Abstract
Several gene-expression signatures predict survival in diffuse large B-cell lymphoma (DLBCL), but the lack of practical methods for genome-scale analysis has limited translation to clinical practice. We built and validated a simple model using one gene expressed by tumor cells and another expressed by host immune cells, assessing added prognostic value to the clinical International Prognostic Index (IPI). LIM domain only 2 (LMO2) was validated as an independent predictor of survival and the “germinal center B cell–like” subtype. Expression of tumor necrosis factor receptor superfamily member 9 (TNFRSF9) from the DLBCL microenvironment was the best gene in bivariate combination with LMO2. Study of TNFRSF9 tissue expression in 95 patients with DLBCL showed expression limited to infiltrating T cells. A model integrating these 2 genes was independent of “cell-of-origin” classification, “stromal signatures,” IPI, and added to the predictive power of the IPI. A composite score integrating these genes with IPI performed well in 3 independent cohorts of 545 DLBCL patients, as well as in a simple assay of routine formalin-fixed specimens from a new validation cohort of 147 patients with DLBCL. We conclude that the measurement of a single gene expressed by tumor cells (LMO2) and a single gene expressed by the immune microenvironment (TNFRSF9) powerfully predicts overall survival in patients with DLBCL.
Introduction
The most common subtype of non-Hodgkin lymphoma, diffuse large B-cell lymphoma (DLBCL), is clinically heterogeneous. Currently, standard regimens such as R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone) induce complete remission rates in DLBCL exceeding 75%.1,2 Nevertheless, current long-term event-free survival ranges from 50%-60%, and 30%-40% of patients eventually succumb to their disease.1-5 Predictive indices that capture such clinical heterogeneity might guide better therapeutic strategies. For example, molecular risk assignment could be used to predict responses to specific therapies6 or to allow risk-adapted stratification within clinical trials, thereby improving their statistical power.7
Traditional stratification schemes based on clinical characteristics such as the International Prognostic Index (IPI) have provided prognostic guidance in the management of patients with DLBCL.8 Despite the ease in its implementation, IPI does not fully represent disease heterogeneity.9 Therefore, efforts have shifted to broad molecular profiles that model and stratify risks of adverse outcomes. Using genome-scale expression profiles, we previously defined 2 distinct subtypes of DLBCL with different normal counterparts and clinical outcomes.10 With their distinct biologic and clinical features independently validated,10-14 these 2 DLBCL groupings, “germinal center B cell–like” (GCB-like) and “activated B cell–like” (ABC-like), are recognized as DLBCL subtypes in the current World Health Organization classification.15 However, the current lack of standardized methods for routine clinical use of expression profiles and the requirement for fresh or frozen tissues has limited their clinical utility.5,16,17
To develop a practical clinical risk tool, we examined associations between genome-wide expression profiles and outcomes at the single-gene level. Our goal was to construct prognostic models that integrate clinical and molecular indices in the current therapeutic era, and then to test and validate them in a simple assay amenable to routine clinical practice.
Methods
Study cohorts
Institutional review board approval was obtained from all participating institutions for inclusion of coded and de-identified patient data in this study. Gene-expression and clinical data were analyzed for 787 adult patients with DLBCL, including 2 cohorts treated with R-CHOP (DLBCL1 and DLBCL4) and 2 with CHOP (DLBCL2 and DLBCL3), as detailed in Table 1.12,18,19 These included 3 previously described multinational cohorts comprising 545 patients whose frozen tumors were profiled using microarrays (ie, DLBCL1-DLBCL3), and an independent cohort of 147 patients whose fixed, paraffin-embedded tumors were profiled using quantitative real-time PCR (DLBCL4). An additional 95 patients were assessed for cell-surface expression of CD137 by flow cytometry or immunohistochemistry, with biopsies obtained at Stanford University (Stanford, CA) or the Norwegian Radium Hospital (Oslo, Norway). Normal tonsils were obtained from routine tonsillectomies of 4 pediatric patients at Lucille Packard Children's Hospital (at Stanford University), and the peripheral blood of 22 healthy donors was used as a source of PBMCs. Additional details are described in Table 1 and in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Statistical methods
Univariate associations between expression profiles and survival were assessed by Cox regression using the coxph function from the R statistical software package. Differences between survival curves were assessed by the Kaplan-Meier product limit method. Statistical significance was measured using the log-likelihood statistic for continuous association between expression and outcome, and the log-rank method for discretely stratified patient groupings as described previously.20 Data from DLBCL1 were used to build survival models, with the DLBCL2-DLBCL4 cohorts used exclusively for validation. Bivariate combinations of genes with LIM domain only 2 (LMO2) were tested for their ability to predict survival using multivariate Cox regression. Detailed statistical and experimental methods are described in supplemental Methods.
Immunohistochemistry
Serial 4-μm sections were cut from formalin-fixed, paraffin-embedded (FFPE) specimens, deparaffinized in xylene, and hydrated in a series of graded alcohols. A mouse monoclonal anti-CD137 (clone BBK-2; Neomarkers) was used at a dilution of 1:15. Antigen retrieval by microwave pretreatment was performed in 10mM citric acid buffer at pH 6.0 for 10 minutes. Detection was performed using the EnVision system (Dako). Images of CD137 immunohistologic staining were acquired using a Nikon Eclipse E400 microscope and a Nikon digital camera (DS-L1), using 20×/0.50, 40×/0.75, or 60×/0.85 Plan Fluor objective lenses. Digitized images were processed using Adobe Photoshop CS3 image processing and manipulation software. Among 75 DLBCL cases assessed, all were negative for CD137 staining in tumor cells, with scattered infiltrating cells staining positively for CD137.
