Abstract
Protracted inhibition of osteoblast (OB) differentiation characterizes multiple myeloma (MM) bone disease and persists even when patients are in long-term remission. However, the underlying pathophysiology for this prolonged OB suppression is unknown. Therefore, we developed a mouse MM model in which the bone marrow stromal cells (BMSCs) remained unresponsive to OB differentiation signals after removal of MM cells. We found that BMSCs from both MM-bearing mice and MM patients had increased levels of the transcriptional repressor Gfi1 compared with controls and that Gfi1 was a novel transcriptional repressor of the critical OB transcription factor Runx2. Trichostatin-A blocked the effects of Gfi1, suggesting that it induces epigenetic changes in the Runx2 promoter. MM-BMSC cell-cell contact was not required for MM cells to increase Gfi1 and repress Runx2 levels in MC-4 before OBs or naive primary BMSCs, and Gfi1 induction was blocked by anti–TNF-α and anti–IL-7 antibodies. Importantly, BMSCs isolated from Gfi1−/− mice were significantly resistant to MM-induced OB suppression. Strikingly, siRNA knockdown of Gfi1 in BMSCs from MM patients significantly restored expression of Runx2 and OB differentiation markers. Thus, Gfi1 may have an important role in prolonged MM-induced OB suppression and provide a new therapeutic target for MM bone disease.
Introduction
Multiple myeloma (MM), a plasma cell malignancy, is the most frequent cancer to involve the skeleton and is characterized by the formation of lytic bone lesions adjacent to MM cells in approximately 90% of patients.1 The lytic lesions and the diffuse osteopenia seen with MM severely impact the patients' quality of life and survival once fractures occur. New bone formation that normally occurs at sites of previous bone destruction is absent or markedly suppressed.2 As a result, the MM-induced osteolytic lesions rarely heal because of MM-induced suppression of osteoblast (OB) bone-forming activity, even when patients are in prolonged remission and MM cells are no longer detectable. Thus, the mechanism underlying prolonged suppression of OB activity in the absence of MM cells cannot be explained by continued production of OB inhibitors by residual MM cells. Among the possible mechanisms responsible for the lack of bone repair in MM could be depletion of the precursor pool or selective intrinsic epigenetic changes in OB precursors that prevent their response to differentiation signals. We recently reported that bone marrow stromal cells (BMSCs) from MM patients exhibited increased support of MM and osteoclastogenesis that persisted after 3 to 4 weeks of in vitro culture in the absence of MM cells,3 indicating that at least part of the MM-induced BMSC change is intrinsic to the BMSCs (cell autonomous) and suggesting that MM cells induce a permanent change in BMSCs.
Recent studies have identified several soluble inhibitors of OB differentiation produced by MM cells that are elevated in MM marrow, including Dickkopf1 (DKK1) and soluble frizzled-related peptide (sFRP)-2 and sFRP-3, which block OB differentiation by affecting Wnt signaling; and cytokines IL-3, IL-7 and TNF-α.4,5 However, the suppressive effect of these soluble factors should be reversible once the tumor is gone. Moreover, the role of Wnt antagonists in human MM bone disease remains controversial. Giuliani et al reported that, although human MM cells produced DKK1, they do not produce sufficient DKK1 to block human OB differentiation.6 Further, as MM bone disease progresses, both DKK17 and sFRP28 expression is lost. Therefore, other mechanisms, in addition to inhibition of Wnt signaling, must play an important role in the OB suppression characteristic of MM bone disease.
We previously showed that IL-3 acts indirectly via stimulation of monocytes/macrophages to block OB differentiation.9 TNF-α is known to block OB differentiation by inhibition of precursor recruitment from progenitor cells10 and suppression of the expression of critical OB transcription factors Runx2,11 TAZ (a Runx2 coactivator),5 and osterix (Osx).12 Further, TNF-α neutralizing antibody can partially prevent MM cells from suppressing osteogenesis.5 IL-7 can also act as an inhibitor of OB differentiation.13 Importantly, treatment of BMSCs with conditioned media from an MM cell line or MM patient marrow plasma blocked early OB precursor differentiation, which was partially reversed by the addition of a neutralizing antibody to IL-7.14 However, the mechanism for the suppression of Runx2 activity in OB precursors by MM cells expressing IL-7 and TNF-α was not clearly defined in these studies.
To investigate whether MM growth in the bone microenvironment depleted the OB precursor pool or selectively suppressed OB differentiation, we developed an in vivo murine model of MM bone disease using MM cells modified to express green fluorescent protein (GFP) for visualization and thymidine kinase (TK) to make them sensitive to selective killing by ganciclovir. This model allowed us to recover BMSCs that had been exposed to MM cells in vivo, remove the MM cells, and both determine their properties ex vivo and compare them with BMSCs from MM patients. We found that MM cells selectively suppress OB differentiation of early multipotent OB precursors. We looked for MM induction in BMSCs of candidate transcription factors that could mediate epigenetic changes and long-term suppression of OB differentiation. We report that the transcription repressor growth factor independence-1 (Gfi1), which can mediate chromatin remodeling,15-18 is increased in BMSCs and pre-OB by MM exposure and by TNF-α or IL-7. In addition, we found that in untreated MC-4 cells, Gfi1 is cytoplasmic and is induced by MM cells, TNF-α, or IL-7 to translocate into the nucleus. Importantly, we demonstrated that Gfi1 plays a role in MM repression of Runx2 expression in BMSCs from MM patients, resulting in OB suppression.
Methods
Chemicals and antibodies
Cell culture media, penicillin and streptomycin (pen/strep) were from Invitrogen. FCS and trichostatin-A (TSA) were from Sigma-Aldrich. siRNA reagents were from Santa Cruz Biotechnology (SCB). Recombinant human IL-7 and TNF-α, and mouse blocking antibodies against IL-7 and TNF-α were from R&D Systems. Additional antibodies were from the following vendors: anti-Gfi1 from Abcam and Millipore, anti-Runx2 and anti-TFIIB from SCB, anti–β-actin and anti–α-tubulin from Sigma-Aldrich, and anti–rabbit or anti–mouse antibodies conjugated with HRP from GE Healthcare and Cell Signaling. Polyvinyl difluoride and nitrocellulose membranes were from Bio-Rad. All other chemicals were reagent grade.
Human samples and primary BMSC cultures
After obtaining informed consent in accordance with the Declaration of Helsinki, BM aspirates were collected in heparin from 9 normals and 16 patients with MM (Table 1). These studies were approved by the University of Pittsburgh Institutional Review Board. Marrow mononuclear cells were separated by Ficoll-Hypaque density sedimentation and cultured in long-term Dexter-type cultures to obtain stromal cells. Briefly, BM cells were incubated in IMDM-10% FCS overnight and the nonadherent cells removed. The cultures were then continued for 21 days with media changes every 4 days. Cells were detached with trypsin at subconfluence and replated (105 cells/10-cm dish) for use at passages 2 and 3.
Cell lines
Mouse MC3T3-E1 subclone-4 (MC-4) pre-OBs were described previously19,20 and maintained in ascorbic acid-free α-modified Eagle medium (α-MEM)–10% FCS-1% pen/strep (proliferation media).
5TGM1 MM cells were provided by Drs Bobatunde O. Oyajobi and Gregory R. Mundy (Vanderbilt University, Nashville, TN).21 These MM cells can be propagated in vitro, home to the BM to induce osteolytic lesions, and produce the same panel of cytokines associated with MM bone disease in patients.22,23 5TGM1 MM cells were stably transfected with a lentiviral construct containing a fusion gene encoding GFP and TK and a drug resistance cassette for blastocidin, provided by Dr Hee-Yong Chung (Hanyang University, Seoul, Korea).24 The 5TGM1-GFP-TK (denoted as 5TGM1 in some figures) cells were maintained in RPMI1640 10% FCS. Ganciclovir killing curves indicated that 40 ng/mL would selectively eradicate 100% of 5TGM1-GFP-TK cells in vitro within 10 days. Cocultures of 5TGM1-GFP-TK and MC-4 (10:1) were performed in α-MEM/RPMI 1640 (50:50) with 10% FCS. For most cocultures, MM cells were in a transwell (pore size, 0.4-μm) clear polyester Costar insert (Corning Life Sciences).
