survival, PARP1/2 activity, and DNA damage. (A) Effect of ABT-888 on MM-cell survival. Cells were treated with 0-20μM ABT-888 for 72 hours and viability was determined by annexin V/PI staining. Results shown here are representative of 3 independent experiments. (B-C) Protein levels of PAR polymers and γ-H2AX (p-Ser139-H2AX) (B) and p-ATM and ATM (C) after ABT-888 treatment in MM cell lines. Representative blot of PAR, γ-H2AX, p-ATM, and ATM both before and after 5μM ABT treatment is shown. (D-E) Effects of ABT-888 on γ-H2AX foci formation induced by ABT-888 (5μM) over time in OPM2 and MM1S MM cells and detected by immunofluorescent microscopy. Image acquisition was performed with an epifluorescence microscope (BX51; Olympus) and multispectral color camera (Nuance FX; CRi) with a 60× magnification lens and oil immersion.
Effects of ABT-888 on MM-cell survival, PARP1/2 activity, and DNA damage. (A) Effect of ABT-888 on MM-cell survival. Cells were treated with 0-20μM ABT-888 for 72 hours and viability was determined by annexin V/PI staining. Results shown here are representative of 3 independent experiments. (B-C) Protein levels of PAR polymers and γ-H2AX (p-Ser139-H2AX) (B) and p-ATM and ATM (C) after ABT-888 treatment in MM cell lines. Representative blot of PAR, γ-H2AX, p-ATM, and ATM both before and after 5μM ABT treatment is shown. (D-E) Effects of ABT-888 on γ-H2AX foci formation induced by ABT-888 (5μM) over time in OPM2 and MM1S MM cells and detected by immunofluorescent microscopy. Image acquisition was performed with an epifluorescence microscope (BX51; Olympus) and multispectral color camera (Nuance FX; CRi) with a 60× magnification lens and oil immersion.
Inhibition of the 26S proteasome with bortezomib transiently suppresses the promoter activity of several HR genes
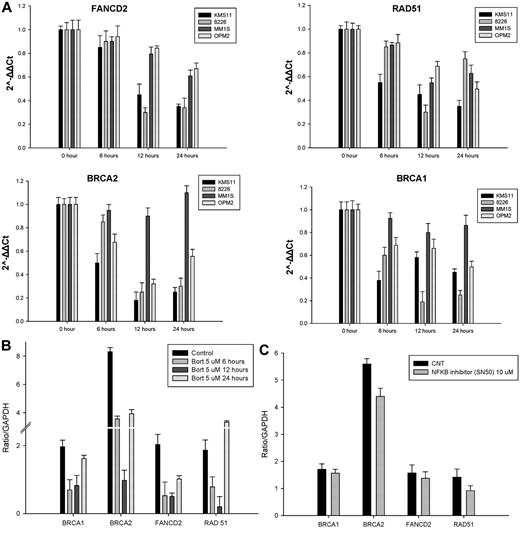
Recent reports have suggested that, in addition to inducing endoplasmic reticulum stress and the unfolded protein response mediating their anti-myeloma effects, proteasome inhibitors also enhance DNA damage via inhibition of DNA-repair mechanisms by reducing FANCD2 baseline expression levels and inhibiting FANCD2 ubiquitylation in response to DNA-damaging agents.27-29 To further assess in MM cells the effect of bortezomib on the HR-mediated DNA-repair pathway at the transcriptional level, we examined the mRNA expression of FANCD2, BRCA1, BRCA2, and RAD51 by qRT-PCR. As shown in Figure 3A, after bortezomib treatment, mRNA levels of FANCD2, BRCA1, BRCA2, and RAD51 significantly decreased in KMS11 and RPMI-8226 MM cell lines, and to a far lesser extent in MM1S and OPM2 cells, in a time-dependent fashion during the first 12 hours of treatment. This was followed by some recovery in their mRNA expression at 24 hours. To further evaluate how bortezomib may regulate HR genes at the transcriptional level, we conducted a promoter luciferase reporter screen. As shown in Figure 3B, within the first 12 hours of treatment, bortezomib induced a down-regulation of the promoter activity of several DDR genes, including BRCA1, BRCA2, RAD51, and FANCD2. Similar to the qRT-PCR mRNA results, the bortezomib-induced suppression of the promoter activity of these genes was transient and recovered to baseline (or higher) by 24 hours of treatment. Furthermore, this transient transcriptional suppression does not appear to be NF-κB mediated because NF-κB inhibition with SN50 did not affect the promoter activity of these genes (Figure 3C).