Flow cytometry
Tumor specimens were obtained at diagnosis and single-cell suspensions were prepared and frozen as described previously.21 Tonsils were similarly handled. PBMCs from healthy individuals (n = 22) were isolated using density gradient separation by Ficoll-Paque Plus (Amersham). Monoclonal antibodies used to stain human primary lymphoma specimens (n = 20), tonsils (n = 4), and PBMCs (n = 22) included: CD4 Pacific blue, CD8 FITC, CD20 APC-Cy7, CD25 PE, and CD45RO PE-Cy7 (all from BD Biosciences), CD3 QD605 (Invitrogen), and CD137 APC (clone 4B4-1; Biosource). Stained cells were detected on a FACSCalibur or an LSR II 3-laser cytometer (BD Biosciences) and analyzed using Cytobank (http://www.cytobank.org).
RNA isolation
For DLBCL4, using consecutive diagnostic fixed archival specimens, total RNA was extracted from two 5-μm-thick slices of FFPE tissue sections, as long as visual inspection found at least 3 mm of embedded tissue within the block, as described previously.19,22 Cases were not selectively excluded (or included) based on the degree of included normal tissue. Of the 137 patients described by Malumbres et al, a total of 98 (72%) had specimen remaining, allowing them to be studied again in the present study. This limitation was related to the small size of the original biopsies and to the prior use of the remaining specimens from the archival tissues for other studies. Quality and yield assessments were made for RNA and cDNA by optical spectra as measured by the OD260/280 ratio and size distribution on Agilent BioAnalyzer 2100 profile as we described previously.22 The yield depends on the quantity of the tissue in the paraffin block, and in the specimens used in this study ranged between 2 and 73.4 μg/sample (median, 8.4 μg).
Quantitative real-time PCR
Total RNA (2 μg) was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems).19 Real-time RT-PCR was performed using an ABI PRISM 7900HT (Applied Biosystems), as described previously.19,20,22,23 Briefly, commercially available Assays-on-Demand, consisting of a mix of unlabeled PCR primers and TaqMan minor groove binder probe (FAM dye-labeled) were used for measurement of expression of LMO2 (Hs00277106_m1) and tumor necrosis factor receptor superfamily member 9 (TNFRSF9; Hs00155512_m1). For endogenous control, we used phosphoglycerate kinase 1 (PGK1) with VIC-dye labeled Pre-Developed Assay Reagent (Applied Biosystems).20,23 PCR reactions were prepared in a final volume of 20 μL, with final concentrations of 1× TaqMan Universal PCR Master Mix (Applied Biosystems) and cDNA derived from 20 ng of input RNA. Thermal cycling conditions included an initial uracil-N-glycosylase incubation at 50°C for 2 minutes, AmpliTaq Gold DNA Polymerase activation at 95°C for 10 minutes, 40 cycles of denaturation at 95°C for 15 seconds, and annealing and extension at 60°C for 1 minute. Each measurement was performed in triplicate, with the results averaged.
Quantitative real-time PCR quality control and reproducibility
The fractional cycle number at which the amount of amplified target reached a fixed threshold (Ct) was determined as described previously. Replicate quantitative RT-PCR measurements were highly concordant, because the range of observed coefficients of variation for LMO2, TNFRSF9, and PGK1 was 0.01%-2.34%, 0.03%-1.92%, and 0.03%-1.64%, respectively. TNFRSF9 and LMO2 mRNA levels were normalized to PGK1 expression and calculated by the Δ-Ct method. For calibration, we used cDNA from the Raji lymphoma cell line (ATCC), obtaining Δ-Δ-Ct values for TNFRSF9 and LMO2 in each sample.22 As described previously,22 this method has high reproducibility on different FFPE sections, because repeated measurements from 3 different sections of the same tumor specimen resulted in R > 0.9 across 20 analyzed genes and also yielded high fidelity of corresponding measurements from fixed and frozen tissues. To assess the robustness of this assay for the 2 genes comprising the 2-gene score (TGS), we repeated various stages of the entire process for 147 patients with available FFPE specimens. Separate assays (including cDNA synthesis and PCR) performed by independent operators were highly concordant (for TNFRSF9, r = 0.99; for LMO2, r = 0.97; and for PGK1, r = 0.95), as were independent assays performed by the same operator (r = 0.97). No patients (0 of 98) had discordant low/high TGS-IPI risk groupings between independent assays, demonstrating the robustness of the method for measurement of these genes from FFPE archival specimens. To prevent erroneous input values, the online calculator for TGS and TGS-IPI was designed to provide users with feedback on whether their input Ct values and the corresponding calculated output results were within the observed ranges across the 4 described studies.
