Abstract
Although microRNAs (miRNAs) are increasingly linked to various physiologic processes, including hematopoiesis, their function in the myeloid development is poorly understood. We detected up-regulation of miR-29a and miR-142-3p during myeloid differentiation in leukemia cell lines and CD34+ hematopoietic stem/progenitor cells. By gain-of-function and loss-of-function experiments, we demonstrated that both miRNAs promote the phorbol 12-myristate 13-acetate–induced monocytic and all-trans-retinoic acid-induced granulocytic differentiation of HL-60, THP-1, or NB4 cells. Both the miRNAs directly inhibited cyclin T2 gene, preventing the release of hypophosphorylated retinoblastoma and resulting in induction of monocytic differentiation. In addition, a target of miR-29a, cyclin-dependent kinase 6 gene, and a target of miR-142-3p, TGF-β–activated kinase 1/MAP3K7 binding protein 2 gene, are involved in the regulation of both monocytic and granulocytic differentiation. A significant decrease of miR-29a and 142-3p levels and an obvious increase in their target protein levels were also observed in blasts from acute myeloid leukemia. By lentivirus-mediated gene transfer, we demonstrated that enforced expression of either miR-29a or miR-142-3p in hematopoietic stem/progenitor cells from healthy controls and acute myeloid leukemia patients down-regulated expression of their targets and promoted myeloid differentiation. These findings confirm that miR-29a and miR-142-3p are key regulators of normal myeloid differentiation and their reduced expression is involved in acute myeloid leukemia development.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous group of blood cancers characterized by increased, uncontrolled proliferation of hematopoietic progenitors, and a blockage in myeloid differentiation is the major characteristic of AML.1 According to the French-American-British classification system, AMLs involving the monocytic and granulocytic lineage account for 85% (M1 to M5 subtypes) of adult patients.2 Under normal conditions, monocytes and granulocytes develop from long-term hematopoietic stem cells (LT-HSCs) in BM under the influence of a complex network of cytokins such as G-CSF, GM-CSF, and M-CSF, and transcription factors such as PU.1, C/EBP, IFN consensus sequence binding protein/IFN regulatory factor 8, Krüppel-like factor 4, c-Maf, and C/EBPϵ.3-6
MicroRNAs (miRNAs) that negatively regulate gene expression at posttranscriptional level7 have also been identified as crucial regulators in normal and malignant myeloid differentiation. Expression and function analyses have unraveled their important regulatory roles during hematopoiesis.8
In a previous study, we demonstrated a significantly decreased expression of miR-29a and 142-3p in the peripheral blood mononuclear cells (PBMNCs) from AML patients (French-American-British M1 to M5 subtypes).9 These 2 miRNAs were also reported to be down-regulated in a variety of tumors and to act as tumor suppressors.10-12 The miR-29a cluster is one of the most studied miRNA clusters. Down-regulation of miR-29a was observed in AML samples with deletions of 7q (del7q).10 Several genes have been reported to be silenced by miR-29, most of which are potential oncogenes, such as SKI, Tcl1, the p53 upstream inhibitors p85a and CDC42, and the Bcl2 family members Bcl2 and Mcl1.11 Meanwhile, miR-142-3p, first identified as being uniquely expressed in hematopoietic system, is aberrantly expressed in T-cell and B-cell leukemia.12 In addition, increased miR-142-3p expression has been observed at different stages of normal granulocytopoiesis.13 Validated targets of miR-142-3p include ADCY9,14 CD133,15 IL-6,16 and the RAC1.17
In this study, we sought to investigate role of these 2 miRNAs in monocytic and granulocytic differentiation (also called myeloid differentiation), and to test whether their down-regulation is related to the differentiation block in AML blasts. Using the leukemia cell lines, NB4, HL-60, and THP-118-23 we observed up-regulation of miR-29a and miR-142-3p expression during all-trans-retinoic acid (ATRA)–induced granulocytic differentiation and phorbol 12-myristate 13-acetate (PMA)–induced monocytic differentiation, and examined effects of overexpression or knockdown of each miRNA on myeloid differentiations. Moreover, we identified targets of both miRNAs and examined direct effect of the target genes on myeloid differentiation. Similar results were also obtained in myeloid induction cultures of CD34+ hematopoietic stem/progenitor cells (HSPCs) derived from normal human umbilical cord blood (UCB) and BM from healthy donors and AML patients. Based on these examinations, we demonstrated that miR-29a and miR-142-3p are important regulators of normal myeloid differentiation and AML development and that they function at least partially though direct suppression of their common target gene, cyclin T2 (CCNT2), and their individual target genes cyclin-dependent kinase 6 (CDK6) and TGF-β activated kinase 1/MAP3K7 binding protein 2 (TAB2), respectively.
Methods
Cell culture and differentiation induction
The human leukemia cell lines NB4, HL-60, and THP-1 and human embryonic kidney cell lines 293T and 293TN were maintained in proper media (supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
The granulocytic differentiation of NB4 and HL-60 was induced with ATRA (Sigma-Aldrich) and the monocytic differentiation of THP-1 and HL-60 with PMA (Sigma-Aldrich; supplemental Methods). The mRNA expression levels of cell surface markers CD11b, CD14, and CSF1R for monocytic and CD11b, CSF3R, and MPO for granulocytic differentiation were determined by real-time PCR. CD11b and CD14 levels were also determined by flow cytometry on a FACSAria (BD Biosciences) or Accuri flowC6 (BD Biosciences). Morphology was evaluated by conventional light-field microscopy of May-Grünwald-Giemsa–stained cytospins using the Olympus BX51 (Japan) optical microscopy. Cells were also stained using the nitroblue tetrazolium assay (supplemental Methods).
RNA extraction, cDNA synthesis of mRNAs and miRNAs, and real-time PCR assays
See supplemental Methods.
PAGE-northern and Western blot
Northern blot analysis of miRNAs and Western blot analysis of proteins were performed as described previously.24 Monoclonal mouse anti-CCNT2 antibody (Abcam), monoclonal mouse anti-CDK6 antibody, monoclonal rabbit anti-TAB2 antibody, polyclonal rabbit anti-pRb antibody (Ser 807/811), and monoclonal mouse anti-Rb antibody (Cell Signaling Technology) were used for Western analysis. The immunoblots were quantified using Gelpro Version 4.0 software.
