Abstract
Type 2A VWD is characterized by the absence of large VWF multimers and decreased platelet-binding function. Historically, type 2A variants are subdivided into group 1, which have impaired assembly and secretion of VWF multimers, or group 2, which have normal secretion of VWF multimers and increased ADAMTS13 proteolysis. Type 2A VWD patients recruited through the T. S. Zimmerman Program for the Molecular and Clinical Biology of VWD study were characterized phenotypically and potential mutations identified in the VWF D2, D3, A1, and A2 domains. We examined type 2A variants and their interaction with WT-VWF through expression studies. We assessed secretion/intracellular retention, multimerization, regulated storage, and ADAMTS13 proteolysis. Whereas some variants fit into the traditional group 1 or 2 categories, others did not fall clearly into either category. We determined that loss of Weibel-Palade body formation is associated with markedly reduced secretion. Mutations involving cysteines were likely to cause abnormalities in multimer structure but not necessarily secretion. When coexpressed with wild-type VWF, type 2A variants negatively affected one or more mechanisms important for normal VWF processing. Type 2A VWD appears to result from a complex intersection of mechanisms that include: (1) intracellular retention or degradation of VWF, (2) defective multimerization, (3) loss of regulated storage, and (4) increased proteolysis by ADAMTS13.
Introduction
VWF is a multimeric plasma glycoprotein that is synthesized in endothelial cells and megakaryocytes and plays a critical role in the clotting process.1,2 VWF is synthesized as a 2813-aa pre-pro-VWF molecule containing a 22-aa signal peptide, a 741-aa propeptide, and a 2050-aa mature VWF protein.3 VWF undergoes extensive intracellular modifications.4 In the endoplasmic reticulum, the signal peptide is removed, disulfide bonds are formed, protein folding occurs, and the pro-VWF forms C-terminal dimers.5,6 In the Golgi, carbohydrate modifications occur, the C-terminal dimers form N-terminal multimers, and the VWF propeptide (VWFpp) is cleaved from the mature VWF by furin.7-10 VWF and VWFpp are then constitutively secreted or packaged into secretory granules—Weibel-Palade bodies in endothelial cells and α-granules in megakaryocytes and platelets.2,11,12 1-Deamino-8-D arginine vasopressin (DDAVP) can induce the release of VWF from these granules into plasma.13
VWF is subjected to extensive posttranslational modifications, and mutations in the VWF gene can affect its intracellular processing.14,15 For example, protein misfolding in the endoplasmic reticulum can lead to VWF retention and degradation or defective VWF multimerization can result in reduced platelet binding.14,16,17 The latter is manifested clinically as type 2A VWD, a qualitative deficiency of VWF characterized by the absence of high-molecular-weight VWF multimers with decreased platelet-dependent function.15 Type 2A VWD is the second most common type of VWD, comprising 10%-15% of cases, and inheritance is usually autosomal dominant.18,19 Phenotypically, type 2A VWD is characterized by a loss of high-molecular-weight VWF multimers and a disproportionate decrease in VWF ristocetin cofactor activity (VWF:RCo) activity relative to VWF antigen (VWF:Ag) levels.20 DDAVP may not be an effective treatment for type 2A VWD, so VWF replacement is usually required.21
Type 2A VWD traditionally has been subdivided into 2 groups according to the original description by Lyons et al22 : group 1 were those variants that had impaired assembly and secretion of VWF multimers and group 2 were those variants that had normal processing and secretion of VWF multimers, but increased susceptibility to proteolysis (now known to be because of cleavage by ADAMTS13, a disintegrin and metalloprotease with a thrombospondin type 1 motif) as it is secreted into plasma.23 However, 2A variants may not clearly fit into one group or another. For example, it has been shown that several group 1 variants have increased ADAMTS13 proteolysis.24 In the present study, 13 mutations spanning the D3, A1, and A2 domains of VWF were identified in type 2A VWD patients recruited through the T. S. Zimmerman Program for the Molecular and Clinical Biology of VWD (ZPMCB-VWD). We examined the homozygous and heterozygous expression of these variants and report herein that type 2A VWD appears to result from the complex intersection of 4 mechanisms: (1) intracellular retention and degradation, (2) defective multimerization, (3) loss of regulated storage, and (4) increased proteolysis by ADAMTS13, and that these type 2A variants can have a negative impact on mechanisms critical for VWF processing and secretion.
Methods
Patients
Subjects with a diagnosis of type 2A VWD were enrolled in the ZPMCB-VWD study. The study was approved by the human research review board at each institution and informed consent was obtained from all patients in accordance with the Declaration of Helsinki. Plasma VWF:Ag, VWF:RCo, VWF multimers, and FVIII levels were determined by the Hemostasis Reference Laboratory at BloodCenter of Wisconsin (Milwaukee, WI) as described previously.25 VWF:Ag was measured using an ELISA-based assay with the mAbs AVW-1 and AVW-5 (Blood Research Institute, Milwaukee, WI) for VWF capture and a polyclonal Ab for VWF detection (Dako). VWF:RCo analysis was performed using the automated BCS system (Dade-Behring) to measure platelet agglutination at a ristocetin concentration of 1 mg/mL. All values were determined using an international plasma standard (SSC/ISTH standard lot number 3). FVIII levels were determined using the BCS analyzer. VWF multimer structure was determined by LiDS-agarose electrophoresis (0.65%), followed by transfer to nitrocellulose and detection using a mAb to VWF and then HRP-conjugated Ab for chemiluminescent autoradiography.
Mutations
Genomic DNA PCR amplification and direct sequencing was performed on all 52 VWF exons, 5′/3′-untranslated regions, and intron/exon boundaries at the Partners Healthcare Center for Genetics and Genomics, Harvard Medical School (Boston, MA). Primer sequences are available on request.
Construction of expression plasmids
The plasmids untagged wild-type VWF (WT-pCIneo) and mycHis-tagged WT-VWF (WT-mycHis) containing WT human full-length VWF cDNA were described previously.26 To generate point mutations in human WT-pCIneo and WT-mycHis, the QuikChange II XL site-directed mutagenesis kit (Stratagene) was used according to the manufacturer's instructions. Primer sequences are available on request. Mutations were verified by sequencing.
Abs
Rabbit anti–c-Myc (Affinity BioReagents) recognizes mycHis-tagged VWF proteins. Monoclonal anti-VWF Abs AVW-1, AVW-3, AVW-5, AVW-15, and 105.4 were produced in the Hybridoma Core Laboratory at BloodCenter of Wisconsin. The epitope for AVW-5 lies on the C-terminal end of VWF and does not recognize VWF with a C-terminus mycHis tag. An anti-VWF polyclonal Ab was purchased from Dako.
Mammalian cell culture and transfection
Two human embryonic kidney cell lines were used in this study: HEK293T cells (David Ginsburg, University of Michigan, Ann Arbor, MI) to examine secretion and/or intracellular retention, and HEK293 cells (CRL 1573; ATCC) to examine regulated storage of VWF. HEK293 cells form Weibel-Palade body-like granules when transfected with VWF, whereas HEK293T cells do not. Both cell lines were cultured at 37°C in a 5% CO2 atmosphere in MEM (Mediatech) supplemented with 10% FBS (Invitrogen). Cells were transfected using TransIT-293 (Mirus) transfection reagent according to the manufacturer's instructions. Forty-eight hours after transfection, cell supernatants and lysates were collected for analysis. The HEK293 cell transfection was similar except that HEK293 cells were plated on single-well glass chamber slides and 48 hours after transfection, supernatants were removed and cells immunostained for confocal microscopy.