Results
LMO2 expression is a robust prognostic determinant in DLBCL
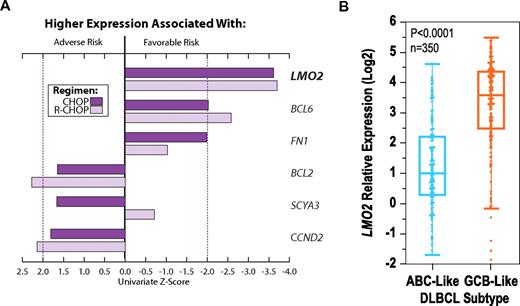
Among a previously described set of 6 genes for which expression is predictive of survival of patients with DLBCL independently of measurement platform,20,24 or use of frozen or fixed biopsy specimens,19 LMO2 was the single gene with the strongest independent prognostic value in distinct therapeutic eras (Figure 1A). By reevaluating univariate associations between overall survival (OS) and expression of nearly the entire transcriptome, LMO2 again emerged as exceptionally prognostic in publicly available data from 414 previously described patients treated by either CHOP or R-CHOP (Table 1 and supplemental Figure 1A).18
LMO2 has prognostic utility in different therapeutic eras independently of DLBCL subtype. (A) LMO2 expression was the best predictor of survival within a 6-gene model in patients treated with CHOP (n = 66)20 or R-CHOP (n = 132).19 Univariate z-scores from Cox regression reflect the magnitude of association for each gene with survival; positive and negative z-scores reflect association between higher expression of a given gene with adverse and favorable risk, respectively. Dotted lines correspond to P = .05. (B) LMO2 overexpression is associated with GCB subtype (P < .0001), yet can stratify outcomes even within this subtype (supplemental Table 1 and supplemental Figure 1D).
LMO2 has prognostic utility in different therapeutic eras independently of DLBCL subtype. (A) LMO2 expression was the best predictor of survival within a 6-gene model in patients treated with CHOP (n = 66)20 or R-CHOP (n = 132).19 Univariate z-scores from Cox regression reflect the magnitude of association for each gene with survival; positive and negative z-scores reflect association between higher expression of a given gene with adverse and favorable risk, respectively. Dotted lines correspond to P = .05. (B) LMO2 overexpression is associated with GCB subtype (P < .0001), yet can stratify outcomes even within this subtype (supplemental Table 1 and supplemental Figure 1D).
As exemplified by LMO2, the prognostic power of most genes tended to be significantly correlated in patients treated with CHOP or with R-CHOP (P < .0001; supplemental Figure 1A). In this cross-therapy analysis, the expression of LMO2 was the top-ranking survival predictor among genes distinguishing tumor cell of origin (Figure 1B and supplemental Figure 1A-C). Indeed, LMO2 expression was significantly higher among the GCB-like subtype (P < .0001; Figure 1B), was strongly correlated with expression of the “germinal center signature”11,18 (P < .0001; supplemental Figure 1B), and was effective as a diagnostic test for capturing tumor cell of origin (sensitivity, 85%; specificity, 73%; supplemental Figure 1C). Nonetheless, LMO2 expression added to cell-of-origin classification in stratifying outcomes in multivariate analyses, carrying prognostic value even among GCB-like DLBCL (supplemental Table 1 and supplemental Figure 1D). We therefore verified LMO2 expression as an exceptional predictor of both cell-of-origin classification and of survival in distinct therapeutic eras (supplemental Figure 1E-G), providing independent prognostic value irrespective of measurement methodology.20,25
TNFRSF9 expression is independently prognostic
Prior studies demonstrated significant prognostic influence derived from stromal signatures in diverse lymphomas.18,26,27 We aimed to capture this contribution by constructing a bivariate survival predictor, evaluating pairwise models including LMO2 and a second gene more highly expressed in nontumor cells. We assessed contribution from the microenvironment by comparing expression of paired sorted tumor (CD19+) and nontumor (CD19−) fractions from DLBCL tumors.18 Among genes more highly expressed in nontumor cells, TNFRSF9, encoding CD137 (also known as 4-1BB), was the best in bivariate combination with LMO2 (supplemental Table 2). Higher TNFRSF9 was also a strong univariate predictor of good outcomes in both therapeutic eras (supplemental Table 2, supplemental Figure 1A, and supplemental Figure 2A-C).
TNFRSF9 expression is limited to a tumor-infiltrating cell subset
We observed higher expression of TNFRSF9 mRNA on nontumor cells than on paired lymphoma B cells (P < .05; Figure 2A). Given its expression as a marker of activated T and NK cells that commonly infiltrate many tumors,28,29 we examined TNFRSF9 expression in publicly available gene-expression data across tumors from diverse histologies (n = 1822), and confirmed significantly higher expression among lymphomas than other tumor types (P < .0001; supplemental Figure 3A).30 Nonetheless, expression of TNFRSF9 similarly predicted clinical outcomes in many other diverse tumors, including adenocarcinomas of the breast, colon, and lung (P < .05; supplemental Figure 3B-D). Within DLBCL, we verified limited distribution of expression of the encoded protein using immunohistochemistry, finding CD137 exclusively within rare tumor-infiltrating cells (Figure 2B). Cell-surface immunophenotyping also demonstrated no significant CD137 expression on tumor cells (mean, 0.4%, range, 0%-1.6%; Figure 2C-D).
TNFRSF9 (CD137) expression is limited to a subset of the DLBCL tumor microenvironment. (A) Within sorted subpopulations, TNFRSF9 is more highly expressed in nontumor (CD19−) than tumor cells (CD19+).18 (B) Immunohistochemical analysis of CD137 expression among rare infiltrating cells in normal tonsil (40×, left) and a representative DLBCL from 75 tumors (200×, right). Insets show high magnification of individual positive infiltrating cells (600×). (C) Flow cytometric evaluation of CD137 expression in a representative DLBCL tumor showing lack of expression among tumor cells but detectable expression in a small proportion (5.9%) of CD4+ T cells and a larger proportion (28.6%) of CD8+ T cells. (D) Analysis of T cells in tonsils, PBMCs, and DLBCL tumors demonstrates CD137 expression on CD4+ and CD8+ T cells in the tumor microenvironment, but not on healthy lymphoid counterparts. (E) Fraction of CD137-expressing T cells is not correlated with total T-cell frequency in DLBCL tumors.
