Abstract
Severe combined immunodeficiency (SCID) and X-linked agammaglobulinemia (XLA) are inborn errors of immune function that require prompt diagnosis and treatment to prevent life-threatening infections. The lack of functional T or B lymphocytes in these diseases serves as a diagnostic criterion and can be applied to neonatal screening. A robust triplex PCR method for quantitation of T-cell receptor excision circles (TRECs) and κ-deleting recombination excision circles (KRECs), using a single Guthrie card punch, was developed and validated in a cohort of 2560 anonymized newborn screening cards and in 49 original stored Guthrie cards from patients diagnosed with SCID, XLA, ataxia-telangiectasia, Nijmegen-breakage-syndrome, common variable immunodeficiency, immunoglobulin A deficiency, or X-linked hyper-IgMsyndrome. Simultaneous measurement of TREC and KREC copy numbers in Guthrie card samples readily identified patients with SCID, XLA, ataxia-telangiectasia and Nijmegen-breakage-syndrome and thus facilitates effective newborn screening for severe immunodeficiency syndromes characterized by the absence of T or B cells.
Introduction
Primary immunodeficiencies (PIDs) comprise a group of more than 200 different diseases.1 The clinical severity ranges from mild to potentially life-threatening. Major efforts are currently being undertaken to develop methods for detection of PIDs in the neonatal period. PCR-based detection of signal joint T-cell receptor excision circles (TRECs), extracted from Guthrie cards, has previously proven to be a valuable tool for identifying patients with severe combined immunodeficiencies (SCIDs).2-4 Recently, a similar method for analysis of κ-deleting excision circles (KRECs) was described, allowing identification of patients with X-linked agammaglobulinemia (XLA).5 The early identification of XLA patients is highly desirable, because the incidence of chronic lung disease in these patients results from delayed diagnosis and is one of the most detrimental factors on the prognosis and quality of life.6 To enable simultaneous screening of T- and B-cell deficiencies, we combined these assays into a novel triplex PCR method and evaluated its potential clinical application.
Patients and methods
Neonatal Guthrie card samples
Single 3.2-mm dried blood spot punches from 2560 freshly collected, anonymized Guthrie cards (903; GE Healthcare) and 28 stored original cards of patients diagnosed with SCID (n = 18), XLA (n = 4), ataxia-telangiectasia (AT; n = 4), or Nijmegen-breakage-syndrome (NBS; n = 2) were included. In addition, Guthrie card samples from patients with common variable immunodeficiency (CVID; n = 4), immunoglobulin A deficiency (IgAD; n = 15), or X-chromosome-linked hyper-IgM-syndrome (X-HIGM, CD40L defect; n = 2) served as disease controls. All the included patients were retrospectively identified based on diagnoses made and none refused allocation of the stored original neonatal screening card. Dried blood spot samples were prepared within the first 72 hours after birth. The KREC level of 1 of the XLA patients has been described in our previous publication.5
DNA elution from dried blood spot punches
On the approval of the institutional review board at the Karolinska University Hospital Huddinge, DNA from a single 3.2-mm punch of the dried blood disks was eluted into 24 μL of Generation DNA Elution Solution (QIAGEN) supplemented with 100 μg/mL yeast tRNA (Ambion), and 8 μL was subjected to real-time quantitative PCR (RT-qPCR) of TRECs, KRECs, and β-actin (ACTB).3 TREC and KREC copy numbers were normalized per microliter of blood, assuming that a 3.2-mm punch contains ∼ 3 μL of whole blood. The triplex RT-qPCR assay was optimized by primer limitation and probe concentration with primers and probes specific for the signal joint of TRECs and KRECs or for ACTB to ensure equal amplification efficacies.3,7 ACTB amplification was used to assess the success of DNA extraction from the Guthrie cards.