Mouse models
SCID NIHIII female mice (3-5 weeks of age) were injected intratibially with 20 μL saline with or without 105 5TGM1-GFP-TK cells. All research protocols were approved by the Veterans Administration Pittsburgh Healthcare System Institutional Animal Care and Use Committee, where this study was conducted. X-ray images of dissected tibias were acquired on a vivaCT 40 scanner (Scanco Medical) at a resolution of 21-μm isotropic, reconstructed and segmented for 3D display using the instruments analysis algorithm software (Sanco Medical Evaluation Program V6). The GFP+ tumor cells in dissected tibias were detected using the LT-9MACIMSYSPLUS Fluorescence imaging system (Lightools Research).
Gfi1Δex1b-3 (Gfi1+/−) mice,25 provided to us by Dr Stuart H. Orkin (Harvard Medical School, Boston, MA), were backcrossed onto a C57Bl/6 background and genotyped as previously published.26,27 Gfi1−/−, Gfi1+/−, and Gfi1+/+ littermates were sacrificed and the long bones shipped in DMEM to the University of Pittsburgh. Animal handling protocols and mouse manipulations were reviewed and approved by the Cincinnati Children's Hospital Research Foundation Institutional Animal Care and Use Committee.
Primary BMSCs were isolated from the bones of mice from both mouse models as previously published.3 BM cells harvested from tibias in DMEM-1% pen/strep were centrifuged, resuspended with 10 mL DMEM-10% FCS-1% pen/strep, and plated in 60 × 15-mm culture dishes. Cultures from mice injected with MM cells (and their controls) also contained ganciclovir (40 μg/mL). Nonadherent cells were removed the next day and the media replaced every other day. Fresh ganciclovir was added every day for 10 days (no fluorescent cells were still visible). Adherent BMSCs were cultured at 37°C for 3 or more weeks before assessing their differentiation potential.
OB differentiation and mineralization assays
Primary murine BMSCs or MC-4 cells were plated (105 cells/mL) in OB differentiation media (α-MEM-10% FCS with 50 μg/mL ascorbic acid, 10μM β-glycerolphosphate) with or without 50 ng/mL bone morphogenetic protein-2 (BMP2), for 1 to 5 days for RNA/protein analyses or 12 to 21 days for mineralization assays; media was changed every other day. Mineralization of primary BMSCs at day 21 was detected after rinsing the cells with PBS and fixing with prechilled 70% ethanol for 1 hour at 4°C, and then staining with alizarin red (40mM alizarin sodium sulfate, Sigma-Aldrich, pH 4.2) for 10 minutes at room temperature. Mineralization of MC-4 cells at 12 days was assessed by von Kossa stain using standard protocols.28
ALP activity assays
Whole cell extracts (200 μL Triton X-100 per well/12-well plates) were used to perform colorimetric alkaline phosphatase (ALP) activity assays using p-nitrophenyl phosphate (Sigma-Aldrich) as a substrate. ALP activity was normalized to total cellular protein assessed by BCA assay (Pierce Chemical) and expressed as units activity/mg protein. One unit of ALP activity is defined as the activity that will liberate 1μM p-nitrophenol/30 minutes at 37°C.
Real-time RT-PCR
Total mRNA was extracted using RNeasy (QIAGEN) or RNABee (Biogenesis) per the manufacturer's protocol and reverse-transcribed using SuperScript II (Invitrogen). Quantitative PCR was performed on an iCycler iQ system (Bio-Rad) using a SYBR Green PCR Core Kit (Applied Biosystems) and cDNA equivalent to 10 ng RNA in a 50-μL reaction according to the manufacturer's instructions. The DNA sequences of mouse and human primers used for quantitative PCR are listed in Table 2. Relative expression was calculated using the comparative 2−ΔΔCt method, with GAPDH used as the internal control.
DNAs
The 5′ deletion mouse Runx2 (mRunx2)–P1 promoter-pGL2 luciferase reporters,29 kindly provided by Dr Patricia F. Ducy (Columbia University, New York, NY), the 5′ deletion rat Runx2 (rRunx2) promoter-pGL3 luciferase reporters,30 kindly provided by Drs Jane B. Lian and Gary S. Stein (University of Massachusetts Medical School, Worcester, MA), and the wild-type (WT) mouse and human Gfi1-pcDNA3.1 as well as MSCV-puro-WT-Gfi1 and MSCV-puro-Gfi1N382S retroviral DNAs were all previously described.31
Transfection of Runx2 reporters
MC-4 cells (4 × 105/well) were plated/well (6-well plates) in α-MEM. The day after, MC-4 cells were transfected according to the manufacturer's instructions in Opti-MEM and 2 μL Lipofectamine2000 with 250 ng Runx2 promoter reporters (pGL2 or pGL3) along with various amounts of Gfi1-pcDNA3.1 and EV-pcDNA3.1 (totaling 250 ng) and 10 ng pTK-Renilla. The cells were incubated with transfection solution for 3 to 5 hours at 37°C in 5% CO2; then 1 mL complete-α-MEM was added and the incubation continued overnight before changing the media followed by another overnight incubation before making 100-μL cleared cell lysates to assay (20 μL) luciferase activity (Promega kit) using a Veritas Luminometer (Turnerbiosystems).
Generation of stably transduced MC-4 cells
Retroviruses produced by cotransfection of MSCV-puro-EV, MSCV-puro-WT-Gfi1, and MSCV-puro-Gfi1N382S with pVSVG into GP2-293 packaging cells (Clontech) were used to stably infect MC-4 cells aided by polybrene. After a 48-hour incubation with virus, the media was changed. After another 48-hour incubation, puromycin (1 μg/mL final) was added for selection and maintenance.
Protein lysates and Western blotting
Protein lysates from patient samples were extracted in RIPA lysis buffer (sc-24948; SCB). Jurkat cell lysates (12-303; Millipore) were used for controls. Proteins separated on SDS-PAGE (10%) were transferred onto nitrocellulose membranes and incubated with appropriately diluted Gfi1 antibody (05-924; Millipore), followed by HRP-conjugated secondary anti–mouse IgG (7076; Cell Signaling) and visualized with an enhanced chemiluminescence kit (GE Healthcare) as previously described.3 Blots were stripped and reprobed with anti–β-actin (A2066) followed by anti–rabbit IgG (7074; Cell Signaling).
Mouse cells were harvested with RIPA lysis buffer containing 1mM EDTA, 1mM EGTA, 50mM NaF, 2.5mM sodium pyrophosphate, 1mM β-glycerolphosphate, 1mM dithiothreitol, and 1mM sodium orthovanadate. Extracts of cytoplasmic and nuclear fractions were prepared using the Nuclear Extract kit from Active Motif. Protein samples (from cell equivalents) were separated by SDS-PAGE (10%), transferred to polyvinyl difluoride membranes, and probed with antibodies to Gfi1 (ab21061; Abcam), Runx2 (sc-10758), α-tubulin (T6074), TFIIB (sc-274), or β-actin (A5316), and visualized using the appropriate HRP-conjugated anti–rabbit (NA934V) or anti–mouse (NA931V) antibodies (GE Healthcare).
Knockdown of Gfi1
Human primary BMSCs, isolated from normals and MM patients, were plated (105/well-12 well plate) in IMDM-10% FCS and incubated overnight at 37°C. Transfection media (800 μL; sc36868) was added to each rinsed well, and 200 μL siRNA complex (5 μg/well) containing either siGfi1 (sc35467) and siControl (sc37007) prepared according to the manufacturer's protocol was added. The cells were incubated for 5 hours with the siRNA, and then 1 mL osteogenic media (IMDM-20% FCS-2% pen/strep with 10μM dexamethasone, 50 μg/mL ascorbic acid, 10μM β-glycerophosphate) was added. The media was changed the next day and then every other day. RNA was collected immediately after siRNA treatment (day 0) and after 5 days in osteogenic media.