Bortezomib transiently repressed the expression of HR genes at the transcriptional level. (A) MM cell lines were treated with 2.5nM bortezomib and harvested at the indicated times. FANCD2, RAD-51, BRCA1, and BRCA2 mRNA levels were determined by qRT-PCR. Data quantification was carried out by the 2−ΔΔCt method. (B-C) MM1S cells transfected with a pSGG_prom vector (SwitchGear genomics) that contains an approximately 3-kb fragment of the human promoter for BRAC1, BRAC2, FANCD2, RAD51, or GAPDH (control) cloned into the MCS and fused to a luciferase reporter gene. Transfected cells were treated with a nonlethal dose(s) of bortezomib alone (B), the NF-κB inhibitor SN50 (C), or vehicle control (CNT) for 24 hours and then assayed for their luciferase activity. Signal was read out on a luminometer.
Bortezomib transiently repressed the expression of HR genes at the transcriptional level. (A) MM cell lines were treated with 2.5nM bortezomib and harvested at the indicated times. FANCD2, RAD-51, BRCA1, and BRCA2 mRNA levels were determined by qRT-PCR. Data quantification was carried out by the 2−ΔΔCt method. (B-C) MM1S cells transfected with a pSGG_prom vector (SwitchGear genomics) that contains an approximately 3-kb fragment of the human promoter for BRAC1, BRAC2, FANCD2, RAD51, or GAPDH (control) cloned into the MCS and fused to a luciferase reporter gene. Transfected cells were treated with a nonlethal dose(s) of bortezomib alone (B), the NF-κB inhibitor SN50 (C), or vehicle control (CNT) for 24 hours and then assayed for their luciferase activity. Signal was read out on a luminometer.
Proteasome inhibition depletes the pool of nuclear ubiquitin and abrogates γH2AX polyubiquitylation and BRCA1 and RAD51 foci formation
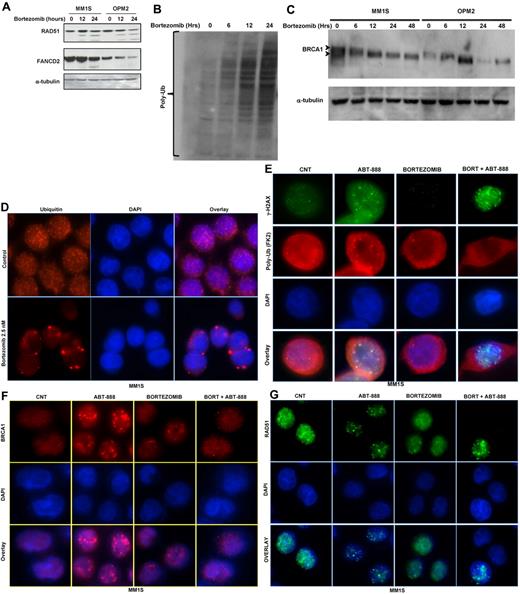
At the posttranscriptional level, and similar to the mRNA changes, treatment of MM cells with bortezomib partially reduced the levels of the FANCD2, RAD51, and BRCA1 proteins (Figure 4A-C). Despite the accumulation of polyubiquitylated proteins in total lysates from cells treated with bortezomib (Figure 4B), we observed a reduction in BRCA1 ubiquitylation (an up-shifted BRCA1 band; Figure 4C). This observation, along with several other reports,32,33 led us to postulate that inhibition of the 26S proteasome in MM cells does result in the accumulation of polyubiquitylated proteins in the cytoplasm, with a resulting depletion of nuclear free ubiquitin required for the Lys63 ubiquitylation of several molecules involved in DNA-damage sensing and repair (eg, γH2AX and BRCA1). As shown in Figure 4D and E and supplemental Figure 2A through D, treatment of OPM2 and MM1S cells with bortezomib did significantly deplete the pools of free nuclear ubiquitin with sequestration of polyubiquitinated proteins in the cytosolic compartment. However, it was unclear whether bortezomib would impair the ubiquitylation of histone H2AX in stressed cells undergoing DNA DSBs. Therefore, we next examined whether ubiquitylation of H2AX does occur in MM cells treated with ABT-888 and, if so, whether it is abrogated by bortezomib. As shown in Figure 4E and supplemental Figure 2B through D, ABT-888 did induce the formation of γH2AX foci (an indicator of DNA DSBs) with colocalization of polyubiquitin (FK2 clone) at the site of DNA DSBs (polyubiquitin foci colocalizing with γH2AX foci). In contrast, treatment with bortezomib did lead to the cytosolic sequestration of the polyubiquitin chains and failure to recruit polyubiquitin chains to the sites of DNA DSBs (γH2AX foci) in ABT-888–treated cells. Furthermore, and because histone H2AX polyubiquitylation by RNF8 and RNF168 is required for the RAP80/BRCA1 complex recruitment and maintenance at the sites of DNA damage, we next examined the effects of bortezomib and ABT-888 on BRCA1 and RAD51 foci formation. As shown in Figure 4F and G and supplemental Figure 2E through H, cotreatment with bortezomib did completely abrogate the BRCA1 and the RAD51 foci induced by ABT-888 treatment. These results provide evidence that bortezomib impairs the ability of MM cells to repair DNA DSBs induced by PARP inhibition.