Cell transfection with miRNA mimics and siRNAs
For cell transfection with miRNA mimics and miRNA inhibitors, or with small interfering RNAs (siRNAs) against the miRNA target mRNAs, see supplemental Methods. For each cell transfection, 2 or 3 replication experiments were performed.
Luciferase miRNA target reporter assay
The full length of the 3′-untranslated regions (UTRs) of human CCNT2, CDK6, and TAB2 mRNAs was each PCR amplified and cloned into the pMIR-REPORT Luciferase Reporter Vector (Ambion) to generate the reporters. Mutations of the predicted seed regions in these 3 mRNA sequences were created using primers, including the mutated sites. The luciferase miRNA target reporter assay is described in supplemental Methods.
Cell viability and cell cycle assay
Cellular proliferation and cell cycle were analyzed using the Cell Counting Kit-8 (Dojindo Laboratories), and intracellular DNA was measured via flow cytometry as described in supplemental Methods.
CD34+ cell separation and granulocytic and monocytic induction culture
Human UCB was obtained from normal full-term deliveries from Beijing Hospital. The peripheral blood samples and BM samples from normal volunteers and AML patients were obtained from the 303 Hospital and the Central Hospital of Xiangtan. The informed consent was obtained from all of the examined subjects in accordance with the Declaration of Helsinki, and the related studies were approved by the ethics committees of the participating hospitals and institute. MNC fractions were isolated from the samples by Percoll density gradient (d = 1.077; GE Healthcare), and the MNC fractions were enriched for CD34+ HSPCs using magnetic-activated cell sorting technology according to the manufacturer's recommendations (Miltenyi Biotec). The CD34+ cells were cultured in granulopoietic and monocytopoietic induction cultures as described in supplemental Methods. The developmental maturation of the differentiated cells was confirmed by observing cytospins as well as the expression of cell surface markers.
Lentiviral production and transduction
The self-inactivating transfer vector pMIRNA1, control plasmid, and the packaging kit System Biosciences were used according to the manufacturer's instructions. DNA fragments 500 bp in size containing the pre-miR-29a or pre-miR-142 were inserted under the CMV promoter in pMIRNA1. After virus packaging, the recombinant lentivirus particles were harvested and titrated as described in supplemental Methods. The CD34+ cells were transduced after 24 hours of expansion with IL-3 and SCF by adding lentivector supernatants (Lenti-miR-29a, Lenti-miR-142, and Lenti-control) to the cells at an MOI of 50 in the presence of polybrene (5 μg/mL). The cells were washed the next day with PBS and plated for colony-forming experiments (supplemental Methods) and liquid cultures.
Results
miR-29a and miR-142-3p expression increases during myeloid differentiation of THP-1, NB4, and HL-60
To investigate whether miR-29a and miR-142-3p participate in myeloid differentiation, we first examined their expression during myeloid differentiation of THP-1, NB4, and HL-60. Cell differentiation was monitored by evaluating the expression of myeloid-specific surface markers CD11b (a marker of both granulocytic and monocytic cell differentiation), and CD14 (a marker of monocytic cell differentiation), by flow cytometry (supplemental Figure 1A). Real-time PCR analysis revealed a gradual increase in the expression of both the 2 miRNAs during PMA-induced monocytic differentiation of THP-1 and ATRA-induced granulocytic differentiation of NB4 cells (supplemental Figure 1B). The same results were also observed in HL-60 cells subjected to monocytic and granulocytic differentiation (supplemental Figure 1B). These expression patterns were also confirmed by Northern blot analysis using the antisense DNA sequences of mature miR-29a and miR-142-3p as probes (supplemental Figure 1C).
miR-29a and miR-142-3p promote ATRA- and PMA-induced myeloid differentiation
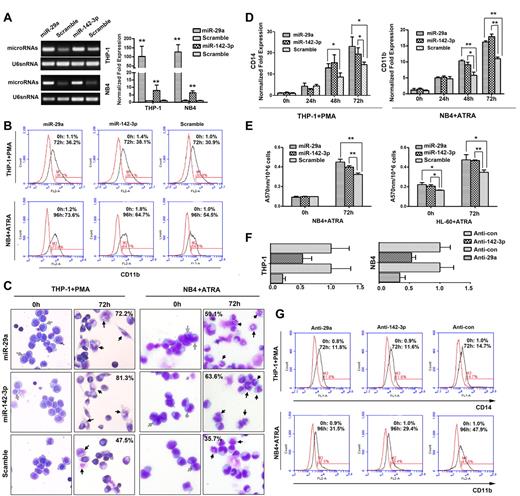
To examine the functional relevance of miR-29a and miR-142-3p with myeloid differentiation, we transiently transfected THP-1, HL-60, and NB4 cells with either miR-29a or miR-142-3p mimics or a scramble control. Transfection efficiency was confirmed by semiquantitative PCR and real-time PCR (Figure 1A; supplemental Figure 2A). After miR-29a or miR-142-3p mimic transfection, flow cytometric analysis revealed a significantly higher percentage of CD11b-positive cells among the PMA-treated THP-1 cells, as well as among the ATRA-treated NB4 cells compared with the cells transfected with the scramble control (Figure 1B; supplemental Figure 2B).