Statistical analyses
Each experiment was repeated at least 3 times, and all quantitative data are presented as mean ± SD except as stated. Statistical differences were determined by Student t test. Results were considered significantly different for P < .05.
Results
In vivo MM mouse model
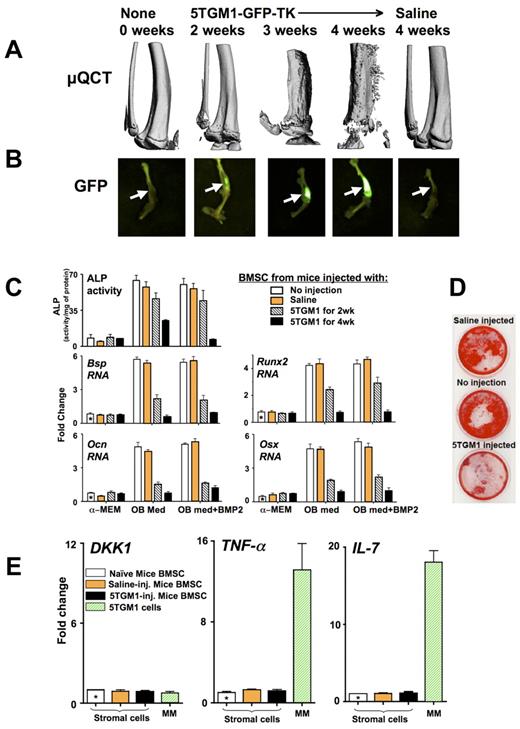
To explore the mechanisms involved in MM-induced OB suppression, we established an in vivo murine model system. In this model, we intratibially injected 5TGM1-GFP-TK cells, a well-characterized murine MM cell line that induces all of the features of MM bone disease in SCID mice.32 These 5TGM1 MM cells were modified to express GFP for visualization and TK for selective sensitivity to ganciclovir. We did not observe any bystander effects of ganciclovir on either OB differentiation or hematopoietic colony formation in vitro (data not shown). The SCID mice were injected intratibially with saline or 5TGM1-GFP-TK cells in saline, and lytic lesions were allowed to develop for 2 to 4 weeks before the mice were killed for analysis (Figure 1). By micro-QCT analysis, mice injected with 5TGM1-GFP-TK cells start developing lytic lesions at 2 weeks after MM cell injection with continued further bone deterioration through the 4 weeks that ultimately involves the entire tibia, leading to animal death from advanced disease (Figure 1A). In contrast, the saline injected controls at 4 weeks were similar to the 0-week time point, demonstrating that the effects detected were not the result of the injection process. Fluoroscopy of the injected tibias demonstrated that an increase in the fluorescent intensity was detected from 2 to 4 weeks, representing increased tumor burden (Figure 1B), and showed an excellent correlation between tumor burden and the amount of lytic lesions. Administration of ganciclovir (20 mg/kg per day subcutaneously) for 2 weeks in vivo was only able to slow tumor growth and bone destruction if started 24 hours after 5TGM1-GFP-TK cells were injected (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Development of lytic lesions in mice injected with 5TGM1-GFP-TK MM cells results in persistent OB suppression after culturing BMSCs in vitro. Mice were injected intratibially with 20 μL saline with or without 105 5TGM1-GFP-TK (5TGM1) cells and compared with uninjected controls. Lytic lesions were allowed to develop for the indicated time periods. At the end of each time point, the tibias were dissected, and micro-QCT and fluorescent images were obtained. (A) Micro-QCT images of right tibiae obtained from mice sacrificed at 0, 2, 3, and 4 weeks after their injection with 5TGM1-GFP-TK cells or at 4 weeks after saline injection. (B) Fluorescent images of the injected tibias taken using the LT-9MACIMSYSPLUS Fluorescence Imaging System. (C-D) BMSCs were recovered from these tibiae, treated with ganciclovir until no GFP+ MM cells were visible (10 days) and expanded (3 weeks), before starting OB differentiation by culturing with or without BMP2 (50 ng/mL) in either α-MEM or OB differentiation medium (OB med). (C) At day 5, protein lysates and RNAs were isolated for measurement of ALP activity and quantitative PCR analysis of Bsp, Ocn, Runx2, and Osx expression relative to the uninjected mice BMSCs (using 2−ΔΔCt analysis). GAPDH, reference gene. (D) At day 21, mineralization was assayed by alizarin red. (E) 5TGM1-GFP-TK cells and BMSCs (with the MM cells removed as in panel C and was photographed using a light box with no magnification) isolated from 4-week injected mice and controls were analyzed by quantitative PCR for expression of TNF-α, IL-7, and DKK1 and the data graphed relative to the BMSCs from uninjected mice using 2−ΔΔCt. GAPDH, reference gene. In 5TGM1-GFP-TK cells, relative to GAPDH using ΔCt analysis, relative fold mRNA expression was: TNF-α (48 ± 7), IL-7 (35 ± 12), and DKK1 (1 ± 0.3).
Development of lytic lesions in mice injected with 5TGM1-GFP-TK MM cells results in persistent OB suppression after culturing BMSCs in vitro. Mice were injected intratibially with 20 μL saline with or without 105 5TGM1-GFP-TK (5TGM1) cells and compared with uninjected controls. Lytic lesions were allowed to develop for the indicated time periods. At the end of each time point, the tibias were dissected, and micro-QCT and fluorescent images were obtained. (A) Micro-QCT images of right tibiae obtained from mice sacrificed at 0, 2, 3, and 4 weeks after their injection with 5TGM1-GFP-TK cells or at 4 weeks after saline injection. (B) Fluorescent images of the injected tibias taken using the LT-9MACIMSYSPLUS Fluorescence Imaging System. (C-D) BMSCs were recovered from these tibiae, treated with ganciclovir until no GFP+ MM cells were visible (10 days) and expanded (3 weeks), before starting OB differentiation by culturing with or without BMP2 (50 ng/mL) in either α-MEM or OB differentiation medium (OB med). (C) At day 5, protein lysates and RNAs were isolated for measurement of ALP activity and quantitative PCR analysis of Bsp, Ocn, Runx2, and Osx expression relative to the uninjected mice BMSCs (using 2−ΔΔCt analysis). GAPDH, reference gene. (D) At day 21, mineralization was assayed by alizarin red. (E) 5TGM1-GFP-TK cells and BMSCs (with the MM cells removed as in panel C and was photographed using a light box with no magnification) isolated from 4-week injected mice and controls were analyzed by quantitative PCR for expression of TNF-α, IL-7, and DKK1 and the data graphed relative to the BMSCs from uninjected mice using 2−ΔΔCt. GAPDH, reference gene. In 5TGM1-GFP-TK cells, relative to GAPDH using ΔCt analysis, relative fold mRNA expression was: TNF-α (48 ± 7), IL-7 (35 ± 12), and DKK1 (1 ± 0.3).
BMSCs isolated from these tibias were then assessed for their osteogenic and adipogenic differentiation capacity in vitro after removal of the MM cells by ganciclovir treatment. BMSCs recovered from these mice and cultured for several weeks in the absence of MM cells demonstrated a marked persistent reduction in OB differentiation, even with added BMP2 (Figure 1C-D), but maintained their capacity to differentiate into adipocytes (supplemental Figure 2). The degree of OB suppression observed by analysis of ALP activity, and mRNA expression of both Runx2 and Osx, critical transcription factors for early and late OB differentiation,33 and OB markers, bone sialoprotein (Bsp) and osteocalcin (Ocn) at 2 and 4 weeks after injection (Figure 1C) correlated with the amount of tumor burden and lytic lesions observed (Figure 1A-B). Mineralization assays further demonstrated the extensive inhibition of OB differentiation caused by the exposure of the BMSCs to 5TGM1-GFP-TK cells in vivo (Figure 1D).
To further determine the mechanisms responsible for the protracted suppression of OB differentiation in the absence of MM cells, both 5TGM1-GFP-TK cells and primary BMSCs isolated from tibia of mice injected with 5TGM1-GFP-TK or saline and depleted of MM cells as in Figure 1C were analyzed for their expression of TNF-α, IL-7, and DKK1, known suppressors of OB differentiation (Figure 1E). The 5TGM1-GFP-TK cells expressed 48-fold more TNF-α and 35-fold more IL-7 than DKK1 mRNA. The BMSCs from the MM-injected mice did not express increased levels of any of these cytokines compared with controls. Further, the BMSCs expressed similar levels of DKK1 mRNA as the 5TGM1-GFP-TK cells.