Cell lysis
Supernatants were removed from HEK293T cell cultures and the cells were washed with PBS. Reporter lysis buffer (Promega) was added to the cells and incubated at room temperature for 15 minutes. The cells then were scraped from the dish and the lysate was transferred to a tube, vortexed, and centrifuged at 4°C to remove cellular debris.
VWF ELISA
VWF protein concentration in HEK293T cell supernatants and lysates was determined by ELISA, as described previously27 with the following modification: multiple mAbs, AVW-1, AVW-3, AVW-5, and 105.4 at a total concentration of 5 μg/mL, were used to capture the VWF protein.
VWF functional assays
VWF binding to platelet glycoprotein GPIb was determined as described previously.28 Briefly, a mAb against human GPIbα was used to capture a gain-of-function recombinant GPIbα (tGPIbα235Y;239V). Recombinant VWF was then added in the absence of ristocetin. A mixture of biotinylated monoclonal anti-VWF Abs (AVW-1 and AVW-15) was used to detect the presence of VWF. Streptavidin-conjugated alkaline phosphatase and p-nitrophenyl phosphate was added and the optical density measured on an ELISA reader. VWF binding to collagen was assessed using human type III collagen at 1 μg/mL (Southern Biotech) for VWF capture and a polyclonal Ab for VWF detection (Dako).
Multimer analysis
VWF in the cell supernatants was analyzed by electrophoresis through a 2% high-gelling temperature agarose-resolving gel (Lonza) containing 1% SDS27 and transferred to Immobilon-FL (Millipore) by electroblotting at 100V for 1 hour in 25mM Tris, 200mM glycine, 20% methanol, and 0.03% (wt/vol) SDS. Membranes were blocked with Odyssey Blocking Buffer (LI-COR), incubated with anti-VWF polyclonal Ab (Dako) followed by IRDye 800–conjugated goat anti–rabbit IgG (LI-COR), and visualized with the Odyssey infrared imaging system (LI-COR). For coexpression studies, membranes were incubated with AVW-5 and rabbit-anti–c-Myc after blocking (Affinity BioReagents), followed by incubation with IRDye 680–conjugated goat anti–mouse IgG and IRDye 800–conjugated goat anti–rabbit IgG (LI-COR).
Immunofluorescent staining
Transfected HEK293 cells were examined for the intracellular localization of VWF with immunofluorescence Ab staining and the Leica TCS SP2 confocal laser scanning imaging system in the Imaging Core at the Medical College of Wisconsin or the Olympus FluoView FV1000 confocal laser scanning microscope in the Imaging Core at the Blood Research Institute of BloodCenter of Wisconsin.29 HEK293 cells were fixed, permeabilized, and blocked as described previously.27 Cells were incubated overnight in the primary Ab polyclonal anti-VWF (Dako), washed, and then incubated in secondary Ab, Alexa Fluor 488–conjugated goat anti–rabbit IgG (H+L [F(Ab')2]; Invitrogen), and then washed with PBS. For coexpression studies, cells were incubated with AVW-5 and rabbit anti–c-Myc (Affinity BioReagents), followed by PBS washes and incubation with Alexa Fluor 568–conjugated goat anti–mouse IgG (H+L) and Alexa Fluor 488–conjugated goat anti–rabbit IgG (H+L) [F(Ab')2]. Cells were coverslipped using Vectashield (Vector Laboratories). Slides were examined with either an HCXAPO 63× oil-immersion objective with a 0.90 numerical aperture on the Leica system or a UPLSAPO 60× water-immersion objective with a numerical aperture of 1.20 on the Olympus system at 22°C. Images were analyzed with LCS Version 2.0 (Leica Microsystems) or FV10-ASW Version 2.0a (Olympus) software for Leica system–acquired and Olympus system–acquired images, respectively.
ADAMTS13 assay
Full-length recombinant ADAMTS13 cDNA was expressed in HEK293T cells and ADAMTS13 protein was then purified from the conditioned medium using a His GraviTrap column (GE Healthcare). Samples were prepared in duplicate, with one the “positive” cleaved sample and the other the “negative” uncleaved sample. For each sample, 50 μL of recombinant ADAMTS13 diluted 1:2 in 5mM Tris, pH 8.0, was combined with 3 μL of Pefabloc SC [4-(2-aminoethyl)-benzene sulfonyl fluoride, hydrochloride] (Roche). To the “positive” samples, 8 μL of 200uM BaCl2 was added; to the “negative” samples, 8 μL of 200mM EDTA was added. Samples were incubated at 37°C to activate ADAMTS13 and then 175 μL of recombinant VWF protein was added. In an airtight container, MF-Millipore Membrane filters were floated on the surface of dialysis buffer (1.5M urea and 5mM Tris, pH 8.0) and the recombinant VWF/ADAMTS13 was transferred to the center of the filters and incubated at 37°C for 15 hours. The samples were removed from the top of the membrane and VWF multimers were analyzed as described in “Multimer analysis.”
Results
Characterization of patients with type 2A VWD
Type 2A VWD patients recruited through the ZPMCB VWD were identified phenotypically based on laboratory parameters (Table 1). The patients in this study had a preexisting diagnosis of type 2 VWD. The first 3 patients represent family members, with patient 1 as the index case, patient 2 the mother of index case (affected), and patient 3 the grandmother of the index case (unaffected). The remaining patients are unrelated index cases. Patients had reduced VWF:RCo and often reduced VWF:Ag. The VWF:RCo/VWF:Ag ratio was decreased and the multimer structure was abnormal (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), which is consistent with a diagnosis of type 2A VWD. VWFpp levels were determined for most patients. The VWFpp/VWF:Ag ratio was slightly to moderately elevated (> 2.0) in some subjects, which may be suggestive of reduced VWF survival.
Identification of mutations
By sequencing genomic DNA (ie, 52 VWF exons, 5′/3′-untranslated regions, and intron/exon boundaries), we identified 13 potentially causative, heterozygous mutations in the VWF gene in the type 2A patients. The changes in nucleotide sequence, predicted amino acid substitution, and domain/exon location are listed in Table 1. The 13 sequence variations spanned the D2, D3, A1, and A2 domains of VWF. Seven sequence variations had been reported previously (denoted by asterisks) and 6 were novel. The M740I and C1190S substitutions were identified in patient 1, whereas the patient's mother (patient 2) had M740I/C1190S/I1380V and the grandmother (patient 3) had only I1380V. All other patients had only 1 heterozygous amino acid substitution.