Effects of bortezomib on ubiquitin cellular distribution and HR molecules at the posttranscriptional level. (A) Immunoblot analysis for FANCD2 and RAD51 protein expression in MM cell lines treated with 2.5nM bortezomib for 12 and 24 hours. (B-C) Protein extracts from MM1S and OPM2 cells treated for the indicated times with 2.5nM bortezomib were subjected to Western blot analysis. As shown, treatment with bortezomib results in the accumulation of polyubiquitin (B) and decreases the ubiquitylation of BRCA1 (C). (D-G) Immunofluorescent microscopy was used to analyze free ubiquitin and polyubiquitin cellular distribution and to detect γ-H2AX, BRCA1, and RAD 51 foci formation induced by bortezomib and/or ABT-888 treatment in OPM2 and MM1S (images for OPM2 cells are included in the supplemental materials). Cells were co-treated with bortezomib (2.5nM) and/or ABT-888 (5μM) for 24 hours. Shown in panel D are the effects of bortezomib on ubiquitin cellular distribution in MM with rapid depletion of the nuclear ubiquitin pool in bortezomib-treated cells. Cy5-labeled Ab was used to detect free ubiquitin and DAPI was selected for nuclear staining. (E) Effects of ABT-888 and/or bortezomib on γH2AX foci formation and polyubiquitin (FK2). Bortezomib did not affect ABT-888 γH2AX foci formation, but did inhibit the polyubiquitylation of γH2AX. (F-G) Effects of ABT-888 and/or bortezomib on BRCA1 (H) and RAD51 (G) foci formation. BRCA1 and RAD1 foci induced by ABT-888 were completely resolved in cells cotreated with bortezomib. Image acquisition was performed with an epifluorescence microscope (BX51; Olympus) and multispectral color camera (Nuance FX; CRi) with a 60× or 100× magnification lens and oil immersion.
Effects of bortezomib on ubiquitin cellular distribution and HR molecules at the posttranscriptional level. (A) Immunoblot analysis for FANCD2 and RAD51 protein expression in MM cell lines treated with 2.5nM bortezomib for 12 and 24 hours. (B-C) Protein extracts from MM1S and OPM2 cells treated for the indicated times with 2.5nM bortezomib were subjected to Western blot analysis. As shown, treatment with bortezomib results in the accumulation of polyubiquitin (B) and decreases the ubiquitylation of BRCA1 (C). (D-G) Immunofluorescent microscopy was used to analyze free ubiquitin and polyubiquitin cellular distribution and to detect γ-H2AX, BRCA1, and RAD 51 foci formation induced by bortezomib and/or ABT-888 treatment in OPM2 and MM1S (images for OPM2 cells are included in the supplemental materials). Cells were co-treated with bortezomib (2.5nM) and/or ABT-888 (5μM) for 24 hours. Shown in panel D are the effects of bortezomib on ubiquitin cellular distribution in MM with rapid depletion of the nuclear ubiquitin pool in bortezomib-treated cells. Cy5-labeled Ab was used to detect free ubiquitin and DAPI was selected for nuclear staining. (E) Effects of ABT-888 and/or bortezomib on γH2AX foci formation and polyubiquitin (FK2). Bortezomib did not affect ABT-888 γH2AX foci formation, but did inhibit the polyubiquitylation of γH2AX. (F-G) Effects of ABT-888 and/or bortezomib on BRCA1 (H) and RAD51 (G) foci formation. BRCA1 and RAD1 foci induced by ABT-888 were completely resolved in cells cotreated with bortezomib. Image acquisition was performed with an epifluorescence microscope (BX51; Olympus) and multispectral color camera (Nuance FX; CRi) with a 60× or 100× magnification lens and oil immersion.
