Abstract
The somatically acquired V600E mutation of the BRAF gene has been recently described as a molecular marker of hairy cell leukemia (HCL). We developed an allele-specific PCR for this mutation and studied 62 patients with HCL, 1 with HCL variant, 91 with splenic marginal zone lymphoma, 29 with Waldenström macroglobulinemia, and 57 with B-cell chronic lymphoproliferative disorders. The BRAF V600E mutation was detected in all HCL cases and in only 2 of the remaining 178 patients. These 2 subjects had B-cell chronic lymphoproliferative disorders that did not fulfill the diagnostic criteria for HCL. Despite the positive PCR finding, the mutation could not be detected by Sanger sequencing in these 2 cases, suggesting that it was associated with a small subclone. We conclude that the BRAF V600E mutation is present in all patients with HCL and that, in combination with clinical and morphologic features, represents a reliable molecular marker for this condition.
Introduction
Hairy cell leukemia (HCL) is a distinct lymphoid neoplasm characterized by splenomegaly and peripheral cytopenias.1-4 Recently, Tiacci et al performed massively parallel sequencing of the whole exome of leukemic and matched normal cells an index HCL patient and identified 5 somatically acquired mutations, including an heterozygous V600E mutation in the BRAF gene. Via the use of targeted resequencing, they detected this mutation in 46 additional patients with HCL but in none of 193 cases of peripheral B-cell lymphoma or leukemia.5
The aforementioned observation is of paramount importance for a clear-cut diagnosis of HCL and also for its differential diagnosis from HCL-mimicking diseases such as splenic marginal-zone lymphoma (SMZL),6,7 splenic lymphoma/leukemia undifferentiated,8 and B-cell chronic lymphoproliferative disorders (B-CLPD).9,10
The aim of this study was to define the incidence of the BRAF V600E mutation in a large series of diverse lymphoid disorders. To this aim, we developed an allele-specific PCR for this mutation and studied a series of 240 mature B-cell lymphoid neoplasms, including 62 cases of HCL.
Methods
Patients
These investigations were approved by the Ethics Committee of the Fondazione IRCCS Policlinico San Matteo, Pavia, Italy, and other local institutional review boards. The procedures followed were in accordance with the Helsinki Declaration of 1975, as revised in 2000, and samples were obtained after subjects provided informed consent.
We studied 62 patients with HCL, 1 with HCL-variant (HCL-V), 91 with SMZL, 29 with Waldenström macroglobulinemia (WM), and 57 with B-CLPD. Diagnosis of HCL and HCL-V was made according to the World Health Organization 2008 classification1 ; no case analyzed in this study was included in the original report on BRAF mutations.5 The diagnosis of SMZL was decided on the basis of the World Health Organization's 2008 classification6 and on the criteria defined by Matutes et al.7 It was determined on the basis of spleen histology in 20 patients and on BM histology combined with immunohistochemistry and flow cytometry findings in 71 cases. The diagnosis of WM was made according to the consensus recommendations from the Second International Workshop on WM.11 Fifty-seven patients were diagnosed with B-CLPD in absence of specific phenotypic and/or genetic alterations (24 cases were CD5-positive, 33 CD5-negative, and 13 HCV-positive)9,10 ; in particular, a diagnosis of chronic lymphocytic leukemia or a leukemic phase of mantle cell lymphoma or follicular lymphoma was ruled out in each subject.
Histology and immunohistochemistry
BM biopsies were formalin-fixed, briefly decalcified (2 hours) with EDTA-HCl solution, and embedded in paraffin. Immunohistochemistry was performed by the streptavidin-biotin peroxidase technique, with enzymatic or microwave oven pretreatments when needed.
DNA extraction
In patients with HCL and in the HCL-V case, genomic DNA was extracted from BM biopsies by the use of the QuickGene DNA tissue kit S (DT-S; Life Science) on the FUJIFILM QuickGene-810 extraction platform (Fujifilm) according to the manufacturer's instructions.