TNFRSF9 (CD137) expression is limited to a subset of the DLBCL tumor microenvironment. (A) Within sorted subpopulations, TNFRSF9 is more highly expressed in nontumor (CD19−) than tumor cells (CD19+).18 (B) Immunohistochemical analysis of CD137 expression among rare infiltrating cells in normal tonsil (40×, left) and a representative DLBCL from 75 tumors (200×, right). Insets show high magnification of individual positive infiltrating cells (600×). (C) Flow cytometric evaluation of CD137 expression in a representative DLBCL tumor showing lack of expression among tumor cells but detectable expression in a small proportion (5.9%) of CD4+ T cells and a larger proportion (28.6%) of CD8+ T cells. (D) Analysis of T cells in tonsils, PBMCs, and DLBCL tumors demonstrates CD137 expression on CD4+ and CD8+ T cells in the tumor microenvironment, but not on healthy lymphoid counterparts. (E) Fraction of CD137-expressing T cells is not correlated with total T-cell frequency in DLBCL tumors.
Among tumor-infiltrating T cells, significant but varying frequencies of CD8 and CD4 cells expressed CD137, mainly within a minor subset with a memory (CD45RO+) phenotype (Figure 2C). Indeed, this activated memory phenotype distinguished infiltrating T-cell subsets of DLBCL tumors from healthy lymphoid tissues (Figure 2D), suggesting unique interactions within the DLBCL microenvironment. TNFRSF9 expression was not simply a proxy for infiltrating T-cell frequency, because no significant correlation was observed between them (Figure 2E). Rather, variations in TNFRSF9 mRNA in DLBCL likely reflect the frequency of an activated T-cell subset (supplemental Figure 4).
A 2-gene model is a significant determinant of survival
We next examined the prognostic strength of the bivariate model combining expression of the tumor biomarker LMO225,31 with the microenvironment marker TNFRSF9, weighting the 2 genes based on their independent contributions from multivariate Cox regression in the training cohort (DLBCL1). We calculated TGS as follows: TGS = (−0.32 × LMO2) + (−0.29 × TNFRSF9). This score effectively predicted OS in patients treated with R-CHOP (P < .0001; Figure 3A-C). For each incremental unit rise of the TGS, there was a 2.7-fold (95% confidence interval [CI] 2.0-3.8) increase in relative risk of death. Tertiles of the TGS stratified patients with distinct outcomes (supplemental Figure 5), with corresponding 2-year OS rates of 56%, 77%, and 91% (Figure 3A). In multiple independent cohorts (DLBCL2-DLBCL4), we validated the model and previously defined thresholds from the training set (Figure 3F-G and supplemental Figure 9).
A 2-gene model is independently prognostic of survival in DLBCL and a composite model integrating IPI. (A) Performance of the TGS comprising expression of LMO2 and TNFRSF9 as evaluated in the training (DLBCL1) cohort. Tertiles of the TGS define 3 risk groups with distinct Kaplan-Meier estimates of survival (high risk, TGS > −0.91; intermediate risk, −0.91 ≥ TGS > −1.60; and low risk, TGS ≤ −1.60). TGS retains prognostic power within GCB (B) and ABC (C) subtypes of DLBCL treated with R-CHOP. Cases “unclassified” for cell of origin were excluded. (D) Kaplan-Meier estimates of strata using tertiles of a composite risk score integrating TGS and IPI (TGS-IPI). Depicted P values reflect log-likelihood estimates. (E) Distribution of the TGS-IPI and its relationship to survival in the training cohort, with survival modeled as a continuous function of the composite score. Tertiles of the TGS-IPI define strata are depicted in panel D, where means and 95% CI reflect Kaplan-Meier estimates at 2 years. Scores for individual patients are depicted as a “rug” above the x-axis. By applying thresholds derived from tertiles in the R-CHOP–treated training cohort (supplemental Figure 5), the TGS also stratifies OS of patients in 2 test cohorts treated with CHOP (DLBCL2, panel F, P < .0001, HR = 1.8 [1.4-2.3]; and DLBCL3, panel G, P = .0002, HR = 2.0 [1.4-2.9]).
A 2-gene model is independently prognostic of survival in DLBCL and a composite model integrating IPI. (A) Performance of the TGS comprising expression of LMO2 and TNFRSF9 as evaluated in the training (DLBCL1) cohort. Tertiles of the TGS define 3 risk groups with distinct Kaplan-Meier estimates of survival (high risk, TGS > −0.91; intermediate risk, −0.91 ≥ TGS > −1.60; and low risk, TGS ≤ −1.60). TGS retains prognostic power within GCB (B) and ABC (C) subtypes of DLBCL treated with R-CHOP. Cases “unclassified” for cell of origin were excluded. (D) Kaplan-Meier estimates of strata using tertiles of a composite risk score integrating TGS and IPI (TGS-IPI). Depicted P values reflect log-likelihood estimates. (E) Distribution of the TGS-IPI and its relationship to survival in the training cohort, with survival modeled as a continuous function of the composite score. Tertiles of the TGS-IPI define strata are depicted in panel D, where means and 95% CI reflect Kaplan-Meier estimates at 2 years. Scores for individual patients are depicted as a “rug” above the x-axis. By applying thresholds derived from tertiles in the R-CHOP–treated training cohort (supplemental Figure 5), the TGS also stratifies OS of patients in 2 test cohorts treated with CHOP (DLBCL2, panel F, P < .0001, HR = 1.8 [1.4-2.3]; and DLBCL3, panel G, P = .0002, HR = 2.0 [1.4-2.9]).