Bortezomib impairs HR-mediated repair of DNA DSBs in MM cells
To examine directly the effect of bortezomib on HR-mediated repair of DNA DSBs, MM1S cells were transfected to stably express the DR-GFP reporter plasmid, followed by infection with adenovirus AdNGUS24i expressing the I-SceI yeast endonuclease (Figure 5A) in the presence or absence of bortezomib (1.25-5nM) for 12 hours. In this assay, the DSBs induced by the restriction enzyme I-SceI in the mutant GFP gene (Sce-GFP) is repaired by HR between the 2 tandemly located mutant GFP genes in the DR-GFP plasmid. After 12 hours, the adenovirus and bortezomib were washed and MM1S cells were assayed by flow cytometry for GFP expression as an indicator of their ability to conduct HR-directed DNA repair. As shown in Figure 5B and C, after ectopic expression of I-SceI endonuclease, approximately 20% of surviving MM1S-DR-GFP (PI− cells) became GFP+ compared with 0% in the controls. In contrast, bortezomib significantly diminished the percentage of GFP+ cells, confirming its ability to impair the HR-mediated repair. To exclude the possibility that this severe impairment of HR was caused by reduced cell viability or by an experimental artifact, we examined the effect of bortezomib on HR in the presence of the caspase inhibitor Z-VAD and repeated the same experiment in cells treated with melphalan at concentrations resulting in the same cell killing as bortezomib. As shown in Figure 5D and E, the bortezomib-induced suppression of the HR effect remained evident in the presence of the caspase-inhibitor Z-VAD. Interestingly, inhibition of HR was not observed in cells treated with the alkylating agent melphalan, but rather an increase in the number of GFP+ cells was observed compared with control or bortezomib. This observation is in agreement with the notion that alkylating agents may induce the DDR repair machinery and possibly promote further genomic instability in MM cells.23 Our findings indicate that 26S proteasome inhibition with bortezomib does impair the HR machinery at the posttranscriptional level, resulting in a state of BRCAness that may sensitize MM cells to PARP inhibitors.
Bortezomib inhibits HR-mediated repair of DNA DSBs. (A) Schema representing the phenotypic DR-GFP assay to measure HR-mediated DNA DSB repair. (B-C) Representative flow cytometric profile to measure the proportion of viable (PI−) and GFP+ MM1S-DR-GFP cells after infection with adenovirus control or AdNGUS24i adenovirus expressing the I-Sce1 endonuclease and incubation for 12 hours with or without bortezomib (1.25-5nM). Shown in panel C are the percentage of GFP+/PI− cells calculated from 3 independent experiments. (D) A representative flow cytometric profile to measure the percentage of PI− and GFP+ MM1S-DR-GFP cells after AdNGUS24i expression of the I-Sce1 endonuclease and treatment with bortezomib (5nM) alone or with Z-VAD (20μM) and melphalan (5μM). (E) Histograms representing the percentage of PI+ and GFP+ cells treated as described above. SD was calculated from 3 independent experiments.
Bortezomib inhibits HR-mediated repair of DNA DSBs. (A) Schema representing the phenotypic DR-GFP assay to measure HR-mediated DNA DSB repair. (B-C) Representative flow cytometric profile to measure the proportion of viable (PI−) and GFP+ MM1S-DR-GFP cells after infection with adenovirus control or AdNGUS24i adenovirus expressing the I-Sce1 endonuclease and incubation for 12 hours with or without bortezomib (1.25-5nM). Shown in panel C are the percentage of GFP+/PI− cells calculated from 3 independent experiments. (D) A representative flow cytometric profile to measure the percentage of PI− and GFP+ MM1S-DR-GFP cells after AdNGUS24i expression of the I-Sce1 endonuclease and treatment with bortezomib (5nM) alone or with Z-VAD (20μM) and melphalan (5μM). (E) Histograms representing the percentage of PI+ and GFP+ cells treated as described above. SD was calculated from 3 independent experiments.
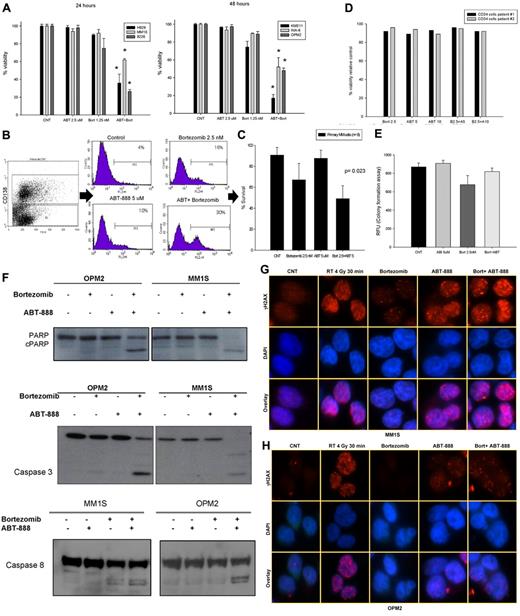
In vitro effects of ABT-888 and bortezomib in MM cell lines, primary MM cells, and CD34+ cells
To evaluate whether the BRCAness induced by the inhibition of the 26S proteasome in MM cells was able to sensitize them to PARP inhibitors, we first tested the effects of ABT-888 alone or in combination with bortezomib on MM-cell survival. As shown in Figure 6A and supplemental Figure 3, the combination of ABT-888 with bortezomib led to a significant reduction in survival compared with each drug alone. Similar results were also observed in primary CD138+ MM cells (Figure 6B-C). In addition, we examined the effects of ABT-888 ± bortezomib on CD34+ peripheral blood stem cell viability and their function in a CFU assay (GM-CFU). As shown in Figure 6D and E, neither the viability of CD34+ hematopoietic stem cells (measured by annexin V/PI staining), nor their GM-colony forming ability was affected by the combination of ABT-888 with bortezomib, which is consistent with a potentially safe therapeutic window of this combination.