Transfection of THP-1 and NB4 cells with miRNA mimics or antisense inhibitors for miR-29a and miR-142-3p affects myeloid differentiation. (A) The expression levels of miR-29a and miR-142-3p were detected by RT-PCR (left) and real-time PCR (right) in THP-1 and NB4 cells that were transiently transfected with miRNA mimics or mimic scramble control. U6 snRNA was used as an internal control. The values of each group were expressed as mean ± SD for 3 real-time PCR assays. (B) Expression of CD11b in THP-1 and NB4 cells was analyzed by flow cytometry. Numbers in the graphs represent the percentages of positively stained cells at 72 hours of PMA or 96 hours of ATRA treatment (black histograms) compared with untreated cells (red histograms). A representative experiment of 3 is presented. The differences are statistically significant (see supplemental Figure 2B). (C) Representative May-Grünwald-Giemsa staining of THP-1 and NB4 cells transfected with microRNA mimics or scramble, and treated with PMA or ATRA for 72 hours. Images were captured at room temperature using 20× objective with numeric aperture 0.5 and acquired through CCD DP72 camera and Cellsens Standard Version 1.2.1 software (Olympus). The mild differentiated cells were annotated by gray arrows. Mature macrophages and segmented neutrophils after 72 hours of differentiation were annotated by black arrows, and their percentages were marked. More mature monocytes show bluish-gray cytoplasm and a saddle-shaped nucleus. More mature granulocytic cells show polylobular nuclei, a decreased ratio of nuclear area to cytoplasmic area, and decreased cytoplasm staining, corresponding to band cells and metamyelocytes. (D) Expression levels of CD14 or CD11b mRNA were analyzed by real-time PCR. Comparative real-time PCR was performed in triplicate and normalized to GAPDH mRNA. Error bars represent SD. The expression of the mRNA in untreated scramble-transfected cell was normalized as 1. (E) Nitroblue tetrazolium assay in NB4 and HL-60 cells transfected with miR-29a mimic, miR-142-3p mimic, or mimic control. (F) Real-time PCR analysis of the expression levels of miR-29a and miR-142-3p in THP-1 and NB4 cells transfected with miR-29a inhibitor (anti-29a), miR-142-3p inhibitor (anti–142-3p), or inhibitor control (anticontrol). U6 snRNA was used as an internal control. (G) Expression levels of CD14 in THP-1 cells and CD11b in HL-60 cells were analyzed by FACS. A representative experiment of 3 is presented. The differences are statistically significant (see supplemental Figure 2F). *P < .05 (Student t test). **P < .01 (Student t test).
Transfection of THP-1 and NB4 cells with miRNA mimics or antisense inhibitors for miR-29a and miR-142-3p affects myeloid differentiation. (A) The expression levels of miR-29a and miR-142-3p were detected by RT-PCR (left) and real-time PCR (right) in THP-1 and NB4 cells that were transiently transfected with miRNA mimics or mimic scramble control. U6 snRNA was used as an internal control. The values of each group were expressed as mean ± SD for 3 real-time PCR assays. (B) Expression of CD11b in THP-1 and NB4 cells was analyzed by flow cytometry. Numbers in the graphs represent the percentages of positively stained cells at 72 hours of PMA or 96 hours of ATRA treatment (black histograms) compared with untreated cells (red histograms). A representative experiment of 3 is presented. The differences are statistically significant (see supplemental Figure 2B). (C) Representative May-Grünwald-Giemsa staining of THP-1 and NB4 cells transfected with microRNA mimics or scramble, and treated with PMA or ATRA for 72 hours. Images were captured at room temperature using 20× objective with numeric aperture 0.5 and acquired through CCD DP72 camera and Cellsens Standard Version 1.2.1 software (Olympus). The mild differentiated cells were annotated by gray arrows. Mature macrophages and segmented neutrophils after 72 hours of differentiation were annotated by black arrows, and their percentages were marked. More mature monocytes show bluish-gray cytoplasm and a saddle-shaped nucleus. More mature granulocytic cells show polylobular nuclei, a decreased ratio of nuclear area to cytoplasmic area, and decreased cytoplasm staining, corresponding to band cells and metamyelocytes. (D) Expression levels of CD14 or CD11b mRNA were analyzed by real-time PCR. Comparative real-time PCR was performed in triplicate and normalized to GAPDH mRNA. Error bars represent SD. The expression of the mRNA in untreated scramble-transfected cell was normalized as 1. (E) Nitroblue tetrazolium assay in NB4 and HL-60 cells transfected with miR-29a mimic, miR-142-3p mimic, or mimic control. (F) Real-time PCR analysis of the expression levels of miR-29a and miR-142-3p in THP-1 and NB4 cells transfected with miR-29a inhibitor (anti-29a), miR-142-3p inhibitor (anti–142-3p), or inhibitor control (anticontrol). U6 snRNA was used as an internal control. (G) Expression levels of CD14 in THP-1 cells and CD11b in HL-60 cells were analyzed by FACS. A representative experiment of 3 is presented. The differences are statistically significant (see supplemental Figure 2F). *P < .05 (Student t test). **P < .01 (Student t test).
miR-29a or miR-142-3p mimic transfection also induced morphologic changes in the cells after monocytic and granulocytic differentiation. May-Grünwald-Giemsa staining showed slightly morphologic changes in untreated cell populations of THP-1 and NB4 cell populations transfected with miR-29a or miR-142-3p mimic. However, after induction for 72 hours with PMA, THP-1 cells transfected with miR-29a or miR-142-3p mimic showed a significantly greater fraction of more mature monocytes (Figure 1C left). Similarly, after induction for 72 hours with ATRA, NB4 cells transfected with miR-29a or miR-142-3p showed a significantly greater fraction of more mature granulocytic cells (Figure 1C right). These morphologic changes were corroborated by examining stained HL-60 cells transfected with miR-29a mimic, miR-142-3p mimic, or scramble control and treated with PMA or ATRA for 72 hours (supplemental Figure 2C).
We next analyzed the effect of miR-29a and miR-142-3p on the mRNA levels of differentiation markers by real-time PCR (Figure 1D). The results were consistent with flow cytometry and morphologic observation. In summary, transfection with miR-29a or miR-142-3p mimic resulted in an elevated expression of CD11b, CD14, CSF1R (supplemental Figure 2D), and CSF3R (supplemental Figure 2E) mRNAs. In contrast, a decreased level of myeloperoxidase mRNA, which is expressed only during the late myeloblast and promyelocyte stages and disappears during later stages of neutrophilic development, was observed in the cells transfected with miR-29a or miR-142-3p mimic (supplemental Figure 2E). The acceleration of maturation resulting from miR-29a or miR-142-3p transfection was also confirmed by nitroblue tetrazolium staining (Figure 1E), which showed enhanced oxidative ability in ATRA-treated NB4 and HL-60 cells. These results suggested that the enforced expression of miR-29a and miR-142-3p promotes the PMA-induced monocytic and ATRA-induced granulocytic differentiation.