TGS carries independent prognostic value
When assessed within DLBCL subtypes, high TGS scores identified patients with relatively adverse outcomes among the more favorable GCB-like variant (Figure 3B). Conversely, within the less favorable ABC-like tumors, low scores identified patients with superior outcomes (Figure 3C). Within specific IPI risk groups, the TGS similarly identified patients with discordant outcomes in both therapeutic eras (supplemental Figure 7). In multivariate analyses, the TGS was also independent of the IPI and a “stromal” model comprising 381 genes integrating cell-of-origin classification (supplemental Table 3).18 Furthermore, the combination of the 2 genes compared favorably to other previously described models comprising 6 genes20 or 381 genes,18 both of which also included LMO2 but not TNFRSF9 (supplemental Figure 8).
A composite model integrating TGS with IPI
Given that the TGS added significantly to IPI, we constructed a combined model integrating both indices. We derived a composite score (TGS-IPI) for patients treated with R-CHOP in the training cohort, weighting the IPI score (on a 0-5 scale) and TGS based on their independent contributions from multivariate Cox regression as follows: TGS-IPI = (0.93 × TGS) + (0.6 × IPI) + 4. This composite score significantly outperformed the TGS or IPI alone in predicting survival in independent cohorts. Tertiles of the TGS-IPI also separated patients into 3 significantly different strata (Figure 3D-E). Patients in the corresponding risk groups—high risk, TGS-IPI > 4.51; intermediate risk, 4.51 ≥ TGS > 3.47; low risk, TGS ≤ 3.47—had estimated 2-year OS rates of 51%, 78%, and 95%, respectively. By applying the previously defined model and thresholds, these findings were validated in independent cohorts (supplemental Figure 6C). To visualize how these groups related to the distribution of continuous TGS-IPI, we modeled risk of death as a function of the TGS-IPI (Figure 3E).
External clinical validation of survival model
Whereas the TGS and TGS-IPI were validated in multiple cohorts when LMO2 and TNFRSF9 were measured using microarrays, this technique is not yet widely available in clinical laboratories. Therefore, we used quantitative real-time PCR to measure the expression of LMO2 and TNFRSF9 in diagnostic FFPE samples from an independent set of 147 patients with DLBCL treated with R-CHOP (DLBCL4). We also created a publicly available calculator to simplify estimation of risk using the TGS and TGS-IPI on fixed specimens (available at http://tgs.stanford.edu).
In univariate and multivariate analyses of this cohort, LMO2 and TNFRSF9 expression remained prognostic of both OS and progression-free survival (PFS; supplemental Figure 9A-D) and, when combined, the previously defined thresholds of the TGS and TGS-IPI stratified groups with distinct OS and PFS (Figure 4C-D and supplemental Figure 9E-F). Within this validation cohort, the TGS and IPI remained independent predictors of OS and PFS (supplemental Table 4). When combined as a continuous score, the TGS-IPI was predictive of both OS (P < .0001; hazard ratio [HR] 2.8; 95% CI 2.0-3.8) and PFS (P < .0001; HR 2.6; 95% CI 1.9-3.4).
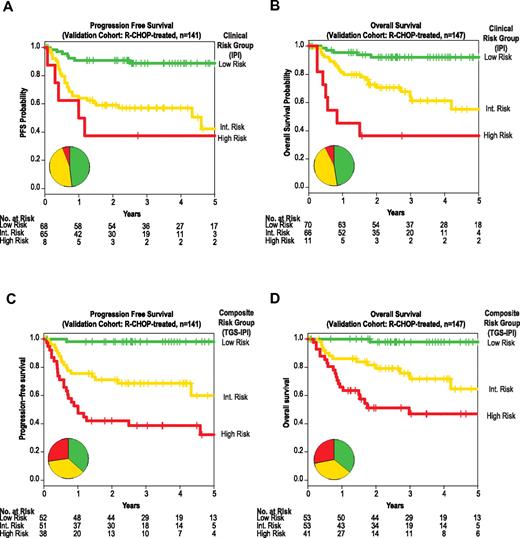
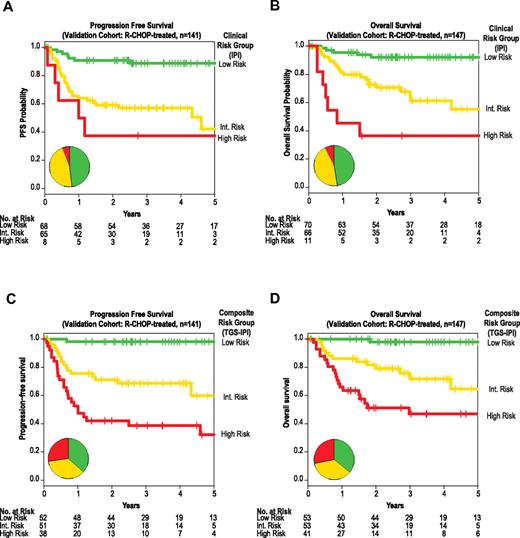
External validation of the composite predictor in fixed samples by quantitative real-time PCR. Kaplan-Meier estimates of PFS (panels A and C), and OS (panels B and D) for strata defined by the IPI (panels A and B) or by prospectively defined thresholds of a composite model integrating the TGS and IPI (TGS-IPI; panels C and D). The 3 TGS-IPI strata correspond to a priori–defined thresholds depicted in Figure 3E (high risk, TGS-IPI > 4.51; intermediate risk, 4.51 ≥ TGS > 3.47; and low risk, TGS ≤ 3.47). Depicted P values reflect log-likelihood estimates. All depicted associations were also significant by log-rank product limit tests of Kaplan-Meier strata, including between TGS-IPI and PFS (P < .0001, HR 3.9, 95% CI 2.4-6.3; panel C) and between TGS-IPI and OS (P < .0001, HR 3.7, 95% CI 2.2-6.2; panel D). Inset pie charts reflect distribution of risk groups as defined by the IPI or TGS-IPI, with color coding corresponding to Kaplan-Meier curves.