In vitro effects of ABT-888 and bortezomib in MM cells. (A) Cells were cultured in control medium or in the presence of ABT-888 (2.5μM) and/or bortezomib (1.25nM) for 24-48 hours. Viability was determined by annexin V/PI staining. Results shown are means ± SD of 3 independent sets of experiments. *P < .05. (B) Effects of ABT-888 and bortezomib on CD138+ primary myeloma cells. BM mononuclear cells isolated by Ficoll gradient from the BM aspirate of MM patients and then cultured in 20% medium with or without ABT-888 (5μM) and/or bortezomib (2.5nM) for 24 hours. MM cells were identified by CD138 staining and flow cytometric analysis. Viability of CD138+ cells was determined by annexin V/PI staining. A representative experiment is shown here. (C) Effects of the combination of ABT-888 and bortezomib in 8 different MM patients. Viability was determined by annexin V/PI staining. (D) Effects of ABT-888 and bortezomib on CD34+ peripheral blood stem cells. Cells were cultured in 20% medium with or without ABT-888 (5-10μM) and/or bortezomib (2.5nM) for 24 hours. Viability was determined by annexin V/PI staining. Two representative samples are shown here. (E) Effects of the combination of ABT-888 and bortezomib on CD34+ peripheral blood stem cell function using a GM-CFU assay (CytoSelect; Cell Biolabs). At 10 days, colonies were quantified using a fluorescence plate reader (485/520 nm filter set) according to the assay protocol. (F) Cleavage of PARP, caspase 3, and caspase 8 was examined after exposure of MM cells to bortezomib (2.5nM) with or without ABT-888 (5μM) for 16 hours. (G-H) Effects of ABT-888 and bortezomib on γ-H2AX foci formation in MM cells treated with ABT-888 (5μM) and bortezomib (2.5nM) for 24 hours. Treatment with radiation (4 Gy for 30 minutes) was used as positive control for γ-H2AX foci formation in response to DNA damage.
In vitro effects of ABT-888 and bortezomib in MM cells. (A) Cells were cultured in control medium or in the presence of ABT-888 (2.5μM) and/or bortezomib (1.25nM) for 24-48 hours. Viability was determined by annexin V/PI staining. Results shown are means ± SD of 3 independent sets of experiments. *P < .05. (B) Effects of ABT-888 and bortezomib on CD138+ primary myeloma cells. BM mononuclear cells isolated by Ficoll gradient from the BM aspirate of MM patients and then cultured in 20% medium with or without ABT-888 (5μM) and/or bortezomib (2.5nM) for 24 hours. MM cells were identified by CD138 staining and flow cytometric analysis. Viability of CD138+ cells was determined by annexin V/PI staining. A representative experiment is shown here. (C) Effects of the combination of ABT-888 and bortezomib in 8 different MM patients. Viability was determined by annexin V/PI staining. (D) Effects of ABT-888 and bortezomib on CD34+ peripheral blood stem cells. Cells were cultured in 20% medium with or without ABT-888 (5-10μM) and/or bortezomib (2.5nM) for 24 hours. Viability was determined by annexin V/PI staining. Two representative samples are shown here. (E) Effects of the combination of ABT-888 and bortezomib on CD34+ peripheral blood stem cell function using a GM-CFU assay (CytoSelect; Cell Biolabs). At 10 days, colonies were quantified using a fluorescence plate reader (485/520 nm filter set) according to the assay protocol. (F) Cleavage of PARP, caspase 3, and caspase 8 was examined after exposure of MM cells to bortezomib (2.5nM) with or without ABT-888 (5μM) for 16 hours. (G-H) Effects of ABT-888 and bortezomib on γ-H2AX foci formation in MM cells treated with ABT-888 (5μM) and bortezomib (2.5nM) for 24 hours. Treatment with radiation (4 Gy for 30 minutes) was used as positive control for γ-H2AX foci formation in response to DNA damage.