Antisense inhibitors directed against either miR-29a or miR-142-3p (anti-29a or anti–142-3p) were then transfected into THP-1 and NB4 cells, and knockdown of the targeted miRNA was confirmed (Figure 1F). The THP-1 cells were then allowed to undergo PMA treatment for 72 hours, and reduced expression of CD14 antigen was observed in anti–miR-29a– and anti–miR-142-3p–treated cells (Figure 1G top; supplemental Figure 2F). The percentage of CD11b-positive cells was also modestly decreased in ATRA-induced NB4 cells transfected with miR-29a or miR-142-3p inhibitors (Figure 1G bottom; supplemental Figure 2F). Taken together, the reduced expression of miR-29a and miR-142-3p suppresses the myeloid differentiation of THP-1 and NB4 cells.
We next verified whether transfection of the 2 miRNAs simultaneously could enhance their ability to promote differentiation. As determined by flow cytometry, cotransfection of the 2 miRNAs exhibited a more obvious effect on the differentiation of both lineages than single transfection before induced differentiation, but after 72 hours treatment with PMA or ATRA the enhanced effect of promoting differentiation by the cotransfection is not significant (supplemental Figure 3).
CCNT2 was identified as a common target of miR-29a and miR-142-3p
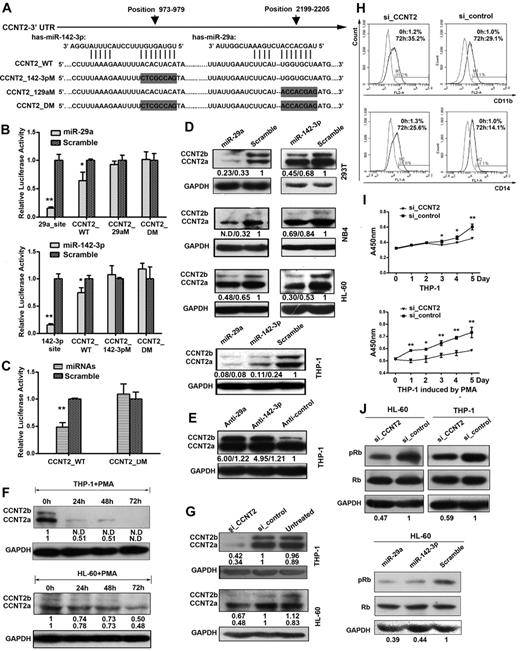
We identified several potential targets of miR-29a or miR-142-3p using the prediction programs TargetScan and Pictar, including the CCNT2 mRNA, which contains sequence motifs that match with the “seed” sequence of miR-142-3p and miR-29a in its 3′-UTR (Figure 2A). To test whether miR-29a and miR-142-3p are able to regulate CCNT2 directly, we made a reporter construct carrying the wild-type CCNT2 3′-UTR (pCCNT2_WT), constructs carrying the CCNT2 3′-UTR sequences with mutation in the putative miR-29a and miR-142-3p recognition elements (pCCNT2_142-3pM and pCCNT2_29aM, respectively), and a construct with both the mutations (pCCNT2_DM). When miR-29a or miR-142-3p mimic was transfected into 293T cells the luciferase activity of pCCNT2_WT was reduced by 30%. Meanwhile, single mutation and double mutations abolished the repression by miR-29a or miR-142-3p, demonstrating that miR-29a and miR-142-3p could specifically target their binding sites in the 3′-UTRs of CCNT2 (Figure 2B). Interestingly, transfection with both microRNAs simultaneously reduced the reporter luciferase activity more significantly, and this reduction could be abolished by mutations of the both sites, indicating that miR-29a and miR-142-3p function independently in targeting the CCNT2 3′-UTR (Figure 2C).
CCNT2 is a target gene of both miR-29a and miR-142-3p, and CCNT2 plays a role in inhibiting PMA-induced monocytic differentiation in THP-1 and HL-60 cells. (A) The nucleotide sequences of miR-29a, miR-142-3p, and their complementary sequences in CCNT2 mRNA. The predicted miR-29a and miR-142-3p binding sites in the CCNT2 3′-UTR and the mutated nucleotides (gray boxes) of the 3′-UTR site are shown. CCNT2_WT is a reporter construct containing the entire wild-type 3′-UTR sequence of CCNT2. CCNT2_29aM, CCNT2_142-3pM, and CCNT2_DM are reporter constructs containing mutations in the miR-29a binding site, the miR-142-3p binding site mutation, and both sites. (B) Luciferase activity in 293T cells cotransfected with one of the 4 reporter constructs and either one of the miRNA (miR-29a or miR-142-3p) mimics or scramble control, as indicated. The reporter constructs with perfect complementary sites for miR-29a and miR-142-3p (p29a_site and p142-3p site, respectively) were used as positive controls. The activities were calculated as a ratio of firefly to renilla luciferase activity, and are expressed as mean ± SD of 3 separate experiments. *P < .05 (Student t test). **P < .01 (Student t test). (C) Luciferase activity in 293T cells cotransfected with either the wild-type or double mutant, and both miR-29a and miR-142-3p mimics simultaneously or scramble control. (D) Western blot analysis of CCNT2 expression in 293T and myeloid cell lines THP-1, HL-60, and NB4 transfected with miR-29a mimic, miR-142-3p mimic, or scramble control. Densitometric values normalized on the basis of GAPDH expression are indicated below the corresponding lanes and shown as fold changes. N.D indicates a densitometric value cannot be given because the band is almost undetectable. (E) Western blot analysis of CCNT2 expression in monocytic cell line THP-1 transfected with an inhibitor for miR-29a (anti-29a), an inhibitor for miR-142-3p (anti–142-3p), or an inhibitor control. (F) Western blot analysis showing expression of the 2 CCNT2 isoforms during PMA-induced monocytic differentiation in THP-1 and HL-60 cells; GAPDH was detected for checking equal protein loading. (G) Western blot analysis showing the expression of CCNT2 in HL-60 and THP-1 cells that were transfected with CCNT2 siRNA (si_CCNT2) or siRNA control (si_control) and subsequently treated with PMA for 72 hours. (H) Flow cytometric analysis of CD11b in THP-1 cells transfected with si_CCNT2 or si_control. Numbers in the graphs represent the percentages of positively stained after 72 hours of differentiation (the right peak) compared with untreated cells (the left peak). A representative experiment of 3 is presented. (I) Cell proliferation assay of THP-1 cells transfected with si_CCNT2 or si_control using cell counting kit 8. The cells were cultured in medium with or without PMA. (J) Western blots showing pRb and Rb protein levels in THP-1 cells after transfection with si_CCNT2 or si_control, and in HL-60 cells after transfection with miR-29a mimic, miR-142-3p mimic, or scramble control.