External validation of the composite predictor in fixed samples by quantitative real-time PCR. Kaplan-Meier estimates of PFS (panels A and C), and OS (panels B and D) for strata defined by the IPI (panels A and B) or by prospectively defined thresholds of a composite model integrating the TGS and IPI (TGS-IPI; panels C and D). The 3 TGS-IPI strata correspond to a priori–defined thresholds depicted in Figure 3E (high risk, TGS-IPI > 4.51; intermediate risk, 4.51 ≥ TGS > 3.47; and low risk, TGS ≤ 3.47). Depicted P values reflect log-likelihood estimates. All depicted associations were also significant by log-rank product limit tests of Kaplan-Meier strata, including between TGS-IPI and PFS (P < .0001, HR 3.9, 95% CI 2.4-6.3; panel C) and between TGS-IPI and OS (P < .0001, HR 3.7, 95% CI 2.2-6.2; panel D). Inset pie charts reflect distribution of risk groups as defined by the IPI or TGS-IPI, with color coding corresponding to Kaplan-Meier curves.
Stratification into low-, intermediate-, and high-risk groups using predefined TGS-IPI thresholds separated groups with distinct PFS and OS (Figure 4C-D). Patients in corresponding risk groups had estimated 2-year PFS rates of 98%, 71%, and 42%; the 2-year OS rates for these groups were 98%, 79%, and 51%, respectively (P < .001). For both PFS and OS, TGS-IPI captured a larger subset of patients at extremes of risk, particularly for those at high risk of death after R-CHOP chemotherapy (Figure 4).
Discussion
Despite significant scientific progress made possible by the Human Genome Project, corresponding advances in clinical medicine have been relatively modest.32,33 Current genome-scale studies have provided a rich source of molecular data that can be correlated with outcomes. Prior efforts to leverage transcriptome profiles have often used average representations of coregulated genes as composite signatures or “metagenes.”10-14,18,34 However, the need to measure many genes poses practical barriers to external validation and clinical application.20,35 Further, the requirement for unfixed diagnostic tissues has limited the clinical utility of such methods.16,17
In the case of DLBCL, variation in outcomes suggests that clinical features cannot fully account for underlying biologic heterogeneity. Molecular-profiling studies have attempted to capture this diversity, for example by defining subtypes relating to cell of origin, such as the GBC-like and ABC-like subtypes.10,15 Multivariate predictive models integrating these subtypes have been proposed, capturing additional features from the tumor microenvironment.11,14,18 Unfortunately, surrogate methods using fewer genes36,37 have had conflicting performance in their prognostic influence, and such methods are unable to classify 15%-50% of patients with DLBCL, limiting clinical utility.38
We used an alternative strategy, leveraging existing knowledge and available data to identify LMO2 and TNFRSF9 as 2 key genes whose expression each provides independent prognostic value. Expression of the LMO2 transcription factor is a marker for the GCB differentiation stage.31 Overexpression of LMO2 among nearly 10% of T-cell acute lymphoblastic leukemias can be ascribed to several recurrent genetic alterations,39 although no significant corresponding prognostic value has been observed in T-cell acute lymphoblastic leukemia.40 Nevertheless, overexpression of LMO2 is associated with induction and promotion of self-renewal within committed lymphocytes en route to leukemia in mice,41 and in humans,42 suggesting a similar role in lymphomagenesis.
TNFRSF9 encodes the costimulatory receptor CD137, with expression largely limited to activated T and natural killer cells, in which it plays important roles in immunologic memory formation and effector functions.28,29 Our observation of higher CD137 expression on a minor subset of infiltrating nonneoplastic cells of DLBCL tumors likely reflects unique interactions within the local microenvironment. Indeed, resting immune effector cells from the peripheral blood could be induced to express high levels of CD137 after contact with tumor cells, an effect that could be augmented by rituximab.43 We found no significant relationship between the expression of TNFRSF9 on infiltrating T cells and reciprocal regulatory molecules on tumor cells, including its ligand TNFSF9 or class II genes from the major histocompatibility loci, both of which have been implicated previously in lymphomas.11,44,45
Recent descriptions of stromal-1 and -2 signatures in DLBCL18 are associated with risk of favorable and adverse outcomes, respectively. The stromal-1 signature reflects vigorous extracellular-matrix deposition and infiltration by cells of the monocytic lineage and not infiltration by T cells, as we found for TNFRSF9 to be a marker of a distinct activated T-cell subset. The stromal-2 signature reflects endothelial cell density imparting a risk of adverse outcomes, not favorable ones, as we found for TNFRSF9. Among genes with higher expression in the tumor microenvironment, we found TNFRSF9 expression to be distinctly prognostic and, when combined with LMO2, obviated the need for measurement of cell-of-origin or other stromal signatures. Nonetheless, the basis for observed variations in frequency of CD137-expressing infiltrating T cells in DLBCL is unclear.