Mechanistically, apoptotic events (caspase 3, caspase 8, and PARP cleavage) were also markedly enhanced by the combination of ABT-888 and bortezomib compared with either drug alone (Figure 6F). Furthermore, using immunofluorescent microscopy to measure DNA DSBs, γH2AX foci formation was notably increased and sustained at 24 hours in MM cells treated with the combination of ABT-888 with bortezomib compared with treatment with ABT-888 alone (Figure 6G-H).
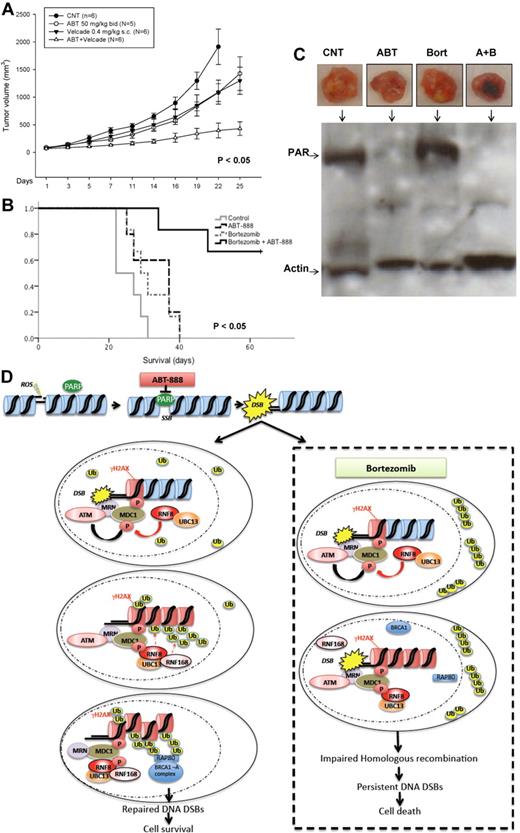
PARP1/2 inhibition potentiates the cytotoxic effects of bortezomib in MM xenograft model
Finally, we evaluated the in vivo activity of the combination of ABT-888 and bortezomib in a murine xenograft model of human MM.41 Cohorts of SCID mice bearing SC MM1S human xenografts were treated with bortezomib ± ABT-888 or vehicle alone. In a first cohort, ABT-888 was given via IP injection once daily for 3 weeks. Significant inhibition of tumor growth (P < .005) and improved survival (P < .05) was noted in mice treated with the combination compared with bortezomib alone or control-treated mice (supplemental Figure 4A-B). In a second cohort, ABT-888 was administered via oral gavage twice daily, a dosing schedule and route of delivery more consistent with the drug pharmacokinetics and pharmacodynamics and its clinical use. Similar results were also observed in this second cohort, with a more significant reduction in tumor growth and improved survival of mice treated with the combination compared with either drug alone or control (Figure 7A-B). When given twice daily, ABT-888 also exhibited some antitumor activity as a single agent. This effect is likely explained by the inhibition of HR under hypoxic conditions,44 a hypothesis that we are currently exploring in vitro under hypoxic culture conditions. No toxicity (eg, weight loss, ruffled coats, or paralysis) was observed in mice treated with the combination in either cohort. In addition to the direct measurement of tumor growth and mice survival, we also assessed the pharmacodynamic effects of ABT-888, bortezomib, or their combination on PAR synthesis in postmortem tumor sections using Western blotting. As shown in Figure 7C, we confirmed in vivo the depletion of PAR levels in the xenografted tumors taken from animals treated with ABT-888. These data demonstrate that the combination of ABT-888 and bortezomib exerts a significant in vivo antimyeloma activity that is superior to that of bortezomib alone and has no apparent additional toxicity, supporting our hypothesis of a contextual synthetic lethality effect between these 2 drugs that is selectively exhibited in MM cells. A summary of the postulated mechanisms of bortezomib-induced BRCAness is schematically represented in Figure 7D.
In vivo effects of ABT-888 and bortezomib in MM xenograft model. (A) CB-17 SCID mice were injected SC with 3 × 106 MM1S cells. After the detection of a measurable tumor, animals were treated with vehicle alone or bortezomib (0.4 mg/kg SC every Monday and Thursday) alone or ABT-888 alone (50 mg/kg by oral gavage twice per day) or the combination of bortezomib and ABT-888. Tumor volume was assessed in 2 dimensions every 2 days until the day of killing. (B) Survival was evaluated from the first day of treatment until death (Kaplan-Meier method). (C) Protein extracts from MM1S-xenografted tumors into SCID mice treated with vehicle alone (CNT), ABT-888 (ABT) 50 mg/kg by oral gavage twice daily and bortezomib (Bort) 0.4 mg/kg SC every Monday and Thursday or the combination of ABT-888 and bortezomib were subjected to SDS-PAGE, blotted with anti-PAR Ab, and reprobed with anti-actin Ab. (D) Schematic summary of the postulated mechanisms mediating effects of ABT-888 and bortezomib in MM cells.