MM soluble factors repress Runx2 expression in BMSCs
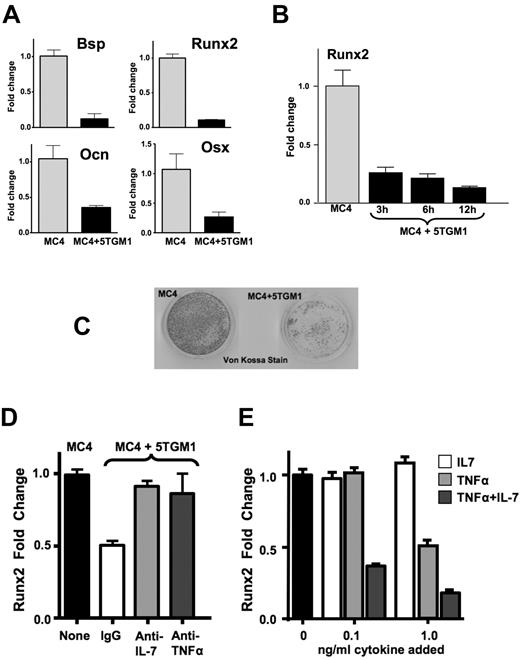
Coculture of 5TGM1-GFP-TK MM cells in a transwell with MC-4 cells, a mouse calvarial pre-OB cell line that when treated with ascorbic acid elevates Runx2 expression as well as the OB differentiation markers, Col1A1, Bsp, Ocn, and ALP, and can mineralize,20 revealed that OB suppression of MC-4 cells was mediated by a soluble factor(s) (Figure 2). Coculture of MC-4 cells with 5TGM1-GFP-TK cells in a transwell insert for 24 hours resulted in marked suppressed expression of Runx2, Osx2, Bsp, and Ocn, similar to BMSCs from MM-injected mice (Figure 2A). Time course studies revealed that MM cell suppression of Runx2 in MC-4 cells was rapid, with decreased Runx2 mRNA levels observed as early as 3 hours after MM cells were added (Figure 2B). In contrast, MM cells had no effect on ATF4 mRNA expression (data not shown), another transcription factor involved in OB differentiation.34 Further, coculture of MC-4 cells with 5TGM1-GFP-TK cells added 5 days after the initiation of differentiation inhibited mineralization by MC-4 cells in vitro (Figure 2C), revealing that the MM cells could halt the ongoing differentiation program.
Effects of 5TGM1-GFP-TK MM cells on MC-4 cell expression of OB markers, transcription factors, and mineralization. MC-4 cells were cocultured with either empty transwells or transwells containing 5TGM1-GFP-TK (5TGM1) cells for either (A) 12 hours or (B) 3, 6, and 12 hours before removing the transwells and isolating RNA from the MC-4 cells for quantitative PCR analysis of Bsp, Ocn, Runx2, and Osx expression, as indicated, relative to expression in the absence of 5TGM1-GFP-TK. (C) The MC-4 cells (2 × 105/60 mm dish) were cultured in α-MEM plus ascorbic acid for 12 days (the media was changed every 2 days), and 5TGM1 cells (2 × 106) were added at 5 days to half the wells. The 5TGM1 cells were removed before the MC-4 cells were stained with von Kossa for mineralization and were photographed using a light box with no magnification. (D) MC-4 cells (1 × 105/well 24-well plate) were pretreated with either 0.01 μg of IgG or neutralizing antibodies to IL-7 and TNF-α for 2 hours and then cocultured with 5TGM1 cells in a transwell for 4 hours, followed by RNA isolation from the MC-4 cells and quantitative PCR analysis of Runx2 expression relative to MC-4 cells cultured with an empty transwell. Higher doses of the antibodies did not have additional effects (data not shown). (E) MC-4 cells were treated with the indicated concentrations of IL-7 and TNF-α (when both added, amount indicated is for each) for 6 hours, and Runx2 expression was analyzed by quantitative PCR.
Effects of 5TGM1-GFP-TK MM cells on MC-4 cell expression of OB markers, transcription factors, and mineralization. MC-4 cells were cocultured with either empty transwells or transwells containing 5TGM1-GFP-TK (5TGM1) cells for either (A) 12 hours or (B) 3, 6, and 12 hours before removing the transwells and isolating RNA from the MC-4 cells for quantitative PCR analysis of Bsp, Ocn, Runx2, and Osx expression, as indicated, relative to expression in the absence of 5TGM1-GFP-TK. (C) The MC-4 cells (2 × 105/60 mm dish) were cultured in α-MEM plus ascorbic acid for 12 days (the media was changed every 2 days), and 5TGM1 cells (2 × 106) were added at 5 days to half the wells. The 5TGM1 cells were removed before the MC-4 cells were stained with von Kossa for mineralization and were photographed using a light box with no magnification. (D) MC-4 cells (1 × 105/well 24-well plate) were pretreated with either 0.01 μg of IgG or neutralizing antibodies to IL-7 and TNF-α for 2 hours and then cocultured with 5TGM1 cells in a transwell for 4 hours, followed by RNA isolation from the MC-4 cells and quantitative PCR analysis of Runx2 expression relative to MC-4 cells cultured with an empty transwell. Higher doses of the antibodies did not have additional effects (data not shown). (E) MC-4 cells were treated with the indicated concentrations of IL-7 and TNF-α (when both added, amount indicated is for each) for 6 hours, and Runx2 expression was analyzed by quantitative PCR.
Because 5TGM1-GFP-TK cells produce large amounts of TNF-α and IL-7 (Figure 1E), MC-4 cells were pretreated with neutralizing antibodies to both cytokines before exposure to 5TGM1-GFP-TK cells (Figure 2D). Both anti–TNF-α and anti–IL-7 neutralizing antibodies individually blocked MM-induced repression of Runx2 in MC-4 cells, whereas addition of control IgG did not prevent Runx2 repression. Interestingly, when TNF-α and/or IL-7 was added to MC-4 cells, TNF-α dose-dependently inhibited Runx2 expression by MC-4 cells, whereas IL-7 had minimal effects (Figure 2E). This is in contrast to the finding that the anti–IL-7 neutralizing antibody blocked Runx2 repression by MM cells. However, addition of low levels of IL-7 (0.1 ng/mL) with a suboptimal dose of TNF-α (0.1 ng/mL) markedly increased the Runx2 repression compared with TNF-α alone, indicating that IL-7 potentiates the effects of TNF-α. Therefore, if the effective dose of TNF-α during coculture of MM cells and MC-4 cells is suboptimal, IL-7 potentiation of TNF-α may be required to generate Runx2 repression, which explained the effectiveness of the anti–IL-7 neutralizing antibody.