Peripheral blood (PB) or BM mononuclear cells were separated by Ficoll-Hypaque gradient centrifugation. Genomic DNA was extracted from BM in 90 cases (33 SMZL, 29 WM, and 28 B-CLPD) and from PB in 88 cases (1 HCL, 58 SMZL, and 29 B-CLPD) using the Puregene Blood DNA isolation kit (QIAGEN) according to the manufacturer's recommendations. DNA concentration was quantitated by A260 absorbance with a BioPhotometer (Eppendorf). Genomic DNA (100 ng/sample) was used as template.
PCR amplification and sequencing of exon 15 of BRAF gene
BRAF gene exon 15 sequencing was performed to identify positive controls (c.T1860A) for allele-specific oligonucleotide (ASO) PCR. Two primers (forward, 5′-TACCTAAACTCTTCATAATGCTTGC-3′; reverse, 5′-GTAACTCAGCAGCATCTCAGGG-3′) were used to amplify a 256-bp fragment. PCR was performed in a final volume of 50 μL containing 1X reaction buffer, 0.2μM of each primer, 200μM dNTPs, 2mM MgCl2, and 2.5 U of HotStarTaq (QIAGEN). PCR consisted of an initial denaturation step of 15 minutes at 95°C, followed by 40 cycles of 94°C for 30 seconds, 59°C for 30 seconds, and 72°C for 60 seconds, with a final extension step of 10 minutes at 72°C.
The amplification product was separated on 3% agarose gel and visualized by SYBR safe staining (Invitrogen, Inc). The PCR products were purified and sequenced with the use of BigDye Terminator Version 3.1 Cycle Sequencing Kit and an ABI 3500 automatic sequencer (Applied Biosystems). Healthy donors were used as negative controls.
ASO-PCR assays
Two different forward primers with substitution of a single base at the end of the primer (forward wild-type 5′-TAGGTGATTTTGGTCTAGCTACCGT-3′ and forward mutated 5′-TAGGTGATTTTGGTCTAGCTACCGA-3′) were designed to amplify the wild-type allele or BRAF T1860A transversion mutation, respectively. To prevent the amplification of the nonmatching primer, an additional nucleotide mismatch (A > C) located 3 bases from the 3′ termini of the allele-specific primers was incorporated. The sequence of the reverse primer was the same as used before. Mutated or wild-type sequences from PB or BM samples were specifically amplified in a noncompetitive PCR as described previously. BM biopsies were amplified in a final volume of 50 μL containing 1μM of each primer, 200μM dNTPs, 1.5mM of MgCl2, and 1 U of AmpliTaq Gold DNA polymerase, with the reaction buffer supplied by the manufacturer (Applied Biosystems). PCR consisted of an initial denaturation step of 15 minutes at 95°C, followed by 38 cycles of 95°C for 40 seconds, 58°C for 60 seconds, and 72°C for 20 seconds, with a final extension step of 10 minutes at 72°C. All PCRs were performed in triplicate.
Results and discussion
Clinical features of the patients with HCL studied are reported in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The BRAF V600E mutation was found in all the 62 HCL cases studied. In 61 cases, DNA derived from BM biopsies with a hairy cell infiltration ranging from 15% to 95%, whereas in the remaining one, it derived from PB with 2% of hairy cells detected by flow cytometry immunophenotyping. Sanger sequencing in patients with heavily infiltrated BM showed an heterozygous pattern for the mutation. Detailed description of HCL samples is reported in supplemental Table 2.