Expression of VWF mutations in mammalian cells
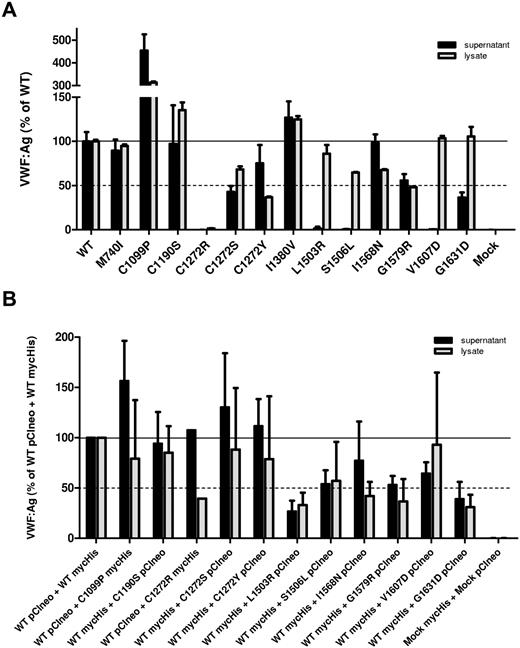
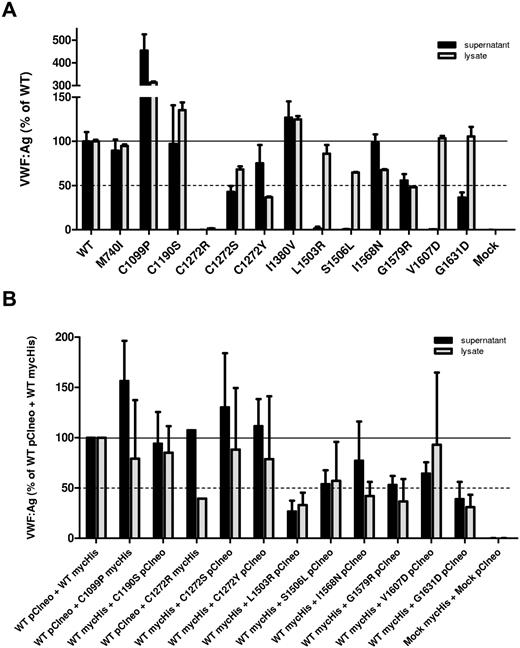
To characterize the effect of the identified mutations on VWF processing, regulated storage, and degradation, individual mutations were introduced into a full-length VWF expression vector. For M740I/C1190S, 3 expression vectors were constructed with each variation individually and 1 with both mutations. The I1380V variation was expressed separately because this was suspected to be a polymorphism on a separate allele. Culture supernatants and cell lysates were harvested from transfected HEK293T cells and the VWF concentration was assessed by ELISA (Figure 1A). Many of the 2A variants demonstrated normal secretion, exhibiting similar, if not higher, levels of VWF:Ag in the culture supernatant than WT-VWF. The variants L1503R, S1506L, and V1607D demonstrated severely impaired secretion with nearly undetectable levels of secreted VWF, although VWF was detected in the cell lysates. C1272S, G1579R, and G1631D had moderately reduced secretion. C1272R was not secreted and very little VWF was found in the cell lysate.
Secretion/intracellular retention of type 2A VWD variants. (A) Homozygous expression of variant VWF in HEK293T cells. (B) Heterozygous expression (1:1 ratio) of variant VWF and WT-VWF. The concentration of VWF in the conditioned medium (black bars) and cell lysate (gray bars) was assessed by VWF ELISA for each VWF variant and compared with WT-VWF. VWF:Ag levels are expressed as a percentage of WT. Each column represents the mean ± SD of at least 3 separate experiments. Secretion of several VWF variants is reduced markedly.
Secretion/intracellular retention of type 2A VWD variants. (A) Homozygous expression of variant VWF in HEK293T cells. (B) Heterozygous expression (1:1 ratio) of variant VWF and WT-VWF. The concentration of VWF in the conditioned medium (black bars) and cell lysate (gray bars) was assessed by VWF ELISA for each VWF variant and compared with WT-VWF. VWF:Ag levels are expressed as a percentage of WT. Each column represents the mean ± SD of at least 3 separate experiments. Secretion of several VWF variants is reduced markedly.
We also examined the secretion of variant VWF coexpressed with WT-VWF in a 1:1 ratio to recapitulate heterozygous patient expression. To identify each allele in subsequent multimer and confocal microscopy analysis, we expressed a mycHis-tagged VWF with untagged VWF. Total VWF in the culture supernatants and cell lysates was assessed by ELISA. WT pCIneo coexpressed with WT mycHis was used as a positive control, and heterozygously expressed variants were normalized to this control. Coexpression of L1503R, S1506L, G1579R and G1631D with WT-VWF appeared to inhibit the secretion of total VWF (Figure 1B). Coexpression with WT-VWF did not improve total VWF secretion of variants C1190S, L1503R, or G1631D; however, WT-VWF coexpression did appear to decrease the amount of VWF retained within the cell, suggesting a marginally improved interaction with intracellular chaperones.
Multimer structure of recombinant type 2A VWF variants
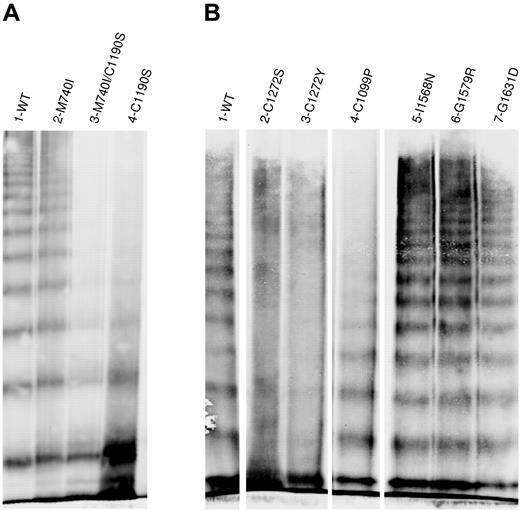
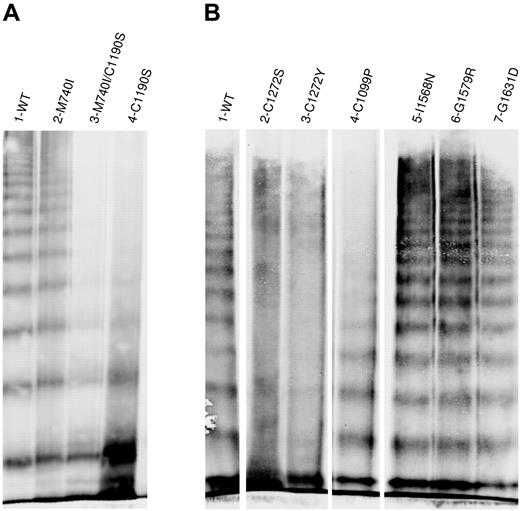
The multimer distribution of the recombinant “homozygously” expressed type 2A variants demonstrated a range of structural abnormalities (Figure 2). C1190S lacked mid- and high-molecular-weight multimers (Figure 2A). M740I had a normal multimer structure, whereas the multimer structure of M740I/C1190S appeared to be identical to C1190S alone, indicating that C1190S is the causative mutation in this patient. C1272S and C1272Y had “smeary” multimers, and C1099P lacked high-molecular-weight multimers (Figure 2B). I1380V (not shown), I1568N, G1579R, and G1631D were multimerized normally. Data for L1503R, S1506L, and V1607D are not shown because these variants lacked multimers due to severely impaired secretion. The normal secretion and multimer structure observed for M740I and I1380V suggests that they are benign polymorphisms.