Gfi1 is a transcriptional repressor of Runx2
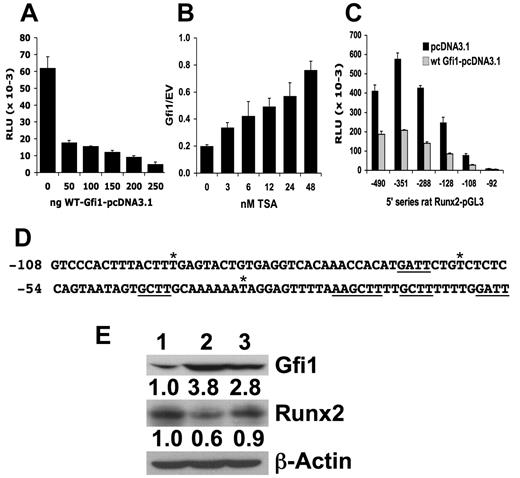
To further delineate the mechanisms involved in MM-induced suppression of Runx2 in OB precursors, MC-4 cells transfected with 5 mouse Runx2-P1 promoter deletion reporter constructs, ranging from −2800/+111 to −79/+111, were cocultured with 5TGM1 cells (data not shown). The presence of 5TGM1 cells resulted in approximately 50% repression of reporter activity in MC-4 cells expressing all the Runx2 deletion constructs except −79/+111, which has little activity indicating that the region from −992 to +111 contains the sites responsible for the repression. This region was previously reported to be responsive to down-regulation by TNF-α.11 Transcription factor binding site analysis of this 1003-bp segment of the Runx2 promoter indicated that within this region there are 29 putative core [AA(T/G)C] binding sites for the transcriptional repressor Gfi1 along with putative sites for other possible Runx2 repressors. Gfi1, a 50-kDa zinc finger protein containing member of the SNAG transcription repressor family that includes Gfi1b, Snail, Slug, IA-1, and Mlt1,35 is a complex protein that can mediate chromatin remodeling.15-18 To examine whether Gfi1 could regulate the Runx2 promoter, we cotransfected a Gfi1 expression plasmid with the −992/+111 mRunx2-P1 prom-pGL2 into MC-4 cells. Gfi1 dose-dependently repressed the mouse Runx2 promoter (Figure 3A). Because Gfi1-mediated repression has been reported to be inhibited by the histone deacetylase (HDAC) inhibitor TSA,17 we added various doses of TSA to MC-4 cells transfected with the Runx2 promoter reporter and either EV or a Gfi1 expression plasmid. TSA dose-dependently blocked Gfi1-mediated repression of the Runx2 promoter (Figure 3B). Cotransfection of Gfi1 with a more restricted deletion series of the rat Runx2 promoter from −490/+1 to −92/+1 demonstrated that the region from −108 to +1 containing 6 putative Gfi1 binding sites was important for Gfi1 regulation of the Runx2 promoter (Figure 3C). This region is 100% homologous between mouse and rat Runx2 and has only 3 nucleotide alterations from rodent to human (Figure 3D). To determine whether Gfi1 overexpression repressed the endogenous expression of Runx2, MC-4 cells were stably transduced with WT Gfi1 and Gfi1N382S (this mutation blocks DNA binding36 ) as well as empty retrovirus. There was a 3- to 4-fold increase in Gfi1 protein expression in both the WT Gfi1 and the Gfi1N382S stably transduced MC-4 cells compared with the cells transduced with the empty retrovirus, but only overexpression of WT Gfi1 decreased Runx2 protein expression to approximately 50% (Figure 3E).
Gfi1, a transcriptional repressor, can repress the Runx2 promoter. (A) Various amounts of expression plasmid encoding wild-type mouse Gfi1 (WT mGfi1) were cotransfected (0-250 ng), balanced by empty pcDNA3.1 vector (EV; to a total of 250 ng) along with the −992/+111 mRunx2 promoter-pGL2 (250 ng) and pRL-TK Renilla (10 ng) into MC-4 cells using Lipofectamine. The relative luciferase units (RLU) of 3 independent transfections were determined, averaged, and plotted with the SD. We did not use the Renilla activity as a reference because it was sensitive to Gfi1. (B) Various amounts of TSA (as indicated) were added to MC-4 cells transfected with 250 ng p976 mRunx2 promoter-pGL2 and 250 ng of either WT mGfi1 or empty vector as in panel A. The ratios (Gfi1/EV) of the RLU expressed at each amount of added TSA were averaged from 3 independent transfections (each set containing independent duplicates) and the SD calculated. (C) Similarly, either WT mGfi1 or empty vector was cotransfected with a set of rat Runx2 5′-deletion series in pGL3 as indicated (3′ end of all is +1). Three independent transfections were averaged and the SD calculated. (D) The −108 to −1 mouse/rat Runx2-P1 promoter sequence is shown with putative Gfi1 cores underlined (the longer one contains 2 overlapped cores in opposite directions, thereby forming a palindrome) and asterisks above denoting the 3 base differences with the human. (E) Western blot of protein extracts from MC-4 cells stably transduced with puromycin-selectable retrovirus containing cDNA for (lane 1) empty, (lane 2) WT mGfi-1, and (lane 3) mGfi-1N382S were analyzed for Gfi-1 and Runx2 expression as well as β-actin. The ratio of Gfi-1 or Runx2 relative to β-actin calculated for each sample and compared with the ratio exhibited by cells transduced with the empty retrovirus and denoted below each blot.
Gfi1, a transcriptional repressor, can repress the Runx2 promoter. (A) Various amounts of expression plasmid encoding wild-type mouse Gfi1 (WT mGfi1) were cotransfected (0-250 ng), balanced by empty pcDNA3.1 vector (EV; to a total of 250 ng) along with the −992/+111 mRunx2 promoter-pGL2 (250 ng) and pRL-TK Renilla (10 ng) into MC-4 cells using Lipofectamine. The relative luciferase units (RLU) of 3 independent transfections were determined, averaged, and plotted with the SD. We did not use the Renilla activity as a reference because it was sensitive to Gfi1. (B) Various amounts of TSA (as indicated) were added to MC-4 cells transfected with 250 ng p976 mRunx2 promoter-pGL2 and 250 ng of either WT mGfi1 or empty vector as in panel A. The ratios (Gfi1/EV) of the RLU expressed at each amount of added TSA were averaged from 3 independent transfections (each set containing independent duplicates) and the SD calculated. (C) Similarly, either WT mGfi1 or empty vector was cotransfected with a set of rat Runx2 5′-deletion series in pGL3 as indicated (3′ end of all is +1). Three independent transfections were averaged and the SD calculated. (D) The −108 to −1 mouse/rat Runx2-P1 promoter sequence is shown with putative Gfi1 cores underlined (the longer one contains 2 overlapped cores in opposite directions, thereby forming a palindrome) and asterisks above denoting the 3 base differences with the human. (E) Western blot of protein extracts from MC-4 cells stably transduced with puromycin-selectable retrovirus containing cDNA for (lane 1) empty, (lane 2) WT mGfi-1, and (lane 3) mGfi-1N382S were analyzed for Gfi-1 and Runx2 expression as well as β-actin. The ratio of Gfi-1 or Runx2 relative to β-actin calculated for each sample and compared with the ratio exhibited by cells transduced with the empty retrovirus and denoted below each blot.
Gfi1 is up-regulated in BMSCs in both murine MM models and in MM patient samples
When we examined BMSC RNA samples from the in vivo MM model described in Figure 1, we found that MM exposure in vivo resulted in long-term up-regulation of Gfi1 expression by quantitative PCR (Figure 4A). Exposure of MC-4 cells to 5TGM1-GFP-TK cells in a transwell for 6 to 72 hours demonstrated that the MM cells up-regulated Gfi1 RNA within 6 hours, which continued to rise 12-fold over 48 to 72 hours (Figure 4B). Similarly, a 6-hour exposure to TNF-α or IL-7 also up-regulated Gfi1 RNA (Figure 4C). Western blot analysis of whole cell extracts revealed that MM cells, TNF-α, and IL-7 induced higher Gfi1 protein levels (Figure 4D) compared with no treatment. Further, MM cells, TNF-α, and IL-7 also induced nuclear localization of Gfi1 in MC-4 cells, whereas under basal conditions Gfi1 was predominantly cytoplasmic. DKK1 (0.1-100 ng/mL) did not increase either Gfi1 RNA or protein (supplemental Figure 3). Most importantly, BMSCs isolated from 11 patients with MM bone disease (stage 3) had significantly higher Gfi1 RNA levels than BMSCs from 4 normals (Figure 4E). In addition, Western blot analysis showed that the Gfi1 protein level was also strongly elevated in the BMSCs from 8 MM patients compared with 5 normals (Figure 4F).