Agonistic monoclonal antibodies against CD137 have potent immunoregulatory properties and can eradicate tumors in multiple preclinical models including lymphomas.30,46 Therapeutic targeting of this molecule is the subject of ongoing clinical trials. We identified TNFRSF9 expression as predictive of clinical outcomes in multiple diverse tumors, including adenocarcinomas of the breast, colon, and lung. Therefore, the biologic basis for variation in TNFRSF9 expression is likely to have a general basis across human tumors. The higher expression of CD137 observed among lymphomas compared with solid tumors may represent a unique therapeutic opportunity in this disease. In addition, CD137 expression on infiltrating immune cells in DLBCL might serve as a predictive biomarker for response to a monoclonal antibody, such as anti-CD137, that triggers effector antitumor functions. Because higher expression of CD137 confers a good prognosis for DLBCL patients, such therapeutic targeting with agonistic monoclonal antibodies could be a means of limiting chemotherapy with its attendant toxicities for this subgroup of patients.
Whereas the contribution of gene-expression signatures from the tumor microenvironment has previously been recognized as an important prognostic factor among diverse lymphomas,18,26,27 to our knowledge, this is the first such report for TNFRSF9. TNFRSF9 was not included on Lymphochip DNA microarrays used in earlier DLBCL-profiling studies.10,11 A more recent study21 from the Leukemia/Lymphoma Molecular Profiling Project also did not identify TNFRSF9 within the stromal signatures, perhaps because TNFRSF9 does not tightly cocluster with other genes in signatures that are associated with outcomes. Finally, the TGS and TGS-IPI are distinguished from prior prognostic models11,18,20,34 in that their weighting of component risk factors was both derived and validated in the current therapeutic era that includes rituximab.
The newly described bivariate model and a composite index integrating the IPI were reproducible within validation studies, including a simple assay using routinely obtained diagnostic pathologic FFPE specimens. Whereas IPI captures a significant portion of attributable risk for adverse outcomes even in the current era,5 the TGS carried independent value and obviated the need for more complex multigene indices. Both the TGS and the IPI added to each other within the TGS-IPI and, most significantly, captured a larger fraction of patients with adverse outcomes.
Although originally devised before the introduction of rituximab, the components of the IPI can be used to predict extremes of outcomes in the current therapeutic era.4,5 For example, patients with zero IPI risk factors have 4-year survivals estimated at 95% in the current era, but comprise only 10%-15% of unselected cohorts.4,47 In comparison, the TGS-IPI identified more than 30% of patients exhibiting similar outcomes after therapy with R-CHOP. Conversely, whereas patients with adverse risk as defined by TGS-IPI comprise at least 25% of the R-CHOP–treated patients, the corresponding IPI high-risk group captures only ∼ 10% of patients. Therefore, by capturing more patients at low and high risk for progression and death, this composite index allows better risk assignment than either the IPI or the TGS alone.
Because fewer than half of patients within the high TGS-IPI risk group are cured, novel strategies to improve their outcomes are urgently needed, incorporating additional therapies or alternative regimens to R-CHOP. Conversely, given their highly favorable risk profile, patients with good risk features are unlikely to benefit from therapies adding to R-CHOP. Similar strategies could be used to guide trials aiming to minimize toxicities from chemotherapy in this latter group.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful for the generosity and courage of patients and physicians who provided specimens, raw microarray data, and clinical covariates, including the Leukemia & Lymphoma Molecular Profiling Project, the Molecular Mechanisms in Malignant Lymphomas Network Project (MMMLNP), and the Stanford Lymphoma Biomarkers Consortium; to Shideh Chinchinian for data collection; to members of the Levy laboratory for helpful discussions; and to Shoshana Levy and Gilbert Chu for critical reading of the manuscript.
This work was supported by funding from the National Cancer Institute (P01CA34233 [R.L.], U54CA149145 and U56-CA112973 [S.K.P.], and CA109335 [I.S.L.]); by the Dwoskin Family, Bankhead-Coley, and Fidelity Foundations (I.S.L); by a Terry Fox Foundation New Frontiers Program Project Grant (019001 [R.D.G.]; and by the Fondation de France and the Association pour la Recherche sur le Cancer (R.H.). A.A.A. is a special fellow of the Leukemia & Lymphoma Society. R.L. is a clinical research professor of the American Cancer Society.