Real-time quantitative triplex PCR
The RT-qPCR reactions were performed in a final volume of 20 μL containing 1× TaqMan Gene Expression Master Mix, 20μM TREC primers, 25μM KREC primers, 7.5μM ACTB primers, 15μM 6FAM-labeled MGB TREC- and NED-labeled MGB ACTB probes, 17.5μM VIC-labeled MGB KREC probe (all from Applied Biosystems), and 0.8 μL of 10 mg/mL BSA (New England Biolabs). The 96-well plate reactions were carried out on ABI 7500 and ViiA7 real-time PCR systems (Applied Biosystems), with an initial cycle at 50°C for 2 minutes and a heating cycle at 95°C for 10 minutes, followed by 45 cycles of 30 seconds at 95°C and 30 seconds at 60°C. An individual cycle threshold for TREC, KREC, or ACTB was fixed for automated data collection and analysis of the amplification during the exponential phase. Calibration curves were generated by 10-fold serial dilution using a TREC-KREC-TRAC (TCRα subunit constant gene) construct containing plasmid and a β-actin sequence containing plasmid. All analyzed RT-qPCR assays fulfilled the quality requirements of similar slopes and R2 values > 0.97.
Sequence analysis of PID genes
Genomic DNA, eluted from the dried blood spot punches of previously identified SCID patients, was used to amplify the coding regions of the RAG1, RAG2, IL2RG, IL-7RA, or AK2 genes to verify mutations.8 Subsequent to gel electrophoresis, PCR products were purified (MSB Spin PCRapace; Stratec) and sent for direct sequencing (IZKF, University of Leipzig).
Results and discussion
Cutoff values for reliable identification of SCID and XLA patients
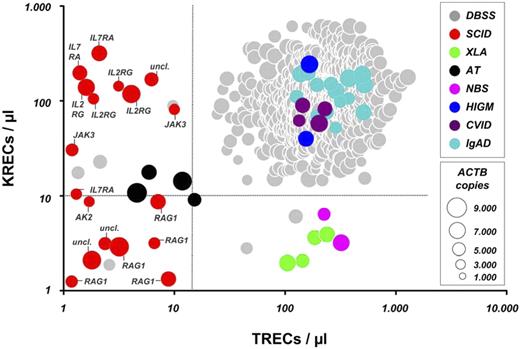
Based on the KREC and TREC copy numbers of the included SCID and XLA patients and repeat testing of all 2560 anonymized samples, suitable diagnostic cutoff scores were established at 15 TRECs/μL and 10 KRECs/μL. Given screening demands, cutoff scores were optimized to correctly identify the included samples from patients diagnosed with SCID or XLA with a sensitivity of 1.0 at expense of the specificity of the test (Figure 1). By using these cutoffs, the disease control samples from patients with CVID, IgAD, and X-HIGM, as expected, all fell within the normal range. However, because the diagnosis of these antibody deficiency syndromes is often delayed in childhood, leading to development of lung damage and lymphoid proliferative disease, and given that effective treatment strategies exist in pediatric patients, efforts are indicated to allow an early-as-possible diagnosis of these diseases.9,10 Of interest, all 4 patients with AT showed a marked reduction of both TRECs and KRECs, and both patients with NBS were identified based on solely low KREC copy numbers (Figure 1). The TREC and KREC levels on retesting of the AT and NBS patients and the available clinical and routine laboratory information are given in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). This implies that the method also might identify some patients with chromosome instability syndromes that are commonly not recognized before the onset of symptoms in early childhood. Repeat testing of all included PID patients, by using either the original DNA eluate or DNA from a second dried blood spot, provided consistent results (data not shown).
TREC and KREC copy numbers in dried blood spot samples (DBSS) from anonymized Guthrie cards and retested samples and in patients diagnosed with SCID, XLA, AT, NBS, X-HIGM, CVID, or IgAD. Dot size correlates with the amount of ACTB per sample. Dashed lines represent cutoff values for TRECs/μL and KRECs/μL, respectively. Proven molecular defects in the shown SCID patients are depicted as follows: RAG1 indicates recombination activating gene 1; IL2RG, interleukin 2 receptor γ chain (X-SCID); AK2, adenylate kinase 2; IL7RA, interleukin 7 receptor subunit α; JAK3, Janus kinase 3; and uncl., unclassified defect.