Multimeric analysis of expressed VWF. VWF in the conditioned medium of transiently transfected HEK293T cells was analyzed by electrophoresis in 2% SDS-agarose gels and immunoblotted as described in “Multimer analysis.” Panels A and B represent 2 individual gels. In panel B, lanes were removed for clarity as shown by the separation. (A) Multimeric structure of WT-VWF (lane 1), M740I-VWF (lane 2), M740I/C1190S-VWF (lane 3), and C1190S-VWF (lane 4). (B) Multimeric composition of WT-VWF (lane 1); cysteine mutations C1272S-VWF, C1272Y-VWF, and C1099P-VWF (lanes 2-5); I1568N-VWF (lane 6); G1579R-VWF (lane 7); and G1631D-VWF (lane 8). VWF variants involving mutation of cysteines results in abnormal multimer structure.
Multimeric analysis of expressed VWF. VWF in the conditioned medium of transiently transfected HEK293T cells was analyzed by electrophoresis in 2% SDS-agarose gels and immunoblotted as described in “Multimer analysis.” Panels A and B represent 2 individual gels. In panel B, lanes were removed for clarity as shown by the separation. (A) Multimeric structure of WT-VWF (lane 1), M740I-VWF (lane 2), M740I/C1190S-VWF (lane 3), and C1190S-VWF (lane 4). (B) Multimeric composition of WT-VWF (lane 1); cysteine mutations C1272S-VWF, C1272Y-VWF, and C1099P-VWF (lanes 2-5); I1568N-VWF (lane 6); G1579R-VWF (lane 7); and G1631D-VWF (lane 8). VWF variants involving mutation of cysteines results in abnormal multimer structure.
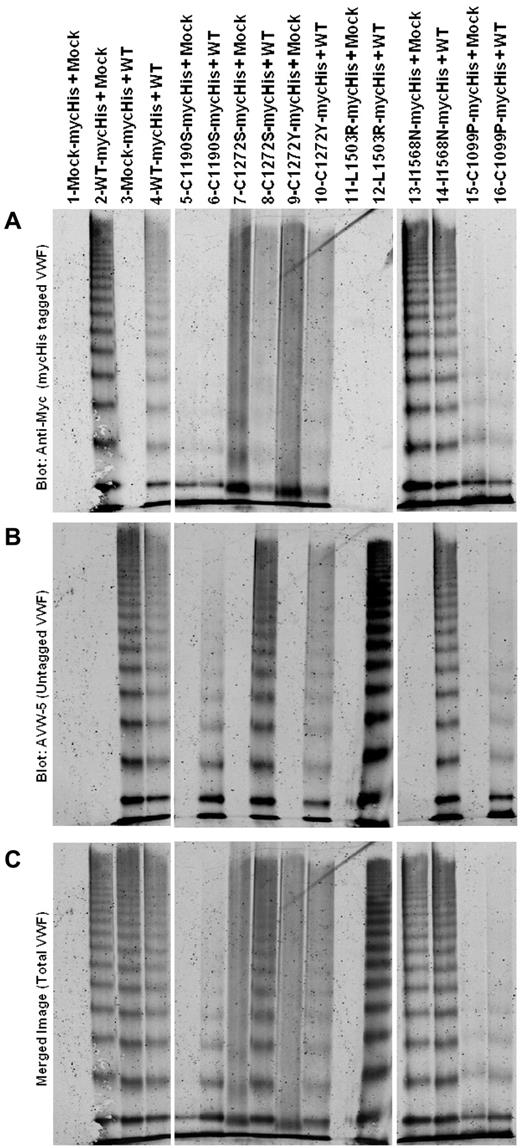
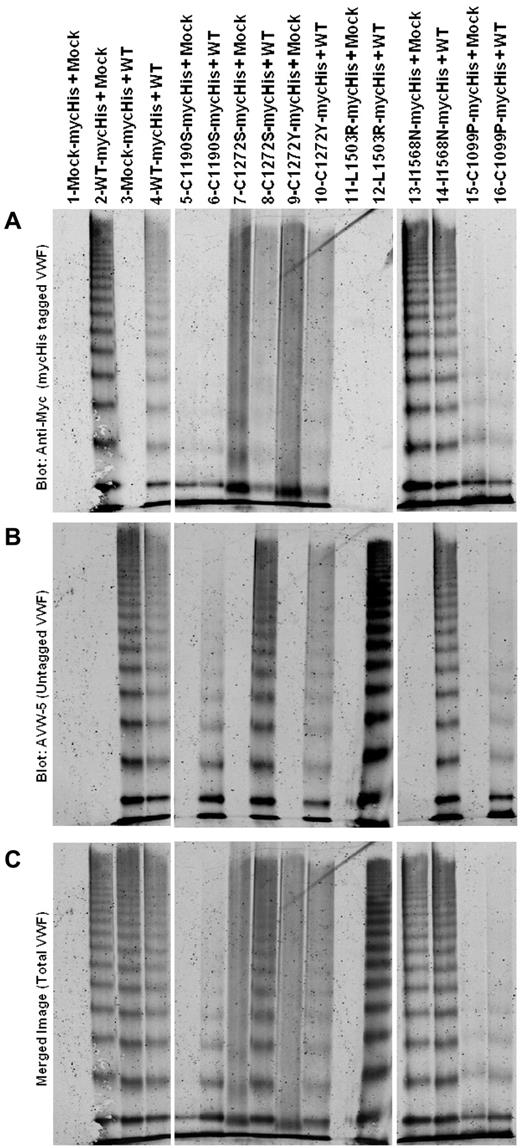
Multimer analysis of heterozygous expression of variants (1:1 ratio) is shown in Figure 3. Homozygous expression is also shown (variant + mock). Differential Ab recognition was used to identify each allele. Staining for mycHis-tagged VWF is shown in Figure 3 panel A, untagged VWF in panel B, and total VWF in panel C (a merge of the 2). AVW-5 has a C-terminal VWF epitope and does not recognize mycHis-tagged VWF (Figure 3B lane 2). Polyclonal anti-His does not identify untagged VWF (Figure 3A lane 3). Data are shown in pairs (Figure 3A lanes 5-16), with the first lane homozygous mycHis variant VWF (coexpressed with mock vector) and the second lane mycHis variant VWF coexpressed with WT-VWF. C1190S is not multimerized (Figure 3A lane 5) and coexpression with WT does not correct C1190S multimerization (Figure 3A lane 6). C1190S expression affects WT-VWF multimerization (Figure 3B lane 6) and results in an overall loss of high-molecular-weight VWF multimers (Figure 3C lane 6). C1272S and C1272Y both have “smeary” multimers (Figure 3A lanes 7 and 9), but only C1272S has a negative effect on WT-VWF multimerization (Figure 3B lanes 8 and 10) and total VWF, resulting in overall “smeary” multimers (Figure 3C lane 10). L1503R is not well secreted, and heterozygous expression at a 1:1 ratio appears to result mainly in the secretion of multimers from the WT allele (Figure 3A-C lanes 11 and 12). Similar results were obtained for S1506L and V1607D (not shown). Heterozygous I1568N expression does not have a negative effect on multimerization (Figure 3A-C lanes 13 and 14). Similar results were obtained with G1579R and G1631D (not shown). Like C1190S and C1272S, heterozygous expression of C1099P had a negative effect on total VWF multimerization (Figure 3A-C lanes 15 and 16). Multimerization deficient variants may have a negative effect on multimerization of coexpressed WT-VWF.