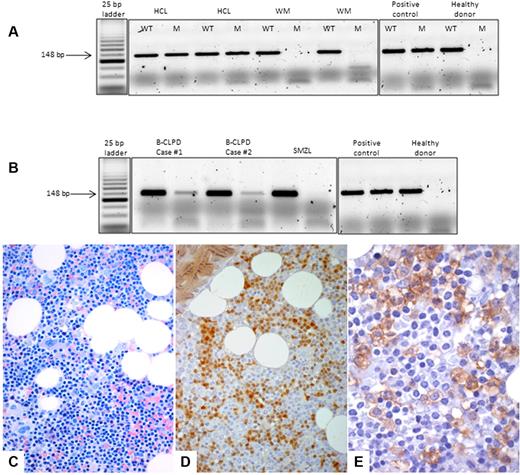
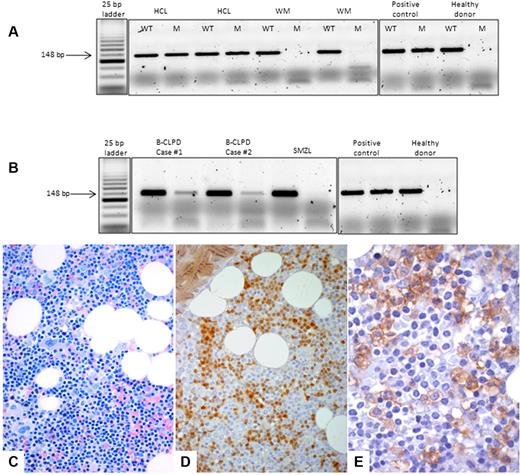
The BRAF V600E mutation was absent in the HCL-V case and in patients with SMZL or WM. Two patients with B-CLPD demonstrated a weak positivity in the BM (Figure 1) by ASO-PCR, but DNA Sanger sequencing did not reveal the mutation in these cases. Histologic and flow cytometry features of these 2 cases are summarized in Table 1.
ASO-PCR assay and bone marrow histology. Top, ASO-PCR assay. PCR products were separated on 3% agarose gel electrophoresis, and for each sample, the wild-type (WT) and the mutant (M) allele were amplified. (A) HCL samples presented the BRAF V600E mutation, whereas the MW cases did not present the mutant allele. (B) Although B-CLPD cases 1 and 2 presented both the wild-type and the mutant allele, the SMZL case did not present the BRAF V600E mutation. An HCL sample with an heterozygous BRAF V600E mutation was used as a positive control, and a healthy donor was used as a negative control. Bottom, BM histology of case 1. (C) On BM biopsy, an abundant (60%) lymphoid infiltrate was observed with interstitial pattern and composed by small lymphocyte-like, centrocyte-like cells, without the typical morphology of hairy cells (Giemsa staining, ×40). (D) By immunophenotyping, either on histologic sections (here) or by flow cytometry on BM aspirate (Table 1), the only similarity between this case and HCL was the expression of cyclin D1/bcl1 oncoprotein (streptavidin-biotic-peroxidase complex method/SABC, DAB chromogen, ×40). (E) ANXA-1, the most specific marker of HCL, was negative on the lymphoid population, and only was expressed by myeloid precursors, which serve as internal control (SABC, DAB chromogen, ×60).
ASO-PCR assay and bone marrow histology. Top, ASO-PCR assay. PCR products were separated on 3% agarose gel electrophoresis, and for each sample, the wild-type (WT) and the mutant (M) allele were amplified. (A) HCL samples presented the BRAF V600E mutation, whereas the MW cases did not present the mutant allele. (B) Although B-CLPD cases 1 and 2 presented both the wild-type and the mutant allele, the SMZL case did not present the BRAF V600E mutation. An HCL sample with an heterozygous BRAF V600E mutation was used as a positive control, and a healthy donor was used as a negative control. Bottom, BM histology of case 1. (C) On BM biopsy, an abundant (60%) lymphoid infiltrate was observed with interstitial pattern and composed by small lymphocyte-like, centrocyte-like cells, without the typical morphology of hairy cells (Giemsa staining, ×40). (D) By immunophenotyping, either on histologic sections (here) or by flow cytometry on BM aspirate (Table 1), the only similarity between this case and HCL was the expression of cyclin D1/bcl1 oncoprotein (streptavidin-biotic-peroxidase complex method/SABC, DAB chromogen, ×40). (E) ANXA-1, the most specific marker of HCL, was negative on the lymphoid population, and only was expressed by myeloid precursors, which serve as internal control (SABC, DAB chromogen, ×60).