CCNT2 is a target gene of both miR-29a and miR-142-3p, and CCNT2 plays a role in inhibiting PMA-induced monocytic differentiation in THP-1 and HL-60 cells. (A) The nucleotide sequences of miR-29a, miR-142-3p, and their complementary sequences in CCNT2 mRNA. The predicted miR-29a and miR-142-3p binding sites in the CCNT2 3′-UTR and the mutated nucleotides (gray boxes) of the 3′-UTR site are shown. CCNT2_WT is a reporter construct containing the entire wild-type 3′-UTR sequence of CCNT2. CCNT2_29aM, CCNT2_142-3pM, and CCNT2_DM are reporter constructs containing mutations in the miR-29a binding site, the miR-142-3p binding site mutation, and both sites. (B) Luciferase activity in 293T cells cotransfected with one of the 4 reporter constructs and either one of the miRNA (miR-29a or miR-142-3p) mimics or scramble control, as indicated. The reporter constructs with perfect complementary sites for miR-29a and miR-142-3p (p29a_site and p142-3p site, respectively) were used as positive controls. The activities were calculated as a ratio of firefly to renilla luciferase activity, and are expressed as mean ± SD of 3 separate experiments. *P < .05 (Student t test). **P < .01 (Student t test). (C) Luciferase activity in 293T cells cotransfected with either the wild-type or double mutant, and both miR-29a and miR-142-3p mimics simultaneously or scramble control. (D) Western blot analysis of CCNT2 expression in 293T and myeloid cell lines THP-1, HL-60, and NB4 transfected with miR-29a mimic, miR-142-3p mimic, or scramble control. Densitometric values normalized on the basis of GAPDH expression are indicated below the corresponding lanes and shown as fold changes. N.D indicates a densitometric value cannot be given because the band is almost undetectable. (E) Western blot analysis of CCNT2 expression in monocytic cell line THP-1 transfected with an inhibitor for miR-29a (anti-29a), an inhibitor for miR-142-3p (anti–142-3p), or an inhibitor control. (F) Western blot analysis showing expression of the 2 CCNT2 isoforms during PMA-induced monocytic differentiation in THP-1 and HL-60 cells; GAPDH was detected for checking equal protein loading. (G) Western blot analysis showing the expression of CCNT2 in HL-60 and THP-1 cells that were transfected with CCNT2 siRNA (si_CCNT2) or siRNA control (si_control) and subsequently treated with PMA for 72 hours. (H) Flow cytometric analysis of CD11b in THP-1 cells transfected with si_CCNT2 or si_control. Numbers in the graphs represent the percentages of positively stained after 72 hours of differentiation (the right peak) compared with untreated cells (the left peak). A representative experiment of 3 is presented. (I) Cell proliferation assay of THP-1 cells transfected with si_CCNT2 or si_control using cell counting kit 8. The cells were cultured in medium with or without PMA. (J) Western blots showing pRb and Rb protein levels in THP-1 cells after transfection with si_CCNT2 or si_control, and in HL-60 cells after transfection with miR-29a mimic, miR-142-3p mimic, or scramble control.
To validate that CCNT2 is a real target of both miR-29a and miR-142-3p, we quantified the expression of the 2 CCNT2 protein isoforms, CCNT2b and CCNT2a, in the cells by Western blotting. A significant reduction of endogenous CCNT2 protein levels was detected in the cells transfected with miR-29a or miR-142-3p mimic (Figure 2D). Inversely, CCNT2 proteins increased as a result of miR-29a or miR-142-3p inhibitor transfection in THP-1 cells (Figure 2E). Noticeably, the CCNT2b protein expression appears to be more sensitive to the expression changes of miR-29a and miR-142-3p than CCNT2a protein.
CCNT2 counteracts monocytic differentiation of PMA-induced HL-60 and THP-1 cells
We then examined whether the function of CCNT2 is linked to myeloid differentiation. By Western blot analysis, we found that the CCNT2 proteins, which are highly expressed in untreated THP-1 cells, were rapidly down-regulated on PMA treatment; meanwhile, this down-regulation was gradual in PMA-induced HL-60 cells (Figure 2F). On the contrary, CCNT2 protein levels are almost unchanged during ATRA-induced NB4 cell differentiation (data not shown).
We also examined the effect of CCNT2 expression levels on monocytic differentiation. Transfection of HL-60 and THP-1 cells with siRNAs targeting CCNT2 mRNA (si_CCNT2) resulted in a significant decrease of endogenous protein levels (Figure 2G). In the presence of PMA, this resulted in a strong increase in the levels of both CD11b and CD14 surface antigens compared with the cells transfected with a nontargeting si_control, as judged by flow cytometry analysis (Figure 2H; supplemental Figure 4A). In both HL-60 and THP-1 cells, the down-regulation of CCNT2 also resulted in increased mRNA levels of CD11b, CD14, and the monocyte terminal differentiation marker CSF1R (M-CSFr; supplemental Figure 4B-C). Morphologic analysis using May-Grünwald-Giemsa staining and a cell adherence assay showed that si_CCNT2 transfection increased the adherence ability of PMA-induced THP-1 cells greatly, indicating that more macrophage cells were generated (supplemental Figure 4D-E).