Gfi1, a transcriptional repressor, is induced in BMSCs and MC-4 cells by MM cells and TNF-α. (A) Primary BMSCs samples, as described in Figure 1, were analyzed for Gfi1 mRNA levels relative to GAPDH by quantitative PCR. (B) RNA from MC-4 cells cocultured with 5TGM1 cells (in transwell inserts) for the indicated times (h) before removal of the 5TGM1 cells and RNA isolation from the MC-4 cells. Expression of Gfi1 was detected by quantitative PCR relative to GAPDH and the “MC-4 alone” sample. (C) MC-4 cells were treated with 1 ng/mL TNF-α or IL-7 for 6 hours, and Gfi1 mRNA expression was determined by quantitative PCR relative to GAPDH. (D) Western blot analysis of Gfi1 protein expression and localization using whole cell, cytoplasmic, and nuclear extracts from 5TGM1 cells, MC-4 cells, and MC-4 cells treated with 5TGM1 cells in a transwell, TNF-α (1 ng/mL), or IL-7 (2 ng/mL). For loading controls, the blots were probed with anti–α-tubulin, TFIIB, or β-actin as indicated. Note that anti–TFIIB did not detect anything in the cytoplasmic extracts and anti–α-tubulin did not detect anything in the nuclear extracts, indicating that there was no significant cross-contamination of the subcellular compartments during their preparation. (E) The BM aspirates from MM patients and normals were cultured overnight; then the adherent cells were CD138-depleted using negative immunoselection (MACS, Miltenyi) and the BMSCs expanded for 3 weeks before use. Stromal cells obtained from 4 normals (●) and 11 MM patients (▴) were analyzed for Gfi1 expression by quantitative PCR relative to GAPDH and the fold change plotted with the SEM. Gfi1 RNA expression was significantly increased with a mean of 8.4-fold in the BMSCs from MM patients compared with the stromal cells from normals (P < .0006) using a 2-tailed unpaired t test with Welch correction. The Gfi1/GAPDH range for the normals was 0.24 to 1.23 and for the MM patients was 0.84 to 8.04 with the fold change MM relative to normal calculated among samples run at the same time. The 11 patients all had bone disease and stage 3 MM; 6 were newly diagnosed and 5 were relapsed, and there was no difference in the Gfi1 levels between these groups. (F) Western blot analysis for human Gfi1 protein expression in the BMSCs from 3 MM patients and 1 normal, along with Jurkat extract as a positive control (20 μg each). The ratio of Gfi1 to β-actin was calculated for each sample and noted below the blot. The ratio of Gfi1 to β-actin for these samples plus data from another 5 MM patients and 4 normals are plotted with the mean and SEM indicated. Gfi1 protein expression was significantly increased with a mean of 18-fold in the BMSCs from MM patients compared with the stromal cells from normals (P < .0194) using a 2-tailed unpaired t test.
Gfi1, a transcriptional repressor, is induced in BMSCs and MC-4 cells by MM cells and TNF-α. (A) Primary BMSCs samples, as described in Figure 1, were analyzed for Gfi1 mRNA levels relative to GAPDH by quantitative PCR. (B) RNA from MC-4 cells cocultured with 5TGM1 cells (in transwell inserts) for the indicated times (h) before removal of the 5TGM1 cells and RNA isolation from the MC-4 cells. Expression of Gfi1 was detected by quantitative PCR relative to GAPDH and the “MC-4 alone” sample. (C) MC-4 cells were treated with 1 ng/mL TNF-α or IL-7 for 6 hours, and Gfi1 mRNA expression was determined by quantitative PCR relative to GAPDH. (D) Western blot analysis of Gfi1 protein expression and localization using whole cell, cytoplasmic, and nuclear extracts from 5TGM1 cells, MC-4 cells, and MC-4 cells treated with 5TGM1 cells in a transwell, TNF-α (1 ng/mL), or IL-7 (2 ng/mL). For loading controls, the blots were probed with anti–α-tubulin, TFIIB, or β-actin as indicated. Note that anti–TFIIB did not detect anything in the cytoplasmic extracts and anti–α-tubulin did not detect anything in the nuclear extracts, indicating that there was no significant cross-contamination of the subcellular compartments during their preparation. (E) The BM aspirates from MM patients and normals were cultured overnight; then the adherent cells were CD138-depleted using negative immunoselection (MACS, Miltenyi) and the BMSCs expanded for 3 weeks before use. Stromal cells obtained from 4 normals (●) and 11 MM patients (▴) were analyzed for Gfi1 expression by quantitative PCR relative to GAPDH and the fold change plotted with the SEM. Gfi1 RNA expression was significantly increased with a mean of 8.4-fold in the BMSCs from MM patients compared with the stromal cells from normals (P < .0006) using a 2-tailed unpaired t test with Welch correction. The Gfi1/GAPDH range for the normals was 0.24 to 1.23 and for the MM patients was 0.84 to 8.04 with the fold change MM relative to normal calculated among samples run at the same time. The 11 patients all had bone disease and stage 3 MM; 6 were newly diagnosed and 5 were relapsed, and there was no difference in the Gfi1 levels between these groups. (F) Western blot analysis for human Gfi1 protein expression in the BMSCs from 3 MM patients and 1 normal, along with Jurkat extract as a positive control (20 μg each). The ratio of Gfi1 to β-actin was calculated for each sample and noted below the blot. The ratio of Gfi1 to β-actin for these samples plus data from another 5 MM patients and 4 normals are plotted with the mean and SEM indicated. Gfi1 protein expression was significantly increased with a mean of 18-fold in the BMSCs from MM patients compared with the stromal cells from normals (P < .0194) using a 2-tailed unpaired t test.
Gfi1 can initiate MM-induced suppression of OB differentiation
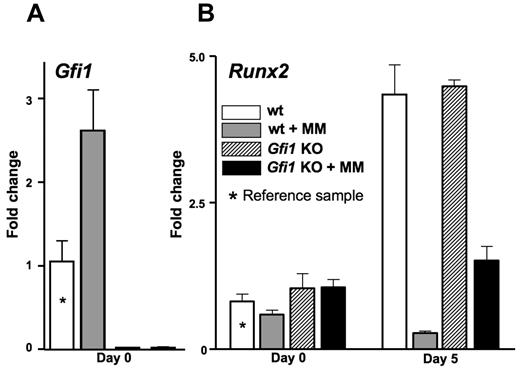
We then investigated whether Gfi1 deficiency before exposure to MM cells could prevent the MM-triggered repression of OB differentiation. Primary BMSCs from Gfi1−/− mice and their littermate controls were cocultured with 5TGM1-GFP-TK cells in a transwell for 24 hours and analyzed for Runx2 mRNA expression before and after 5 days in OB differentiation media (Figure 5). The primary BMSCs from Gfi1+/+ (and Gfi1+/−, not shown) mice up-regulated Gfi1 expression in response to the MM cells (Figure 5A) and failed to up-regulate Runx2 after 5 days in OB differentiation media (Figure 5B), recapitulating what we had observed with primary BMSCs exposed to MM in vivo (Figures 1 and 4) and MC-4 cells exposed in vitro (Figures 2 and 4). The primary BMSCs from Gfi1−/− mice (Figure 5A) up-regulated Runx2 normally in OB differentiation media in the absence of MM cell exposure (Figure 5B). Importantly, the Gfi1−/− primary BMSCs were significantly resistant to Runx2 repression by MM cells (Figure 5B).
Lack of Gfi1 in mouse BMSCs decreases the degree of MM repression of Runx2 in vitro. BMSCs recovered from Gfi1+/+ and Gfi1−/− mice were expanded in culture for 3 to 4 weeks. The BMSCs were then exposed to 5TGM1-GFP-TK cells in a transwell for 48 hours followed by either RNA harvest (day 0) or 5-day incubation in OB differentiation media before RNA harvest (day 5). RNAs were analyzed by quantitative PCR for (A) Gfi1 and (B) Runx2 levels. The Runx2 expression in the MM-exposed Gfi1−/− BMSCs was significantly increased relative to the MM-exposed Gfi1+/+ BMSCs (P < .05). BMSCs from Gfi1+/− mice were also analyzed but were found to behave identically to Gfi1+/+ and were not included for clarity.