Multimeric analysis of heterozygous expression of type 2A VWF variants with WT-VWF. VWF variants with a mycHis tag were coexpressed 1:1 with WT-VWF (heterozygous) or with mock (homozygous). VWF in the conditioned medium of transiently transfected HEK293T cells was analyzed by electrophoresis in 2% SDS-agarose gels and immunoblotted as described in “Multimer analysis.” All samples were run on the same gel and lanes were removed for clarity, as denoted by the white space. (A) Immunoblot for the mycHis tag. (B) Immunoblot using Ab AVW-5 that does not recognize mycHis-tagged VWF. (C) Overlay of panels A and B for total VWF. Lane 1 is supernatant from mock-transfected cells. Lanes 2-4 show WT-VWF proteins with and without myc His tags to demonstrate Ab specificity. In lanes 5-16, data are shown in pairs, with one lane showing the mycHis-tagged type 2A variant cotransfected with mock (homozygous) or with untagged WT-VWF (heterozygous). Some type 2A variants have a negative impact on WT-VWF multimerization.
Multimeric analysis of heterozygous expression of type 2A VWF variants with WT-VWF. VWF variants with a mycHis tag were coexpressed 1:1 with WT-VWF (heterozygous) or with mock (homozygous). VWF in the conditioned medium of transiently transfected HEK293T cells was analyzed by electrophoresis in 2% SDS-agarose gels and immunoblotted as described in “Multimer analysis.” All samples were run on the same gel and lanes were removed for clarity, as denoted by the white space. (A) Immunoblot for the mycHis tag. (B) Immunoblot using Ab AVW-5 that does not recognize mycHis-tagged VWF. (C) Overlay of panels A and B for total VWF. Lane 1 is supernatant from mock-transfected cells. Lanes 2-4 show WT-VWF proteins with and without myc His tags to demonstrate Ab specificity. In lanes 5-16, data are shown in pairs, with one lane showing the mycHis-tagged type 2A variant cotransfected with mock (homozygous) or with untagged WT-VWF (heterozygous). Some type 2A variants have a negative impact on WT-VWF multimerization.
Formation of VWF-containing regulated storage granules
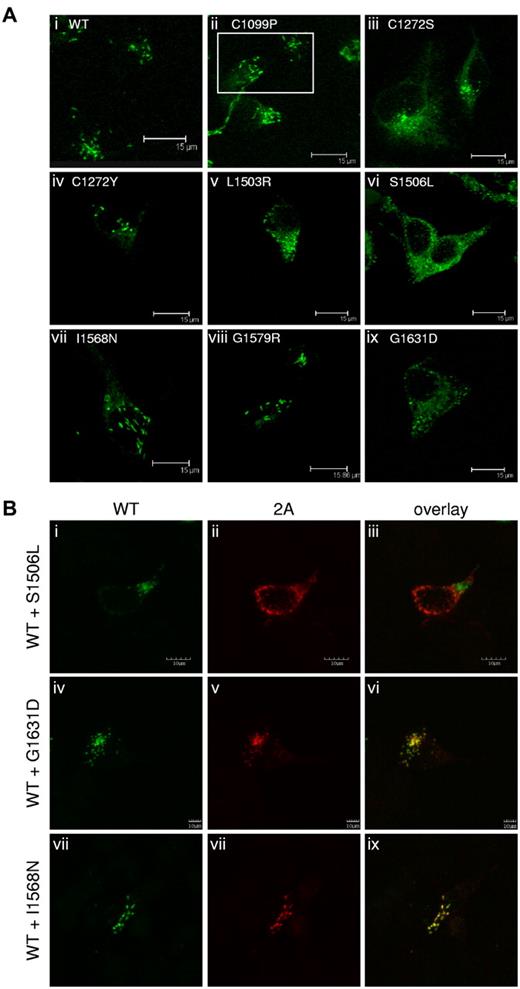
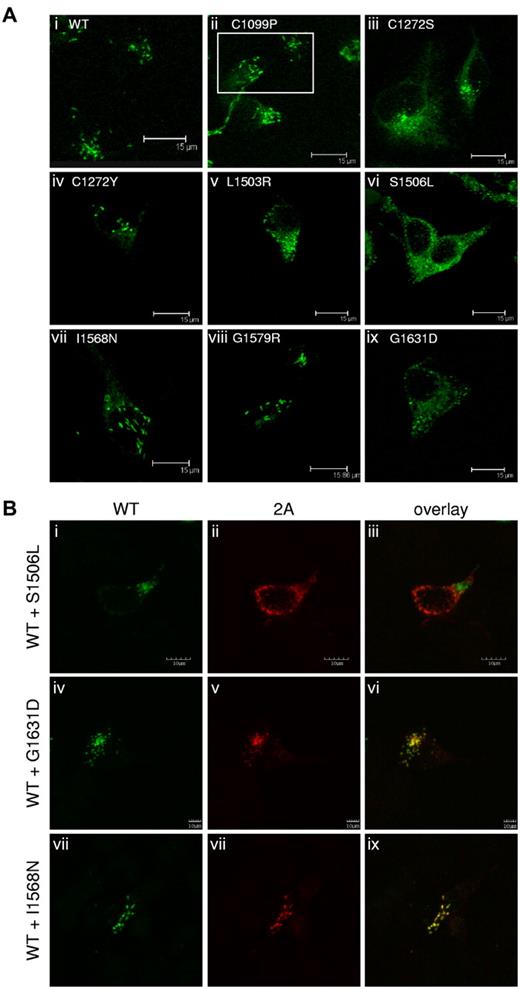
VWF is stored for regulated release in Weibel-Palade bodies of endothelial cells and in α-granules of platelets.2,12 VWF expressed in HEK293 cells can form Weibel-Palade body–like granules.29,30 The 2A variants were expressed in HEK293 cells, immunostained, and examined by confocal microscopy (Figure 4A). Only C1099P, C1190S (data not shown), C1272Y, I1568N, and G1579R (Figure 4Aii,iv,vii,viii) formed elongated, Weibel-Palade body–like storage granules similar to WT-VWF (Figure 4Ai). The other expressed 2A variants demonstrated substantially reduced or a complete loss of granular storage, as evidenced by the lack of elongated storage granules and a mostly diffuse staining pattern. Representative images are shown in Figure 4A subpanels iii, v, vi, and ix.
Regulated storage of type 2A variants in HEK293 cells. (A) Type 2A variants were expressed homozygously in HEK293 cells, immunostained, and examined by confocal microscopy, as described in “Immunofluorescent staining.” C1190S (not shown), C1099P, C1272Y, I1568N, and G1579R (ii, iv, vii, and viii, respectively) formed Weibel-Palade body–like storage granules, similar to WT-VWF (A). Variants C1272S, L1503R, S1506L,G1631D (iii, v, vi, and ix) and V1607D (not shown) demonstrated a loss of regulated storage, as indicated by the lack of storage granules and the diffuse staining pattern. (B) Type 2A variants were coexpressed in a 1:1 ratio with WT-VWF in HEK293 cells, immunostained, and examined by confocal microscopy, as described in “Immunofluorescent staining.” Cells expressing both WT-VWF and S1506L (i-iii) demonstrated a loss of Weibel-Palade body–like granules. Cells expressing WT-VWF and G1631D (iv-vi) formed less-elongated granules. WT-VWF storage was not disrupted when cotransfected with I1568N (vii-ix). Regulated storage was disrupted for some type 2A variants.