National Institutes of Health
Authorship
Contribution: A.A.A. and A.J.G. designed and performed the analyses and wrote the manuscript; A.J.A. performed the RT-PCR experiments; H.E.K., R.H., M.J.G., and J.H.M. conducted the flow cytometry experiments; J.B., I.S.L., R.L., R.H.A., and R.D.G. provided the patient samples and clinical data; S.Z. and Y.N. conducted immunohistochemical evaluations of the samples; C.L.L. implemented the project website; R.J.T. contributed statistical expertise; and A.A.A., R.L., I.S.L., and S.K.P. supervised the project and obtained the funding.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ash A. Alizadeh, MD, PhD, 269 Campus Dr, CCSR 1145c, Stanford, CA 94305-5151; e-mail: arasha@stanford.edu; or Ronald Levy, MD, 269 Campus Dr, CCSR 1105, Stanford, CA 94305-5151; e-mail: levy@stanford.edu.
References
Author notes
A.A.A. and A.J.G. contributed equally to this study.



![Figure 3. A 2-gene model is independently prognostic of survival in DLBCL and a composite model integrating IPI. (A) Performance of the TGS comprising expression of LMO2 and TNFRSF9 as evaluated in the training (DLBCL1) cohort. Tertiles of the TGS define 3 risk groups with distinct Kaplan-Meier estimates of survival (high risk, TGS > −0.91; intermediate risk, −0.91 ≥ TGS > −1.60; and low risk, TGS ≤ −1.60). TGS retains prognostic power within GCB (B) and ABC (C) subtypes of DLBCL treated with R-CHOP. Cases “unclassified” for cell of origin were excluded. (D) Kaplan-Meier estimates of strata using tertiles of a composite risk score integrating TGS and IPI (TGS-IPI). Depicted P values reflect log-likelihood estimates. (E) Distribution of the TGS-IPI and its relationship to survival in the training cohort, with survival modeled as a continuous function of the composite score. Tertiles of the TGS-IPI define strata are depicted in panel D, where means and 95% CI reflect Kaplan-Meier estimates at 2 years. Scores for individual patients are depicted as a “rug” above the x-axis. By applying thresholds derived from tertiles in the R-CHOP–treated training cohort (supplemental Figure 5), the TGS also stratifies OS of patients in 2 test cohorts treated with CHOP (DLBCL2, panel F, P < .0001, HR = 1.8 [1.4-2.3]; and DLBCL3, panel G, P = .0002, HR = 2.0 [1.4-2.9]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/5/10.1182_blood-2011-03-345272/4/m_zh89991175830003.jpeg?Expires=1766076373&Signature=AHrm09Y4Gc9isy1DbgE8MvwIDiSogvg7rtaDTYueAYQY1g3nSIgyclB-wd0gAxQTIo86r97gVm8LiYC22gIcpISOrJwnN2fD3C1rw5ujJB1b7CxRFJw5M4WwakP6D5oQ9TCHrPxM35a2B2OSgPdmhIAJYe8Mz1MX1Jkn2Hqbffl0N~r82H1u6qtqO5ov~dHMdeFUKPL7b1ApLFUWKNDSwPdvFAIF32yK5Dh8TzEwRbav1W3xoMuBSFspXC4hW8iDhTII6cwF77flrO1NwatwmxMWXOr46xgFUedlQZvKB-uZMStOBBaw5rjyTEf1b9EqG9Kd0fAIyu21-GIrNwRVSQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 3. A 2-gene model is independently prognostic of survival in DLBCL and a composite model integrating IPI. (A) Performance of the TGS comprising expression of LMO2 and TNFRSF9 as evaluated in the training (DLBCL1) cohort. Tertiles of the TGS define 3 risk groups with distinct Kaplan-Meier estimates of survival (high risk, TGS > −0.91; intermediate risk, −0.91 ≥ TGS > −1.60; and low risk, TGS ≤ −1.60). TGS retains prognostic power within GCB (B) and ABC (C) subtypes of DLBCL treated with R-CHOP. Cases “unclassified” for cell of origin were excluded. (D) Kaplan-Meier estimates of strata using tertiles of a composite risk score integrating TGS and IPI (TGS-IPI). Depicted P values reflect log-likelihood estimates. (E) Distribution of the TGS-IPI and its relationship to survival in the training cohort, with survival modeled as a continuous function of the composite score. Tertiles of the TGS-IPI define strata are depicted in panel D, where means and 95% CI reflect Kaplan-Meier estimates at 2 years. Scores for individual patients are depicted as a “rug” above the x-axis. By applying thresholds derived from tertiles in the R-CHOP–treated training cohort (supplemental Figure 5), the TGS also stratifies OS of patients in 2 test cohorts treated with CHOP (DLBCL2, panel F, P < .0001, HR = 1.8 [1.4-2.3]; and DLBCL3, panel G, P = .0002, HR = 2.0 [1.4-2.9]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/5/10.1182_blood-2011-03-345272/4/m_zh89991175830003.jpeg?Expires=1766076374&Signature=wDwur5E-QgHPEqS~DTUR9dvRY4FWTaa4DOoV1diHgW5-S-hafn-F5asEF--JoAaprj1xiw9h8Go2yDerTX4a~0OwKBnovug0Io~~e~rEhlLoZ3moW1K~fu6Zw-MEu4nZyIB3STnWmxrOKKKuDbuj5fGTnd0pDOwf7DPRJ814dFtVo0bCek2W4fZv3acT5NODTDJvOuHwxB8J1K3N7kXzVJ84k9A3H7fndRoyCZTsD5HIb7lhjMVlc6cGqpZWFov7zxr7ByvST6MfrVVGEX~2nDH0tQ7PX9AIHp3dWmVr8FTrHMn7idzQ-kbUIMl3xDqC2Xqhw7JVtCHm5N7Q3~fP9w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)