In vivo effects of ABT-888 and bortezomib in MM xenograft model. (A) CB-17 SCID mice were injected SC with 3 × 106 MM1S cells. After the detection of a measurable tumor, animals were treated with vehicle alone or bortezomib (0.4 mg/kg SC every Monday and Thursday) alone or ABT-888 alone (50 mg/kg by oral gavage twice per day) or the combination of bortezomib and ABT-888. Tumor volume was assessed in 2 dimensions every 2 days until the day of killing. (B) Survival was evaluated from the first day of treatment until death (Kaplan-Meier method). (C) Protein extracts from MM1S-xenografted tumors into SCID mice treated with vehicle alone (CNT), ABT-888 (ABT) 50 mg/kg by oral gavage twice daily and bortezomib (Bort) 0.4 mg/kg SC every Monday and Thursday or the combination of ABT-888 and bortezomib were subjected to SDS-PAGE, blotted with anti-PAR Ab, and reprobed with anti-actin Ab. (D) Schematic summary of the postulated mechanisms mediating effects of ABT-888 and bortezomib in MM cells.
Discussion
Ongoing chromosomal instability is believed to be a defining feature of clonal plasma cells, resulting in a perpetual accumulation of large-scale changes to their genomic architecture as the disease progresses from the early stages of monoclonal gammopathy of undetermined significance to intramedullary MM and advanced-stage plasma cell leukemia. Whereas the mechanisms for this karyotypic instability in MM is largely unknown, recent observations have correlated these abnormalities with impaired nonhomologous end-joining DNA-damage repair and increased HR activity.23,24 The deregulation in the maintenance of genomic integrity is a shared feature among all malignancies, and was recently successfully exploited for therapeutic end points, as exemplified by the synthetic lethality of PARP inhibitors in BRCA1- and BRCA2-defective tumors.16,17 In MM, direct evidence of homozygous loss or mutations in BRCA1/2 or other HR genes is lacking. In contrast, Shammas et al23 previously reported an increased HR activity that may contribute to the genomic instability in this disease. Nevertheless, based on the newly identified ubiquitylation events required for the regulation of DNA repair and the role of proteasome inhibitors in MM, we have postulated that inhibition of the 26S proteasome may induce a BRCAness state that sensitizes MM cells to PARP inhibitors.
In the current study, we observed a higher PARP1 mRNA expression in GEP-defined poor-risk MM (MF, MS, and PR) and correlated its expression (at the mRNA and protein levels) with shortened disease survival outcomes. The PARP1 gene is located on chromosome 1q42.12, an area of frequent amplification in MM, and PARP1 was among the 15 genes in the MM high-risk signature previously validated by the Intergroupe Francophone du Myelome.45 Whether PARP1 is an independent prognostic factor and if it is directly implicated in the chromosomal instability in this disease will require further validation in future studies.
Whereas PARP inhibitors have already shown a direct antitumor effect in several HR-deficient malignancies,18 no cytotoxic activity was observed in MM-cell lines and primary cells treated with ABT-888 in our in vitro studies. This was despite the depletion of PAR levels and the transient occurrence of DNA DSBs shown by histone H2AX phosphorylation, γH2AX foci formation, and ATM activation. Therefore, the MM cells were able to cope and repair the DNA DSBs induced by ABT-888 with adequate histone H2AX polyubiquitylation and BRCA1 and RAD51 recruitment to sites of DNA damage. These results are consistent with other studies showing an increased ability to MM cells to cope with genotoxic stress and their heightened HR activity.22,23
In contrast to Lys48-linked polyubiquitin chains targeting proteins to the 26S proteasome for degradation, other ubiquitin linkages (eg, Lys63) are associated with nondegradative ubiquitin signaling (eg, HR mediation of DNA repair).46 In particular, polyubiquitylation of histones (H2A, H2AX, and H2B) upon detection of DNA damage by the 2 ubiquitin ligases RNF8 and RNF168 and the ubiquitin-conjugating enzyme Ubc13 is required for the recruitment of BRCA1 A complex and the initiation of HR-mediated DNA repair. In the current study, we demonstrated that the 26S proteasome inhibition with bortezomib did predominantly alter HR at the posttranscriptional level by impairing the recruitment of BRCA1 and RAD51 to sites of DNA damage. BRCA1 and RAD51 foci failed to accumulate at the sites of DNA DSBs (γH2AX foci) induced by the PARP1/2 inhibitor ABT-888 when MM cells were cotreated with bortezomib. In addition, bortezomib treatment depleted the nuclear pools of