Regulated storage of type 2A variants in HEK293 cells. (A) Type 2A variants were expressed homozygously in HEK293 cells, immunostained, and examined by confocal microscopy, as described in “Immunofluorescent staining.” C1190S (not shown), C1099P, C1272Y, I1568N, and G1579R (ii, iv, vii, and viii, respectively) formed Weibel-Palade body–like storage granules, similar to WT-VWF (A). Variants C1272S, L1503R, S1506L,G1631D (iii, v, vi, and ix) and V1607D (not shown) demonstrated a loss of regulated storage, as indicated by the lack of storage granules and the diffuse staining pattern. (B) Type 2A variants were coexpressed in a 1:1 ratio with WT-VWF in HEK293 cells, immunostained, and examined by confocal microscopy, as described in “Immunofluorescent staining.” Cells expressing both WT-VWF and S1506L (i-iii) demonstrated a loss of Weibel-Palade body–like granules. Cells expressing WT-VWF and G1631D (iv-vi) formed less-elongated granules. WT-VWF storage was not disrupted when cotransfected with I1568N (vii-ix). Regulated storage was disrupted for some type 2A variants.
We also investigated the impact of heterozygosity on regulated storage. The 2A variants were coexpressed with c-Myc–tagged WT-VWF (Figure 4B) and stained with the anti-VWF Ab AVW-5 that does not recognize tagged-VWF (green) and anti–c-Myc Ab (red). C1272S, S1506L, V1607D, and L1503R inhibited the ability of WT-VWF to form storage granules. Confocal images for variant S1506L are shown as representative images for this phenotype (Figure 4B top row). G1631D interfered slightly with the regulated storage of WT-VWF (Figure 4B middle row). C1099P, C1190S, C1272Y, I1568N (Figure 4B bottom row), and G1579R did not disrupt the regulated storage of WT-VWF, and both the variant and WT-VWF appeared to colocalize to the same Weibel-Palade body–like granules. Variants associated with reduced secretion demonstrated reduced secretory granule formation and had a negative impact on the secretion and storage of coexpressed WT-VWF.
The administration of DDAVP results in the release of VWF from Weibel-Palade bodies into the plasma. DDAVP data were available for only 3 patients in this study (Table 2). All 3 patients responded to DDAVP, and this was correlated with expression data documenting the formation of Weibel-Palade body–like granules (C1190S, I1568N, and G1579R). The low VWF:RCo/VWF:Ag ratio after DDAVP administration in patients 2 and 11 suggests that the released VWF was not multimerized normally, it was quickly degraded by ADAMTS13, or a combination of these. Based on VWF:RCo alone, only patients 2 and 10 had a sufficient response to DDAVP.
Susceptibility of type 2A variants to cleavage by ADAMTS13
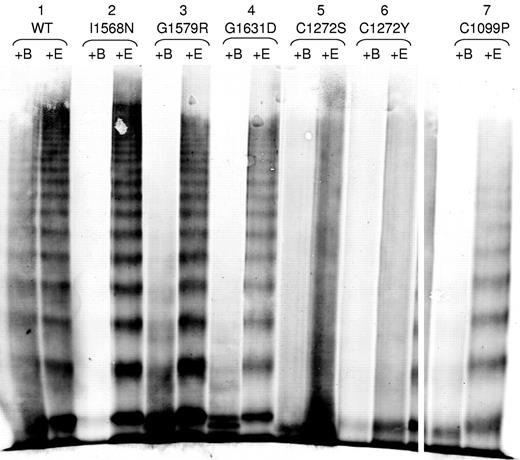
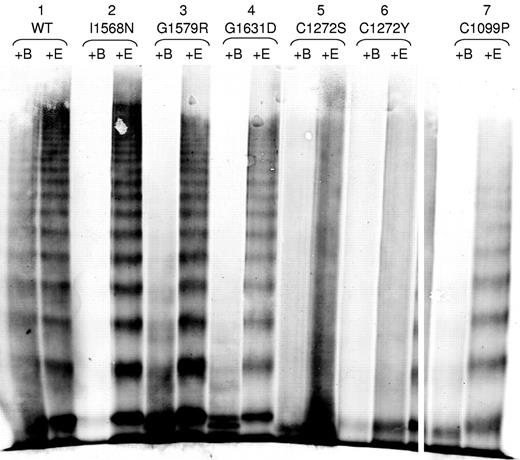
We investigated whether the 2A variants had increased susceptibility to ADAMTS13 proteolysis (Figure 5). BaCl2 is required for the ADAMTS13 cleavage of VWF (+B) and EDTA inhibits the ADAMTS13 activity (+E). I1568N, G1579R, G1631D, and C1099P (Figure 5) had increased susceptibility to cleavage by ADAMTS13 compared with the WT-VWF control, as demonstrated by the substantial loss of high molecular weight multimers after exposure to ADAMTS13 compared with the +EDTA control. C1272S and C1272Y also may have increased susceptibility to proteolysis by ADAMTS13, although this is difficult to assess given the lack of a clear multimer banding pattern (Figure 5). Data are not shown for L1503R, S1506L, and V1607D because they were not secreted well. A subset of type 2A variants demonstrated increased susceptibility to ADAMTS13 proteolysis.
ADAMTS13 degradation of type 2A variant VWF proteins. VWF in the conditioned medium of transiently transfected HEK293T cells was digested with rhuADAMTS13 and analyzed by electrophoresis in 2% SDS-agarose gels and immunoblotted as described in “Multimer analysis.” All samples were run on the same gel and lanes were removed for clarity, as denoted by white space. Each variant was digested with BaCl2 to activate the rhuADAMTS13 (+B), or in the presence of EDTA to prevent activation of huADAMTS13 (+E). Sample set 1 shows WT-VWF, which is cleaved minimally by rhuADAMTS13. Sample sets 2-4 show type 2A mutations that displayed a full range of VWF multimers (I1568N, G1579R, and G1631D) and had increased susceptibility to cleavage by rhuADAMTS13. Sample sets 5-7 show type 2A VWD mutations involving cysteines (C1272S, C1272Y, and C1099P). C1272S and C1272Y may have increased proteolysis, whereas C1099P does have increased proteolysis.
ADAMTS13 degradation of type 2A variant VWF proteins. VWF in the conditioned medium of transiently transfected HEK293T cells was digested with rhuADAMTS13 and analyzed by electrophoresis in 2% SDS-agarose gels and immunoblotted as described in “Multimer analysis.” All samples were run on the same gel and lanes were removed for clarity, as denoted by white space. Each variant was digested with BaCl2 to activate the rhuADAMTS13 (+B), or in the presence of EDTA to prevent activation of huADAMTS13 (+E). Sample set 1 shows WT-VWF, which is cleaved minimally by rhuADAMTS13. Sample sets 2-4 show type 2A mutations that displayed a full range of VWF multimers (I1568N, G1579R, and G1631D) and had increased susceptibility to cleavage by rhuADAMTS13. Sample sets 5-7 show type 2A VWD mutations involving cysteines (C1272S, C1272Y, and C1099P). C1272S and C1272Y may have increased proteolysis, whereas C1099P does have increased proteolysis.
Functional characterization of recombinant VWF variants
Functional analysis of VWD variants expressed under homozygous and heterozygous conditions was assessed by collagen binding and platelet GPIb-binding assays, as summarized in Table 3. Variants with abnormalities in multimer structure demonstrated reduced binding to both collagen and GPIb (C1190S, C1099P, and C1272S/Y), whereas variants with normal multimer structure had normal binding. Variants with normal multimers and increased susceptibility to ADAMTS13 degradation had essentially normal binding function, as would be expected because they had not had exposure to ADAMTS13 under tissue-culture conditions. Heterozygous expression of suspected group 1 variants (L1503R, S1506L, and V1607D) appeared to result in the secretion of mainly WT-VWF, which had normal binding function. With the exception of the suspected group 2 variants, most variants reflected patient plasma functional abnormalities.