TREC and KREC copy numbers in dried blood spot samples (DBSS) from anonymized Guthrie cards and retested samples and in patients diagnosed with SCID, XLA, AT, NBS, X-HIGM, CVID, or IgAD. Dot size correlates with the amount of ACTB per sample. Dashed lines represent cutoff values for TRECs/μL and KRECs/μL, respectively. Proven molecular defects in the shown SCID patients are depicted as follows: RAG1 indicates recombination activating gene 1; IL2RG, interleukin 2 receptor γ chain (X-SCID); AK2, adenylate kinase 2; IL7RA, interleukin 7 receptor subunit α; JAK3, Janus kinase 3; and uncl., unclassified defect.
To assess the overall reproducibility of the assay, 160 duplicate control samples were analyzed on different PCR systems (ABI 7500 and ViiA7). Within the dynamic range of copy numbers with excellent linearity, the intra-assay coefficient of variation was 1.2% for TRECs/μL and 1.4% for KRECs/μL. The interassay variance coefficient of variation was 3.5% for TRECs/μL and 4.2% for KRECs/μL, indicating that our triplex RT-qPCR assay exhibits a robustness and reproducibility that meets diagnostic requirements.
Concordance of SCID immunophenotypes with TREC and KREC copy numbers
To conform with the diagnostic demands of neonatal screening, this study included only samples from the originally stored Guthrie cards of 18 SCID patients. All patients with confirmed mutations in the RAG1 gene demonstrated out-of-range values for both TRECs and KRECs, paralleling the T− B− (NK+) immunophenotype, and none of the SCID patients with copy numbers of more than 10 KRECs/μL was found to have RAG1 mutations (Figure 1).8 In patients with verified IL2RG mutations, KREC copy numbers were comparable with those of healthy newborns, reflecting the T− B+ (NK+) immunophenotype of X-SCID patients.8 In contrast, no mutations in the IL2RG gene were demonstrated in SCID patients with absent KRECs (Figure 1). Thus, our triplex PCR assay could be of value to guide the molecular diagnosis with regard to different types of SCID.
Diagnostic procedures for routine prospective Guthrie card analysis
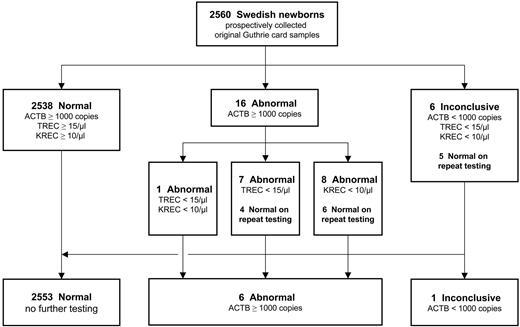
Guthrie card samples with ACTB copy numbers below 1000/μL and concomitant reduction of TRECs and KRECs were referred to as “inconclusive” because of a lack of DNA starting sample. Thus, a second punch was repeatedly tested (n = 6; 0.23% of total), resulting in normal findings for 5 of these samples, which is in the range of previous reports.3,4 Guthrie card samples with TREC or KREC copy numbers below the respective cutoff values were considered “abnormal” and likewise subjected to repeat testing. Seven such samples (0.27% of total) were retested because of low TREC numbers (< 15 TRECs/μL), 8 samples (0.31% of total) because of low KREC numbers (< 10 KRECs/μL), and 1 sample because of a combined reduction of both markers. The repeat testing of a second dried blood disk from the original cards yielded normal results for 10 samples (Figure 2). Because the Guthrie cards used in this study were anonymized, the underlying cause for the T or B lymphopenia in the remaining 6 samples (0.23% of total) is unknown but might be because of patients with the DiGeorge's syndrome (22q11 deletion syndrome), Trisomy 21, and others.11 Furthermore, the presence of congenital abnormalities and complications because of prematurity have been shown to increase the number of abnormal test results from excision circle assays, as will the detection of additional combined immunodeficiencies such as DOCK8 deficiency.12,13 This observation is likely to be expanded to isolated abnormal KREC copy numbers, as presented here for the included samples from NBS patients. The clinical significance of low KREC levels therefore deserves further investigation and will be the subject of 2 large-scale prospective studies in Sweden and Germany.