Lack of Gfi1 in mouse BMSCs decreases the degree of MM repression of Runx2 in vitro. BMSCs recovered from Gfi1+/+ and Gfi1−/− mice were expanded in culture for 3 to 4 weeks. The BMSCs were then exposed to 5TGM1-GFP-TK cells in a transwell for 48 hours followed by either RNA harvest (day 0) or 5-day incubation in OB differentiation media before RNA harvest (day 5). RNAs were analyzed by quantitative PCR for (A) Gfi1 and (B) Runx2 levels. The Runx2 expression in the MM-exposed Gfi1−/− BMSCs was significantly increased relative to the MM-exposed Gfi1+/+ BMSCs (P < .05). BMSCs from Gfi1+/− mice were also analyzed but were found to behave identically to Gfi1+/+ and were not included for clarity.
Gfi1 is required to maintain MM-induced OB repression
To determine whether elevated Gfi1 was required to maintain suppression of OB differentiation of the MM-exposed BMSCs, BMSCs recovered from MM patients and normals were grown in culture for more than 3 weeks and were either not transfected or transfected with siGfi1 or siControl before initiating OB differentiation (Figure 6). Gfi1 was knocked down approximately 70% in the BMSCs from both MM patients and normals. As expected, in untransfected BMSCs and siControl-transfected BMSCs, Gfi1 was increased and Runx2, Ocn, and Bsp were decreased in the MM-exposed BMSCs compared with the normal BMSCs. However, when Gfi1 was knocked down, the capacity for the BMSCs from MM patients to respond to OB-inductive media and increase Runx2, Ocn, and Bsp was partially recovered. A similar experiment was conducted with MC-4 cells exposed to MM cells for 24 hours in vitro in a transwell and, after removal of the MM cells, treated with siRNA before initiating OB differentiation for 5 days. In this model, the knockdown of Gfi1 also permitted Runx2 and Osx RNA levels to partially recover and become elevated in response to OB differentiation signals (supplemental Figure 4).
Knockdown of Gfi1 in BMSCs from an MM patient rescues Runx2, Ocn, and Bsp expression. The BM aspirates from MM patients and normals were as described in Figure 4E. BMSCs from an MM patient and a normal were transfected with either siGfi1 or siControl (untransfected samples were also analyzed). RNAs were collected after transfection (day 0) and after 5 days in OB differentiation media (day 5) and analyzed by quantitative PCR for Gfi1, Runx2, Ocn, and Bsp expression levels. All samples were analyzed relative to the RNA level in the day 0 sample from the normal, and the day 5 data are presented. A 70% knockdown of Gfi1 was detected in BMSCs from both the MM patient and the normal. The Runx2, Ocn, and Bsp expression in the siGfi1 treated BMSCs from the MM patient was significantly increased relative to the siControl-treated BMSCs (P < .05). These data are representative of 2 MM patients analyzed.
Knockdown of Gfi1 in BMSCs from an MM patient rescues Runx2, Ocn, and Bsp expression. The BM aspirates from MM patients and normals were as described in Figure 4E. BMSCs from an MM patient and a normal were transfected with either siGfi1 or siControl (untransfected samples were also analyzed). RNAs were collected after transfection (day 0) and after 5 days in OB differentiation media (day 5) and analyzed by quantitative PCR for Gfi1, Runx2, Ocn, and Bsp expression levels. All samples were analyzed relative to the RNA level in the day 0 sample from the normal, and the day 5 data are presented. A 70% knockdown of Gfi1 was detected in BMSCs from both the MM patient and the normal. The Runx2, Ocn, and Bsp expression in the siGfi1 treated BMSCs from the MM patient was significantly increased relative to the siControl-treated BMSCs (P < .05). These data are representative of 2 MM patients analyzed.
Discussion
In this study, we demonstrated that MM-exposed MC-4 cells and primary BMSCs from our in vivo MM model and from MM patients all expressed elevated levels of the transcriptional repressor Gfi1 as well as decreased Runx2 expression under osteogenic conditions in vitro. The differentiation of OB from BMSCs requires the activity of Runx2.37 The MM-induced elevation of Gfi1 in MC-4 or BMSCs persisted long after the MM cells were removed, matching the protracted block to osteogenic induction of Runx2 and bone formation seen in MM. Gfi1−/− BMSCs were significantly resistant to MM repression of Runx2 induction by OB differentiation media. Importantly, siRNA knockdown of Gfi1 expression partially restored Runx2 expression in primary BMSCs from MM patients and MM-exposed MC-4 cells. Together, these data reveal a significant role for MM induction of Gfi1 expression in the long-term block preventing OB-inductive signals from up-regulating Runx2 and driving OB differentiation.
Gfi1 is a multidomain and multifunctional transcription factor (Figure 7A) that can bind DNA via Zn-finger domains and mediate chromatin remodeling, resulting in long-term changes in gene expression. Gfi1 has been reported to recruit enzymes and corepressors to target genes that result in long-term epigenetic changes in chromatin structure through modification of histone methylation and acetylation states, resulting in gene silencing, such as histone lysine methyltransferase G9a, histone lysine demethylase LSD1, CoREST, ETO proteins, and HDACs.15-18 The Zn-finger domains also are involved in protein-protein interactions, such as binding to the transcription factors Spi-1/PU.1,38 ETS1,39 PDRM5,40 and Miz-1,41 and to ubiquitin E3 ligases Triad142 and Mdm2.43 Gfi1 interaction with other transcription factors can serve to repress their function (ETS1,39 Spi-1/PU.1,38 p65-NFκB44 ) or to act as a corepressor (Miz-141 ), and in the case of PDRM5,40 another repressor, to act together as a transcriptional activator. The intermediate domain is also involved in protein-protein interactions that regulate other transcription factors, such as binding PIAS3, which causes STAT3 activation,45 as well as binding the splicing factor U2AF26,46 which results in modification of RNA splicing of CD45R. Furthermore, the SNAG domain interaction with GATA3 stabilizes GATA3 protein, thereby enhancing it is activating function, which is required for TH2 cell differentiation.43 Thus, there are many possible mechanisms by which elevated Gfi1 may block Runx2 induction by osteogenic signals (Figure 7B).
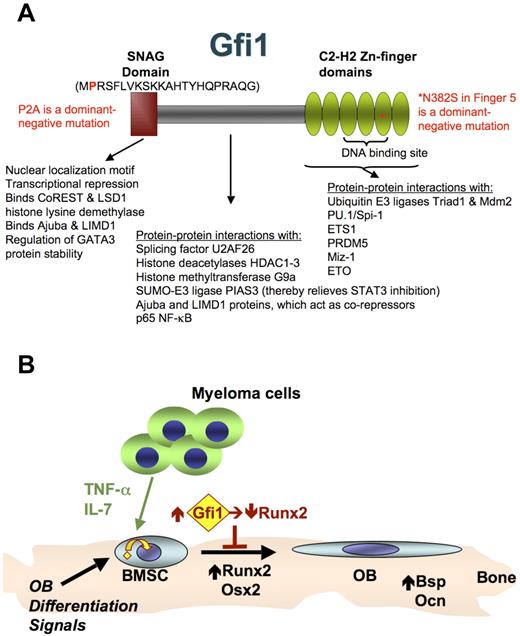
Gfi1 schematic and role in MM-induced block to OB differentiation. (A) Gfi1 is a multifunctional protein that interacts with many other proteins as indicated.49 Nuclear localization signal domains lie within the N-terminal SNAG domain and within the region encoding the 6 C-terminal C2-H2 zinc finger domains.35 Zinc fingers 3, 4, and 5 are required for sequence-specific DNA binding.48 The SNAG domain is required for transcriptional repression by Gfi1 via binding corepressors CoREST and histone lysine demethylase LSD1 (H3K4 demethylation).18 It also binds and stabilizes GATA3, independently of the repressor binding amino acids.43 The intermediate domain between the SNAG and Zn-finger domains is also involved in protein-protein interactions, such as binding the corepressors histone lysine methyltransferases G9a (H3K9 and H3K27 dimethylation/trimethylation),16 histone deacetylases HDAC1-3 (H3K9 deacetylation),16,17 and Ajuba LIM domain protein.50 This region also binds SUMO-E3 ligase PIAS3 (thereby enhancing STAT3 activation),45 the splicing factor U2AF26,46 and the p65 subunit of NF-κB (thereby inhibiting p65-p50 DNA binding).44 The Zn-finger domains also are involved in protein-protein interactions, such as binding to the transcription factors Spi-1/PU.1,38 ETS1,39 PDRM5,40 and Miz-1,41 and the corepressor ETO.17 The DNA-binding domain also interacts with the ubiquitin E3 ligases Triad142 and Mdm2.43 Direct gene repression by Gfi1 requires both binding to the target gene and interaction with corepressor proteins. Mutations in Gfi1 that either block SNAG binding to corepressors (P2A)35 or Zn-finger DNA-binding (N382S)31 both can act as dominant-negatives and relieve Gfi1 target gene repression. (B) Model for Runx2 repression in BMSCs by MM cells. The MM cells secrete TNF-α and other factors (eg, IL-7) that interact with cognate receptors on the BMSCs to increase the expression of Gfi1 and induce its translocation from the cytoplasm to the nucleus. Gfi1 tethered in the cytoplasm could down-regulate Runx2 by mechanisms involving modulation of other transcription factors. Eventual induction of Gfi1 translocation into the nucleus allows direct recruitment of corepressors to target genes, such as Runx2.