CCNT2 inhibits the monocytic differentiation of HL-60 and THP-1 cells by increasing proliferation
CCNT2 is a component of positive transcription elongation factor b (P-TEFb), a complex that is composed of cyclin-dependent kinase 9 and a cyclin T (CDK9/CCNT). CDK9 activation requires binding of a T family cyclin (CCNT1, CCNT2a, or CCNT2b) or CCNK.25-27 CCNT2 was not previously reported to be involved in myeloid differentiation. We performed a proliferation assay on THP-1 cells. PMA induced the cells to differentiation, but an obvious reduction of cell amplification was observed in the cells transfected with CCNT2 siRNA compared with the control cells (Figure 2I); a similar reduction in proliferation was not observed in uninduced cells.
Previous studies demonstrated that CDK9 is able to phosphorylate p56/Rb in vitro,26 and CDK9/CCNT2 can bind to Rb and phosphorylate the Rb region spanning amino acids 793 to 834 in vivo.27 To confirm this interaction between CCNT2 and Rb in THP-1 and HL-60 cells, we detected phosphorylated Rb (pRb) by Western blot analysis. A decreased pRb level was detected in the cells transfected with CCNT2 siRNA compared with the cells transfected with the control, whereas the level of total Rb remained consistent (Figure 2J top), which is consistent with the observation that the ectopic expression of both miRNAs decreased the pRb level (Figure 2J bottom). Given that Rb is well known for its involvement in mediating the cell cycle, we examined intracellular DNA content using flow cytometry and failed to see any obvious G1 arrest induced by si_CCNT2 in the uninduced cells. However, after PMA treatment for 24 hours, a significantly increased percentage of cells in G1 was detected among these cells transfected with si_CCNT2 compared with the cells transfected with si_control (supplemental Figure 5).
These results demonstrated that the CCNT2 down-regulation and the resulting decrease in pRb levels and cell proliferation are one of the pathways by which miR-29a and miR-142-3p promote monocytopoiesis.
Validation of CDK6 as a target of miR-29a in THP-1 and NB4 cells
CDK6 was previously reported as a target of miR-29a in mantle cell lymphoma, in which CDK6, expressed at elevated levels, cooperates with cyclin D1 to further promote cell cycle progression.28 We thus hypothesized that CDK6 may also function as a target of miR-29a in promyelocytic cell lines. We constructed 4 pMIR-REPORT constructs that contained either the wild-type sequence of CDK6 3′-UTR (pCDK6_WT) or the sequences of the 3′-UTR with mutation in the first (pCDK6_MUT1), or second binding site (pCDK6_MUT2) or in both (pCDK6_DM; Figure 3A). As expected, miR-29a mimic transfection decreased the activity of the pCDK6_WT reporter markedly, with little effect on the activity of pCDK6_MUT1 or pCDK6_MUT2 reporters and no effect on the activity of the CDK6_DM reporter (Figure 3B). miR-29a mimic transfection also obviously decreased the CDK6 protein levels in 293T, THP-1, and NB4 cells (Figure 3C top), whereas knockdown of miR-29a by inhibitor transfection resulted in increased CDK6 levels in NB4 and THP-1 cells (Figure 3C bottom). These results demonstrated CDK6 is a direct target of miR-29a in these cell lines.
Validation of CDK6 and TAB2 as target genes for miR-29a and miR-142-3p, respectively. (A) The predicted miR-29a binding sites in the 3′-UTR of CDK6 mRNA are shown. CDK6_WT is the pMIR-REPORT construct containing the entire 3′-UTR sequence of CDK. CDK6_MUT1, MUT2, and DM are the pMIR-REPORT constructs containing mutated nucleotides in the first, second, and both miR-29a binding sites, respectively. (B) Luciferase reporter assays. Relative luciferase levels in 293T cells cotransfected with the CDK6_WT, CDK6_MUT1/2, or CDK6_DM report construct and either miR-29a mimic or scrambled control. *P < .05 (Student t test). **P < .01 (Student t test). (C) Western blot analysis of CDK6 levels in 293T, THP-1, and NB4 cells. miR-29a mimic, inhibitor (anti-29a), or a control was transfected into the indicated cells. A representative Western blot from 3 independent experiments is shown. (D) The predicted miR-142-3p binding site in the 3′-UTR of TAB2 mRNA is shown. TAB2_WT is the reporter construct containing the entire 3′-UTR sequence of TAB2, and TAB2_MUT is the reporter construct containing mutated nucleotides at the miR-142-3p binding site. The mutated nucleotides of the 3′-UTR site are indicated by gray boxes. (E) Relative luciferase expression levels in 293T cells cotransfected with TAB2_WT or TAB2_MUT with either miR-142-3p mimic or a scrambled control. (F) Western blot analysis of TAB2 levels in 293T, THP-1, and NB4 cells. miR-142-3p mimic, a miR-142-3p inhibitor (anti–142-3p), or a control was transfected into the indicated cells. A representative Western blot from 3 independent experiments is shown.
Validation of CDK6 and TAB2 as target genes for miR-29a and miR-142-3p, respectively. (A) The predicted miR-29a binding sites in the 3′-UTR of CDK6 mRNA are shown. CDK6_WT is the pMIR-REPORT construct containing the entire 3′-UTR sequence of CDK. CDK6_MUT1, MUT2, and DM are the pMIR-REPORT constructs containing mutated nucleotides in the first, second, and both miR-29a binding sites, respectively. (B) Luciferase reporter assays. Relative luciferase levels in 293T cells cotransfected with the CDK6_WT, CDK6_MUT1/2, or CDK6_DM report construct and either miR-29a mimic or scrambled control. *P < .05 (Student t test). **P < .01 (Student t test). (C) Western blot analysis of CDK6 levels in 293T, THP-1, and NB4 cells. miR-29a mimic, inhibitor (anti-29a), or a control was transfected into the indicated cells. A representative Western blot from 3 independent experiments is shown. (D) The predicted miR-142-3p binding site in the 3′-UTR of TAB2 mRNA is shown. TAB2_WT is the reporter construct containing the entire 3′-UTR sequence of TAB2, and TAB2_MUT is the reporter construct containing mutated nucleotides at the miR-142-3p binding site. The mutated nucleotides of the 3′-UTR site are indicated by gray boxes. (E) Relative luciferase expression levels in 293T cells cotransfected with TAB2_WT or TAB2_MUT with either miR-142-3p mimic or a scrambled control. (F) Western blot analysis of TAB2 levels in 293T, THP-1, and NB4 cells. miR-142-3p mimic, a miR-142-3p inhibitor (anti–142-3p), or a control was transfected into the indicated cells. A representative Western blot from 3 independent experiments is shown.