Case 1.
In November 2008, a 41-year-old female patient presented with asymptomatic lymphocytosis without any evidence of lymphadenopathy or organomegaly. Laboratory data revealed the following results: hemoglobin 12.9 g/dL, white blood cell count 16 × 109/L (45% circulating clonal B-cells by flow cytometry) and platelets 283 × 109/L. On the BM biopsy (Figure 1), an interstitial lymphoid infiltrate (60% of the whole cellularity) composed by small, lymphocyte/centrocyte-like cells was found and. FISH for t(11;14), performed for cyclin D1 expression, was negative. Immunoglobulin rearrangement was IGHV3-48*02, IGHD7-27*01 IGHJ4*02. In this case, the BRAF V600E mutation also was confirmed in PB. At the last follow-up, lymphocytosis was stable and the patient did not have any need of treatment.
Case 2.
In 2006, a 62-year-old male patient presented with thrombocytopenia and splenomegaly, and a diagnosis of HCL was established in another Hospital. Treatment with cladribine produced only a partial response. We saw this patient in May 2008, when his spleen was palpable 3 cm under the costal margin. A blood cell count revealed hemoglobin 15.4 g/dL, white blood cell count 3.9 × 109/L with 14% circulating clonal B cells by flow cytometry immunophenotyping, and a platelet count of 95 × 109/L. A BM biopsy showed 20% lymphoid infiltrate with interstitial and sinusoidal pattern, composed of small-to medium-sized cells, with evident nucleoli, similar to prolymphocytes. Because this patient was asymptomatic, a watch-and-wait policy was adopted.
The BRAF V600E mutation has been previously found in a significant proportion of solid cancers12-14 and in Langerhans cell histiocytosis,15 whereas BRAF mutations other than V600E have been observed in small proportions of patients with acute lymphoblastic leukemia16,17 or B-cell lymphomas.18
The findings of this study indicate that the allele-specific PCR we developed is able to detect the BRAF V600E mutation in all patients with HCL. The same results have been recently obtained with the use of another PCR approach,19 or high-resolution melting analysis.20 The fact that we successfully analyzed DNA extracted from BM biopsies is of crucial importance because this most often represents the only available material containing hairy cells in patients with HCL.
Within diverse mature B-cell neoplasms, the BRAF V600E mutation was highly specific for HCL. In fact, the allele-specific PCR was positive in only 2 of 57 patients with B-CLPD, who did not fulfill the diagnostic criteria for HCL. Despite the PCR positivity, the mutation could not be detected by Sanger sequencing, suggesting that it was associated with a small subclone. The 2 patients had different features, and no conclusion can be drawn at present concerning the prevalence of BRAF V600E-positive clones in B-CLPD. We conclude that the BRAF V600E mutation is present in all patients with HCL and that, in combination with clinical and morphologic features, represents a reliable molecular marker for the laboratory diagnosis this mature B-cell neoplasm.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Giorgio Croci for technical support.
Authorship
Contribution: L.A. and M.C. designed the research; S.Z. developed the allele-specific PCR and did molecular investigations; E.B., M.L., and M.P. reviewed the histologic diagnosis; R.R. extracted DNA from BM biopsies; A.T. performed flow cytometry analysis; S. Rattotti, M.V., M.L.G., M.M., S. Rizzi, L.M., C.C., and M.C.D.V. collected clinical data; L.A. and M.C. wrote the paper; and all authors critically revised the manuscript and approved the version to be published.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Luca Arcaini, MD, Department of Hematology Oncology, Fondazione IRCCS Policlinico San Matteo, University of Pavia, 27100 Pavia, Italy; e-mail: luca.arcaini@unipv.it.