Gfi1 schematic and role in MM-induced block to OB differentiation. (A) Gfi1 is a multifunctional protein that interacts with many other proteins as indicated.49 Nuclear localization signal domains lie within the N-terminal SNAG domain and within the region encoding the 6 C-terminal C2-H2 zinc finger domains.35 Zinc fingers 3, 4, and 5 are required for sequence-specific DNA binding.48 The SNAG domain is required for transcriptional repression by Gfi1 via binding corepressors CoREST and histone lysine demethylase LSD1 (H3K4 demethylation).18 It also binds and stabilizes GATA3, independently of the repressor binding amino acids.43 The intermediate domain between the SNAG and Zn-finger domains is also involved in protein-protein interactions, such as binding the corepressors histone lysine methyltransferases G9a (H3K9 and H3K27 dimethylation/trimethylation),16 histone deacetylases HDAC1-3 (H3K9 deacetylation),16,17 and Ajuba LIM domain protein.50 This region also binds SUMO-E3 ligase PIAS3 (thereby enhancing STAT3 activation),45 the splicing factor U2AF26,46 and the p65 subunit of NF-κB (thereby inhibiting p65-p50 DNA binding).44 The Zn-finger domains also are involved in protein-protein interactions, such as binding to the transcription factors Spi-1/PU.1,38 ETS1,39 PDRM5,40 and Miz-1,41 and the corepressor ETO.17 The DNA-binding domain also interacts with the ubiquitin E3 ligases Triad142 and Mdm2.43 Direct gene repression by Gfi1 requires both binding to the target gene and interaction with corepressor proteins. Mutations in Gfi1 that either block SNAG binding to corepressors (P2A)35 or Zn-finger DNA-binding (N382S)31 both can act as dominant-negatives and relieve Gfi1 target gene repression. (B) Model for Runx2 repression in BMSCs by MM cells. The MM cells secrete TNF-α and other factors (eg, IL-7) that interact with cognate receptors on the BMSCs to increase the expression of Gfi1 and induce its translocation from the cytoplasm to the nucleus. Gfi1 tethered in the cytoplasm could down-regulate Runx2 by mechanisms involving modulation of other transcription factors. Eventual induction of Gfi1 translocation into the nucleus allows direct recruitment of corepressors to target genes, such as Runx2.
Gfi1 expression vector cotransfection with a series of Runx2-P1 promoter-reporters narrowed the Gfi1-responsive region to the proximal 108 bp of the promoter (−108 to +1), matching the region previously reported to be responsive to TNF-α repression.11 Although this region contains 5 putative Gfi1 binding sites (one of which is a double palindromic core), there are several other transcription factors (both repressors and activators) that bind this region that might be regulated by Gfi1. Of note, TSA blocked Gfi1 inhibition of the cotransfected Runx2 promoter, indicating that Gfi1 recruitment of HDACs, which modify chromatin, plays a significant role. Our studies also revealed that Gfi1 subcellular localization is regulated, with Gfi1 largely cytoplasmic in untreated MC-4 cells. Both MM cell exposure and TNF-α triggered Gfi1 translocation into the nucleus over a 48-hour period. The exact nature of this regulation may be cell-type specific. However, most previous studies examining the subcellular localization of Gfi1 used systems in which Gfi1 was overexpressed and may have swamped an endogenous regulatory system.35,47,48 The mechanisms by which Gfi1 is tethered in the cytoplasm or released to translocate into the nucleus are unknown.
We found that 5TGM1-GFP-TK MM cells in a transwell repressed Runx2 expression in both MC-4 and BMSCs, indicating that soluble factors play a key role in this process. However, this does not preclude a role for cell-cell contact. Guiliani et al14 reported that human MM cells exhibited a stronger inhibitory effect when cell-cell contact was involved (via VLA-4/VCAM-1). We found in this study that TNF-α neutralizing antibody blocked both MM elevation of Gfi1 and repression of Runx2 expression. IL-7 is also elevated in MM marrow14 and can act as an inhibitor of OB differentiation.13 Further, Guiliani et al showed that IL-7 was involved in generating MM-induced inhibition of Runx2 activity and OB differentiation.14 We also observed that addition of IL-7 alone to MC-4 cells did not suppress Runx2 mRNA levels, although IL-7 could elevate Gfi1 and induce nuclear translocation. Because we have shown that MM cells suppress Runx2 expression and OB differentiation via Gfi1, these results indicate that MM cells and TNF-α must induce a Gfi1 modification or partner that IL-7 does not. Moreover, IL-7 was able to potentiate repression of Runx2 expression by suboptimal levels of TNF-α. Further, IL-7 neutralizing antibody blocked MM-induced Runx2 repression. These results suggest that the MM cells in transwell culture conditions secrete TNF-α levels that require IL-7 potentiation to repress Runx2. This could result from the lack of cell-cell interactions between the MM and MC-4 pre-OB cells driving MM cells to produce more TNF-α or simply a problem of dilution in the cell culture well environment. Alternatively, this could indicate that multiple signals from MM cells are required to work together to generate strong inhibition of OB differentiation. Thus, these data support an important role for TNF-α and IL-7 as suppressors of OB differentiation in MM.
Our results demonstrate that MM cells increase Gfi1 expression and its nuclear localization in BMSCs, which blocks increased Runx2 expression under osteogenic conditions, thereby inhibiting osteoblastogenesis and new bone formation in MM bone disease. Thus, these results demonstrate that Gfi1 is responsible, at least in part, for the MM-induced prolonged OB suppression and may provide a new target for treatment of MM bone disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Chung for the GFP-TK-blastocidin-puromycin lentivirus, Drs Oyajobi and Mundy for the 5TGM1 cells, Drs Lian and Stein for rat Runx2 plasmids, Dr Ducy for mouse Runx2 plasmids, Dr Orkin for the Gfi1 knockout mice, Donna Gaspich for excellent secretarial assistance, Leah Furhman and Avinash Baktula for excellent technical assistance, and the Veterans Administration Pittsburgh Healthcare System, Research and Development for use of the facilities.
This work was supported by the Veterans Administration (Merit Award, G.D.R.), the Multiple Myeloma Research Foundation (G.D.R.), and the National Institutes of Health (NIAMS grants AR059647, G.X.; and AR059679, G.D.R. and D.L.G.).
National Institutes of Health
Authorship
Contribution: S.D., D.d.P., S.J., Q.S., A.J.H., F.E.K., B.S., C.-S.H., and S.Y. designed and performed research and analyzed and interpreted data; J.L.A. and K.D.P. performed experiments; C.S.V. and H.L.G. contributed vital new reagents and discussions; G.X. helped design the research; and G.D.R. and D.L.G. designed the research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: G.D.R. is a consultant for Amgen, Celgene, and Millennium. The remaining authors declare no competing financial interests.
Correspondence: Deborah L. Galson, University of Pittsburgh, Veterans Administration Pittsburgh Healthcare System Research and Development, 151-U, Rm 2E-109, University Dr C, Pittsburgh, PA 15240; e-mail: dgalson@gmail.com.
References
Author notes
S.D. and D.d.P. contributed equally to this study.