TAB2 was identified as a target of miR-142-3p in THP-1 and NB4 cells
We also constructed 2 pMIR-REPORT constructs containing either the wild-type sequence of the TAB2 3′-UTR (pTAB2_WT) or a sequence with a mutation in the miR-142-3p binding site (pTAB2_MUT; Figure 3D). Significantly reduced luciferase activity was detected in the cells cotransfected with the pTAB2_WT and miR-142-3p mimics but not in the cells cotransfected with the pTAB2_MUT and miR-142-3p mimics (Figure 3E). Western blot analysis indicated that transfection of miR-142-3p mimic reduced the TAB2 level in the transfected cells (Figure 3F top), whereas knockdown of miR-142-3p by inhibitor transfection increased TAB2 level (Figure 3F bottom). Taken collectively, these data indicated that TAB2 is a direct target of miR-142-3p.
Effects of CDK6 and TAB2 on granulocytic and monocytic differentiation
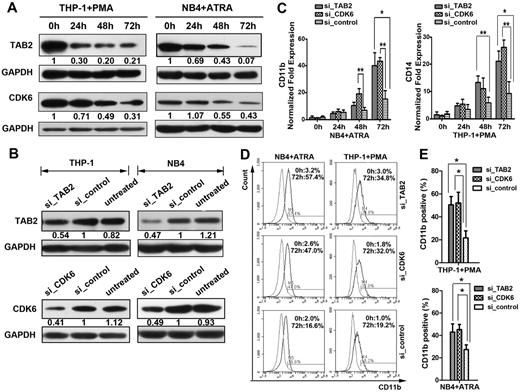
By Western blot analysis, we observed gradually decreased levels of both TAB2 and CDK6 proteins during the ATRA-induced granulocytic differentiation of NB4 and the PMA-induced monocytic differentiation of THP-1. The decrease in TAB2 levels appeared to be more rapid than that of CDK6 (Figure 4A). To further elucidate the roles of TAB2 and CDK6 in myeloid differentiation, we transfected NB4 and THP-1 cells with TAB2-siRNA (si_TAB2), CDK6-siRNA (si_CDK6), or nontargeting siRNA (si_control). The decrease of CDK6 and TAB2 protein levels via RNA interference was confirmed by Western blot (Figure 4B). The suppressed expression of TAB2 or CDK6 promoted both the PMA-induced monocytic differentiation and ATRA-induced granulocytic differentiation, as evaluated by the up-regulation of CD11b and CD14 mRNA expression (Figure 4C), morphologic changes (supplemental Figure 6), and the CD11b antigens levels (Figure 4D-E).
Expression inhibition of CDK6 and TAB2 increases ATRA-induced differentiation in NB4 cells and PMA-induced differentiation in THP-1 cells. (A) THP-1 cells treated with 50 ng/mL PMA (left) and NB4 cells treated with 2μM ATRA (right) were collected at the indicated time points for Western blot analysis of TAB2 and CDK6 protein levels. GAPDH was used as a loading control. (B) THP-1 and NB4 cells were transiently transfected with control siRNA (si_control), TAB2 siRNA (si_TAB2), or CDK6 siRNA (si_CDK6). A representative Western blot detecting TAB2 and CDK6 is shown. (C) Real-time PCR analysis of the CD11b mRNA level in the transfected NB4 cells after ATRA induction, and real-time PCR analysis of the CD14 mRNA level in the transfected THP-1 cells after PMA induction. Error bars represent mean ± SD (n = 3). (D) Flow cytometric analysis of CD11b levels. The cells were transfected with the indicated inhibitors and then treated for 72 hours with ATRA (for NB4 cells) or PMA (for THP-1 cells). A representative experiment of 3 is presented. (E) Statistical analysis of the flow cytometric assays. Data are mean ± SD from 3 independent experiments. *P < .05 (Student t test). **P < .01 (Student t test).
Expression inhibition of CDK6 and TAB2 increases ATRA-induced differentiation in NB4 cells and PMA-induced differentiation in THP-1 cells. (A) THP-1 cells treated with 50 ng/mL PMA (left) and NB4 cells treated with 2μM ATRA (right) were collected at the indicated time points for Western blot analysis of TAB2 and CDK6 protein levels. GAPDH was used as a loading control. (B) THP-1 and NB4 cells were transiently transfected with control siRNA (si_control), TAB2 siRNA (si_TAB2), or CDK6 siRNA (si_CDK6). A representative Western blot detecting TAB2 and CDK6 is shown. (C) Real-time PCR analysis of the CD11b mRNA level in the transfected NB4 cells after ATRA induction, and real-time PCR analysis of the CD14 mRNA level in the transfected THP-1 cells after PMA induction. Error bars represent mean ± SD (n = 3). (D) Flow cytometric analysis of CD11b levels. The cells were transfected with the indicated inhibitors and then treated for 72 hours with ATRA (for NB4 cells) or PMA (for THP-1 cells). A representative experiment of 3 is presented. (E) Statistical analysis of the flow cytometric assays. Data are mean ± SD from 3 independent experiments. *P < .05 (Student t test). **P < .01 (Student t test).
Expression inhibition of the target genes partially rescues the anti–miR-29a– and anti–miR-142-3p–mediated myeloid differentiation block
To test whether the regulation of myeloid differentiation by miR-29a and miR-142-3p is mediated via the targets identified in the preceding 5 paragraphs, NB4 and THP-1 cells were transfected with either CCNT2 or CDK6 siRNA after anti–miR-29a transfection, or with CCNT2 or TAB2 siRNA after anti–miR-142-3p transfection. As shown in supplemental Figure 7A, transfection of the target siRNAs repressed the up-regulation of target expression that should have been induced by anti–miR-29a or anti–miR-142-3p transfection. Flow cytometric analysis showed that the expression up-regulation of myeloid differentiation markers was partially rescued by knocking down the target genes (supplemental Figure 7B). These data further demonstrate that miR-29a and miR-142-3p promote myeloid differentiation via directly targeting their common target CCNT2 and individual targets, CDK6 and TAB2, respectively.