Classification of type 2A variants
A summary of data derived from homozygous and heterozygous expression studies can be found in Table 4. Based on the current subgroup classification for type 2A, variants C1272R, L1503R, S1506L, and V1607V fit the criteria for group 1, having substantially reduced secretion and a lack of mid- and high-molecular-weight multimers. Variants I1568N, G1579R, and G1631D could be tentatively classified as group 2 variants with increased susceptibility to ADAMTS13 degradation, although variant G1631D also had moderately reduced secretion and reduced Weibel-Palade body formation, leading us to label this variant as “unclassified.” The remaining variants, C1099P, C1190S, C1272Y, and C1272S, do not fit clearly into either group, but would be best labeled “unclassified.”
Discussion
In the present study, we identified 13 potentially causative sequence variations in patients with type 2A VWD and examined the homozygous and heterozygous expression of these variants. Our results showed that: (1) some variants fit into the traditional group 1 or 2 categories, whereas others did not clearly fall into either category; (2) substantially reduced VWF secretion was associated with a loss of Weibel-Palade body formation; (3) type 2A group 2 variants were associated with VWF A2 domain mutations and normally regulated storage; (4) mutations involving cysteines may or may not affect VWF secretion, but are likely to cause abnormalities in multimer structure; and (5) when coexpressed with WT-VWF, type 2A variants negatively affect 1 or more mechanisms important for normal VWF processing.
The results from the present study revealed the limitations of the group 1 or 2 subclassification in type 2A VWD. The variants C1272R, L1503R, S1506L, and V1607D demonstrated severely impaired secretion and defective multimerization, which are consistent with a subclassification of group 1.22,24 However, C1272R did not appear to be expressed efficiently. When C1272R-transfected cells were stained and examined by confocal microscopy, it was difficult to find VWF-positive cells. This suggests that the primary processing defect may lie upstream at the point of transcription, as reported previously by Nichols et al, and may be similar to the previously reported type 2A variant N528S-VWF.29,31 In contrast, several variants were secreted efficiently, but had various degrees of multimer abnormalities, including C1099P, C1190S, C1272S, and C1272Y. These variants were placed in an “unclassified” group because they did not seem to fit into either of the 2 traditional groups for type 2A VWD. Whereas I1568N, G1579R, and G1631D were all multimerized normally and demonstrated increased susceptibility to ADAMTS13-mediated degradation, only I1568N and G1579R were secreted normally, making them possible group 2 candidates. Conversely, G1631D had modestly reduced secretion and reduced formation of Weibel-Palade body–like granules, which are uncharacteristic of group 2. Therefore, this variant was labeled “unclassified.” These data indicate that many type 2A variants do not fit the criteria for the historical group 1 or 2 classification scheme.
Two of the sequence variations identified in our type 2A patients were found to be polymorphisms. When this study was initiated, I1380V was associated with type 2A in the VWF database (http://www.VWF.group.shef.ac.uk/), but is currently listed as a polymorphism. We identified this sequence variation in several healthy controls recruited through the ZPMCB-VWD study. This sequence variation had no effect on VWF secretion, multimerization, granule formation, or ADAMTS13-mediated degradation. We also identified an M740I variation in conjunction with C1190S. Our expression studies demonstrated the causative nature of C1190S, whereas M740I had no effect on VWF processing. In addition, we identified M740I in 6.3% of all healthy controls and, more specifically, in 18% of healthy African-American controls (Bellissimo et al, manuscript submitted, 2011). M740I has also been identified in conjunction with the causative R1205H mutation in some type 1 (Vicenza) subjects.32 M740I appears to be a polymorphism.
Whereas some variants investigated in this study have been shown previously to be associated with a reduced secretion phenotype (ie, L1503R, S1503L, and V1607D), previous studies did not examine regulated storage.22,24,33 The impact of mutations in VWF on formation of storage granules has been investigated for few variants.17,26,29,30,34-36 A loss of regulated storage was characteristic of the “classic” group 1 variants, which did not form Weibel-Palade body–like storage granules and had only a diffuse staining pattern in HEK293 cells. The group 2 variants G1579R and I1568N formed storage granules. As for the variants that fell into the “unclassified” group, some formed storage granules (ie, C1099P, C1190S, and C1272Y), whereas others did not (ie, C1272S and G1631D). Although the D3 domain is also involved in the formation of WPB, the D3 domain mutants in our study also formed WPB.37,38 We did not find a link between VWF multimerization and regulated storage. Variants C1190S and C1099P have multimer abnormalities yet formed storage granules, whereas variant G1631D normally multimerized and did not form storage granules. This is consistent with previous studies demonstrating that VWF multimerization and storage are independent processes.17,27,30,35,37 Three patients in our study with mutations that formed Weibel-Palade body–like granules in HEK293 cells were found to release sufficient amounts of VWF after DDAVP administration. A loss or reduction in endothelial cell Weibel-Palade bodies may explain the diminished DDAVP response observed in some type 2A VWD patients.21
Of the variants examined in the present study, 5 involved mutation of cysteine residues (ie, C1099P, C1190S, and C1272R/S/Y). Four of these variants were secreted efficiently, whereas C1272R was expressed inadequately. All variants had some degree of multimer abnormality with loss of HMW multimers or a smeary multimer pattern, suggesting that cysteine mutations are likely to cause multimer abnormalities. Both C1099P and C1190S are located in the D3 domain, which is critical in VWF multimerization through the formation of disulfide bridges. Several D3 domain mutations have been shown to cause defective multimerization of VWF.6,35,39,40
Purvis et al reported that C1099 and C1142 are essential for proper VWF multimer assembly in the Golgi,41 showing that C1099A mutation hindered the formation of multimers and suggesting that C1099 forms C1099-C1099 intersubunit bonds. If that cysteine is essential for proper multimer assembly, then the C1099P mutation that we have presented herein may impair the formation of multimers in a similar manner.