Flow chart of the triplex RT-qPCR assay, including results of 2560 freshly collected, anonymized regular Guthrie cards from Swedish newborns. Repeat testing was carried out using a second dried blood spot punch from the same Guthrie card.
Flow chart of the triplex RT-qPCR assay, including results of 2560 freshly collected, anonymized regular Guthrie cards from Swedish newborns. Repeat testing was carried out using a second dried blood spot punch from the same Guthrie card.
To ensure the diagnostic suitability of the retesting procedure, all Guthrie card samples with previously “normal” results were repeatedly tested using the original DNA eluate (data not shown). None of these samples reversed into either the abnormal or inconclusive category, indicating that the result of the analysis is inherently determined by the starting DNA sample and not by the triplex PCR method. In the light of the screening purpose of our method, and given that results in tested PID patients did not change on reanalysis, the repeat testing of a second Guthrie card disk is a reliable and meaningful procedure that is also warranted in view of the low percentage of abnormal and inconclusive samples (0.86% of total).
The observation that patients with the chromosome instability disorders AT or NBS could be detected, based on out-of-range levels for TRECs or KRECs in the original Guthrie cards, requires further considerations to conform with the tracking process of neonatal screening programs. Most importantly, these diseases should be taken into account on clinical assessment of potential patients, because features such as microcephaly or bird-like faces can be initial hallmarks.14,15 Although genotyping common founder mutations in NBS is a useful diagnostic approach, the detection of elevated α-fetoprotein serum levels or truncations of the ATM protein is more practicable in AT.16-18 The therapeutic perspective for AT and NBS patients has made considerable progress during recent years, providing evidence for successful stem cell transplantation in NBS and slowdown of neurodegeneration in AT using antioxidants and poly(ADP-ribose) polymerase inhibitors.19,20 Novel treatment strategies also might arise from the use of translational read-through compounds to correct the ATM gene function, whereas the substitution of immunoglobulin preparations is commonplace practice to treat the humoral immunodeficiency in NBS and to improve the clinical outcome.21
In summary, a triplex RT-qPCR measuring the levels of TRECs and KRECs provides a suitable screening for the majority of severe immunodeficiency diseases characterized by T or B lymphopenia in newborns.
There is an Inside Blood commentary on this article in this issue.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are indebted to John M. Routes and Mei W. Baker for helpful discussions and excellent technical guidance and support. They also thank Alessandra Sottini and Luisa Imberti for supplying a TREC-KREC-TRAC plasmid, and William J. Grossmann and Donna K. Mahnke for providing a β-actin sequence containing plasmid. They are grateful to Kerstin Krist for excellent technical skills in genomic sequence analysis.
This work was supported in part by the European Research Council (242551-ImmunoSwitch), the Swedish Research Council, the German National Academic Foundation (S.B.), the German Federal Ministry of Education and Research (BMBF, PtJ-Bio, 0315883), the Saxon State Ministry of Social Affairs (SMS), and the Jeffrey Modell Foundation (M.B. and L.H.).
Authorship
Contribution: S.B. performed and analyzed the research, created the figures, and wrote the paper; N.W. and M.J. performed experiments and analyzed data; U.v.D., A.F., J.W., and M.B. provided Guthrie card samples and were in charge for the diagnosis and treatment of involved PID patients; U.S. and Q.P.-H. designed the research; and L.H. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephan Borte or Lennart Hammarström, Division of Clinical Immunology F79, Karolinska Institutet at Karolinska University Hospital Huddinge, SE141-86 Stockholm, Sweden; e-mail: stephan.borte@ki.se or lennart.hammarstrom@ki.se.