ubiquitin and abrogated the polyubiquitylation (lack of colocalization polyubiquitin FK2 foci with γH2AX), but not the phosphorylation and foci formation of histone H2AX in ABT-888–treated cells. Therefore, our results demonstrate that bortezomib does not alter the initial phase of DNA-damage sensing (MRN complex recruitment, ATM and H2AX phosphorylation, and MDC1 activation), but does impair the Lys63 polyubiquitylation of γH2AX, a required modification for the second wave of DNA-repair protein recruitment and retention of BRCA1 and RAD51 at the sites of DNA damage (Figure 7D). It is likely that this effect is merely the result of the depletion of the pools of free nuclear ubiquitin with the sequestration of polyubiquitylated proteins in the cytosol. However, other possible mechanisms may include a bortezomib-induced impairment in the ubiquitin ligases (RNF8 or RNF168) or in the function of the ubiquitin-conjugating enzyme Ubc13 or possibly an increase in the activity of the deubiquitylating enzyme USP3. Our results are consistent with initial studies demonstrating the role of RNF8 in H2AX ubiquitylation.32,33 In these studies, and similar to RNF8 silencing, treatment with the 26S proteasome inhibitor MG132 depleted of the pools of free ubiquitin and impaired BRCA1 foci formation in irradiated U-2-OS osteosarcoma and 293T human embryonic kidney cells. Furthermore, 2 prior studies28,29 also showed delayed or impaired 53BP1, NBS1, BRCA1, FANCD2, and RAD51 foci formation in irradiated HeLa cells pretreated with proteasome inhibitors. More recent wok by Yarde et al27 showed that NF-κB inhibition with BMS-345541 (an inhibitor of IκB kinase) or with the proteasome inhibitor bortezomib reduced FA/BRCA mRNA levels and FANCD2 protein expression in MM cells, resulting in enhanced sensitivity to melphalan. In that study, similar to our results, the decrease in the mRNA levels of the HR genes (BRCA1, BRCA2, and RAD51) induced by bortezomib were transient and therefore unlikely to mimic the effects of a homozygous deletion or biallelic mutations of these genes on HR-mediated repair of DNA DSBs. Furthermore, in our studies, the NF-κB inhibitor SN50 did not affect the promoter reporter activity of BRCA1, BRCA2, RAD51, or FANCD2, suggesting that the decrease in the mRNA levels of these genes was instead NF-κB independent.
Using the DR-GFP/I-SceI assay, we also confirmed that bortezomib does impair HR-mediated repair of DNA DSBs in MM1S cells. However, treatment with melphalan did have an opposing effect: an increase in HR-mediated repair. These results do raise the concern, described previously,23 that in MM cells with a heightened HR activity, exposure to DNA-damaging agents (eg, melphalan) without simultaneously targeting their DNA damage-repair machinery may result in further genomic instability and acquisition of drug resistance.
Our mechanistic studies did demonstrate that proteasome inhibition with bortezomib induces a functional state of BRCAness in MM cells and therefore sensitizes them to the activity of PARP inhibitors such as ABT-888. This cytotoxic effect of the combination of ABT-888 and bortezomib was also demonstrated in primary CD138+ MM cells while sparing the viability and function CD34+ stem cells. In addition, we confirmed the anti-MM activity of this combination in vivo in mice xenografted with MM1S. In vivo, ABT-888 alone also demonstrated some antimyeloma activity that was not observed in our in vitro studies. This effect may be the result on the impairment in HR under hypoxic conditions, as described previously.44
In summary, we have shown that inhibition of the 26S proteasome impairs HR-mediated repair of DNA breaks in MM cells and results in a contextual synthetic lethality when combined with PARP inhibitors. These results provide the framework for the future clinical investigation of the safety and efficacy of the combination of proteasome and PARP inhibitors in relapsed and refractory MM patients.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by research grants from the Multiple Myeloma Research Foundation (to P.N.), the Alberta Cancer Foundation (to D.S. and N.J.B.), and by the Terry Fox Foundation (to D.S. and N.J.B.).
Authorship
Contribution: P.N. and N.J.B. designed and performed the research, analyzed the data, and wrote the manuscript; P.N., L.R., K.G., E.S., J.J., A.K., A.M., and P.D. performed the research; and P.N., P.T., D.A.S., S.L., L.H.B., and N.J.B. analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nizar Jacques Bahlis, MD, University of Calgary, Division of Hematology and Bone Marrow Transplant, 1403 29th St NW Rm 681, Calgary, AB T2N 2T9, Canada; e-mail: nbahlis@ucalgary.ca.