Mutations associated with a historic group 2 classification were located in the A2 domain, including variants I1568N and G1579R. Increased ADAMTS13 cleavage may also be the primary defect for the A2 variant G1631D, but it fell into the “unclassified” category because of moderately reduced secretion and lack of WPB-like granules. Other variants may have increased susceptibility to degradation modestly, but this appeared to be secondary to defective multimerization. We did not analyze ADAMTS13 degradation for the group 1 mutations because they were not secreted well. However, Hassenpflug et al demonstrated using truncation mutants that group 1 variants also may have increased ADAMTS13 susceptibility.24 In the context of poorly secreted full-length VWF mutants, this may be of less impact on the type 2A phenotype. Hassenpflug et al also pointed out that mutations in the A2 domain are not restricted to group 2 variants, a conclusion that our present study supports. Whereas increased ADAMTS13 cleavage clearly can result in type 2A VWD, modestly increased cleavage may also play a role in type 1 VWD, suggesting that ADAMTS13 proteolysis may contribute to VWD phenotypes other than the 2A-group 2 variants.42,43
Because type 2A VWD inheritance is usually autosomal dominant, we coexpressed the 2A variants with WT-VWF to mimic heterozygous expression. Type 2A variants appeared to affect negatively one or more mechanisms important for VWF processing. Group 1 variants L1503R and S1506L appeared to inhibit the secretion of WT-VWF. Using differential Ab recognition, we were able to observe the interplay of the 2 alleles. A subset of variants with multimer abnormalities affected WT-VWF multimerization negatively (ie, C1099P, C1190S, and C1272S). Group 1 variants did not affect total VWF multimerization at a 1:1 ratio of expression, similar to previous reports of similar VWF variants.16,26,44 Little information is available on the relative level of expression of 2 alleles in patients in vivo, although variable expression may affect the overall phenotype. In one previous study, the dimeric variant Δ437-442-VWF was incorporated into mid- and high-molecular-weight multimers when coexpressed with WT-VWF.26 In the present study, we did not observe correction of multimer structure when C1190S, C1272S, C1272Y, or C1099P was coexpressed with WT-VWF. In regard to regulated storage, the group 1 variants S1506L, L1503R, and V1607D had a negative impact on WT-VWF storage, inhibiting its ability to form rod-like storage granules, as did C1272S, an unclassified variant. In general, it appears that there was a negative impact on regulated storage if the 2A variant itself had a storage defect.
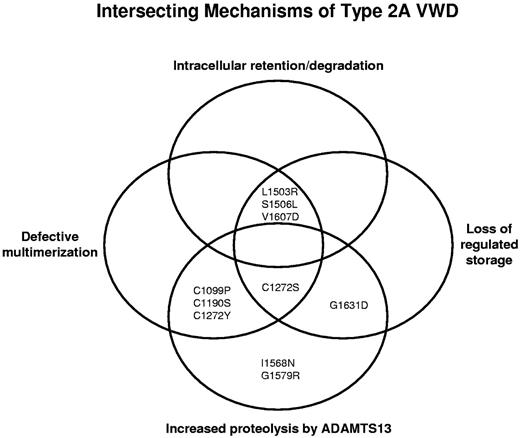
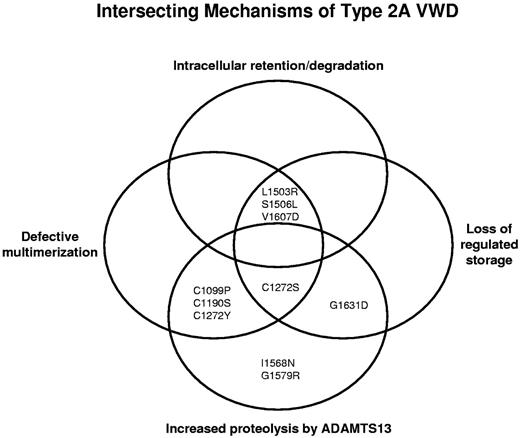
Type 2A VWD appears to result from a complex intersection of mechanisms including: (1) intracellular retention or degradation of VWF, (2) defective multimerization, (3) loss of regulated storage, and (4) increased proteolysis by ADAMTS13 (Figure 6). Traditional group 1 mutations, such as L1503R, S1506L, and V1607D, are intracellularly retained, have no high-molecular-weight multimers, and show loss of regulated storage. The traditional group 2 mutations, I1568N and G1579R, have full-length multimers that are secreted normally, but have increased proteolysis by ADAMTS13 and form storage granules. Still other variants demonstrate an overlap of defective intracellular or extracellular processing. The relative contribution of each mechanism may be influential. If there is a defect in a mechanism upstream, such as intracellular retention/degradation, the abnormalities that are downstream (eg, regulated storage) may have much less of an effect on the overall VWD phenotype.
Proposed complex intersection of mechanisms causing type 2A VWD. Type 2A VWD appears to result from an overlap of mechanisms, including intracellular retention/degradation, defective multimerization, loss of regulated storage, and increased proteolysis by ADAMTS13. Type 2A variants are affected by one or more of these defects in VWF processing.
Proposed complex intersection of mechanisms causing type 2A VWD. Type 2A VWD appears to result from an overlap of mechanisms, including intracellular retention/degradation, defective multimerization, loss of regulated storage, and increased proteolysis by ADAMTS13. Type 2A variants are affected by one or more of these defects in VWF processing.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all of the subjects, physicians, and staff involved in the ZPMCB-VWD study, including the program director, Robert R. Montgomery, Milwaukee, WI, and the directors of the primary clinical centers for the Zimmerman program: T. C. Abshire and A. L. Dunn, Atlanta, GA; J. M. Lusher, Detroit, MI; W. K. Hoots and D. L. Brown, Houston, TX; A. D. Shapiro, Indianapolis, IN; J. A. Di Paola and S. R. Lentz, Iowa City, IA; J. C. Gill, Milwaukee, WI; C. Leissinger, New Orleans, LA; and M. V. Ragni, Pittsburgh, PA. In addition, numerous secondary centers contributed to subject recruitment: J. Hord, Akron Children's Hospital, Akron, OH; M. Manco-Johnson, Mountain States Regional Hemophilia and Thrombosis Center, Aurora, CO; A. Ma, University of North Carolina Chapel Hill, Chapel Hill, NC; L. Valentino, Rush University Medical Center, Chicago, IL; R. Gruppo, Cincinnati Children's Hospital, Cincinnati, OH; B. Kerlin, Nationwide Children's Hospital, Columbus, OH; R. Kulkarni, Michigan State University, East Lansing, MI; D. Green, Northwestern University, Evanston, IL; D. Mahoney, Baylor College of Medicine, Houston, TX; L. Mathias, Loma Linda University Medical Center, Loma Linda, CA; C. Diamond, University of Wisconsin Madison, Madison, WI; A. Neff, Vanderbilt University, Nashville, TN; D. DiMichele, Weill Cornell Medical College, New York, NY; A. Cohen, Newark Beth Israel Medical Center, Newark, NJ; E. Werner, Children's Hospital of the King's Daughters, Norfolk, VA; A. Matsunaga, Children's Hospital & Research Center Oakland, Oakland, CA; M. Tarantino, Comprehensive Bleeding Disorders Center, Peoria, IL; F. Shafer, Drexel University College of Medicine, Philadelphia, PA; B. Konkle, University of Pennsylvania, Philadelphia, PA; P. Kouides, Rochester General Hospital, Rochester, NY; and D. Stein, Toledo Children's Hospital, Toledo, OH.
This work was supported by the National Institutes of Health (grants HL-33721, HL-081588, and HL-44612) and the American Heart Association (grant SDG 0435466N).
National Institutes of Health
Authorship
Contribution: P.M.J. collected and analyzed the data and wrote the manuscript; D.A.J. collected and analyzed the data; J.C.G, V.H.F., and K.D.F. designed the study and collected the data; S.L.H. designed the study, collected and analyzed the data, and wrote the manuscript; and all authors reviewed and approved the final manuscript.
Conflict-of-interest disclosure: J.C.G. is on the advisory boards of Baxter, Bayer, Octapharma, and CSL Behring and is a participant in clinical trials with Baxter, Octapharma, and CSL Behring. The remaining authors declare no competing financial interests.
Correspondence: Sandra L. Haberichter, PhD, BloodCenter of Wisconsin, 638 N 18th St, PO Box 2178, Milwaukee, WI 53201; e-mail: sandra.haberichter@bcw.edu.