Role of miR-29a, miR-142-3p, and their targets in the normal hematopoiesis
To study the role of miR-29a and miR-142-3p in the normal hematopoietic program, we analyzed their levels in monocyte/macrophage and granulocytic induction cultures of UCB CD34+ HSPCs. Figure 5A shows a gradual increase of miR-29a and miR-142-3p during the induction cultures.
Role of miR-29a and miR-142-3p in CD34+ HSPCs derived from UCB. CD34+ cells purified from human UCB samples were infected with Lenti-control, Lenti-29a, or Lenti-142. Cells were cultured for 17 days. Granulocytic differentiation was induced by human G-CSF (20 ng/mL) and rhIL-6 (10 ng/mL), and monocytic differentiation by rhM-SCF (50 ng/mL), rhIL-6 (1 ng/mL), and rhFlt-3L (100 ng/mL). Cells were collected for morphologic, colony formation, and myeloid marker expression analysis. (A) Real-time PCR analysis of miR-29a and miR-142-3p expression during differentiation. Samples were collected at the indicated days after induction. The relative expression of miR-142-3p and miR-29a was normalized to the expression level at day 0. (B) CD14 mRNA levels for monocytic differentiation and CD11b mRNA levels for granulocytic differentiation were analyzed by real-time PCR. (C) Colony formation assay after an 11-day colony formation period. A total of 1 × 104 of infected CD34+ cells were plated in complete methylcellulose medium without erythropoietin. The arrows indicate typical CFU-GM (colony-forming unit-granulocyte/macrophage), CFU-G (colony-forming unit-granulocyte), and CFU-M (colony-forming unit-macrophage; left). Quantification of formation of blood colonies was shown (right). Images were captured with microscopy Nikon eclipse TS100 using 10× phase-contrast objective with numeric aperture 0.3 and acquired through CCD DS-Qi1 camera and NIS-Elements F Version 3.0 software (Nikon) at room temperature. (D) Changes in the morphology of May-Grünwald-Giemsa–stained CD34+ cells infected with the viruses at day 13 of the differentiation culture. Images were captured at room temperature with Olympus BX51 microscope using 20× with numeric aperture 0.5. (E) The number of cells in various differentiation stages. In total, 100 cells were counted per sample. The averages from 2 independent experiments are shown. MB indicates myeloblasts; PM, promyelocytes; MM, metamyelocytes; Band and Seg, band neutrophils and segmented neutrophils for granulocytic differentiation; Mob, monoblasts; PMo, promonocytes; Mo, monocytes; and Ma, macrophage for monocytic differentiation.
Role of miR-29a and miR-142-3p in CD34+ HSPCs derived from UCB. CD34+ cells purified from human UCB samples were infected with Lenti-control, Lenti-29a, or Lenti-142. Cells were cultured for 17 days. Granulocytic differentiation was induced by human G-CSF (20 ng/mL) and rhIL-6 (10 ng/mL), and monocytic differentiation by rhM-SCF (50 ng/mL), rhIL-6 (1 ng/mL), and rhFlt-3L (100 ng/mL). Cells were collected for morphologic, colony formation, and myeloid marker expression analysis. (A) Real-time PCR analysis of miR-29a and miR-142-3p expression during differentiation. Samples were collected at the indicated days after induction. The relative expression of miR-142-3p and miR-29a was normalized to the expression level at day 0. (B) CD14 mRNA levels for monocytic differentiation and CD11b mRNA levels for granulocytic differentiation were analyzed by real-time PCR. (C) Colony formation assay after an 11-day colony formation period. A total of 1 × 104 of infected CD34+ cells were plated in complete methylcellulose medium without erythropoietin. The arrows indicate typical CFU-GM (colony-forming unit-granulocyte/macrophage), CFU-G (colony-forming unit-granulocyte), and CFU-M (colony-forming unit-macrophage; left). Quantification of formation of blood colonies was shown (right). Images were captured with microscopy Nikon eclipse TS100 using 10× phase-contrast objective with numeric aperture 0.3 and acquired through CCD DS-Qi1 camera and NIS-Elements F Version 3.0 software (Nikon) at room temperature. (D) Changes in the morphology of May-Grünwald-Giemsa–stained CD34+ cells infected with the viruses at day 13 of the differentiation culture. Images were captured at room temperature with Olympus BX51 microscope using 20× with numeric aperture 0.5. (E) The number of cells in various differentiation stages. In total, 100 cells were counted per sample. The averages from 2 independent experiments are shown. MB indicates myeloblasts; PM, promyelocytes; MM, metamyelocytes; Band and Seg, band neutrophils and segmented neutrophils for granulocytic differentiation; Mob, monoblasts; PMo, promonocytes; Mo, monocytes; and Ma, macrophage for monocytic differentiation.
The role of miR-29a and miR-142-3p in the myeloid differentiation was validated by ectopic expression of them in induction cultures. Overexpression of the miRNAs was accomplished by infecting CD34+ cells with Lenti-29a or Lenti-142. After 4 days of viral infection, the miR-142-3p level reached 1.9-fold in Lenti-142–infected cells and the miR-29a level reached 2-fold in Lenti-29a–infected cells compared with Lenti-control–infected cells, and this overexpression persisted until the differentiation cultures were terminated. The differentiation status of cells was evaluated by morphologic analysis, colony-forming assay, and real-time PCR analysis, particularly for the appearance and rising expression of monocytic/macrophage (CD11b and CD14) and granulocytic (CD11b and CSF3R) differentiation markers. Compared with the cells infected with the Lenti-control, overexpression of either miR-29a or miR-142-3p resulted in an increased expression of differentiation markers that began 7 days after induction (Figure 5B; supplemental Figure 8A). Colony-forming assay showed that both miR-29a and miR-142 promote the forming of CFU-GM, CFU-G, and CFU-M, increasing both clone number and size (Figure 5C). Giemsa staining also confirmed an increased percentage of more mature cells in the monocytic and granulocytic induction cultures of CD34+ cells infected with either Lenti-29a or Lenti-142 compared with the control cultures (Figure 5D-E; supplemental Figure 8B).
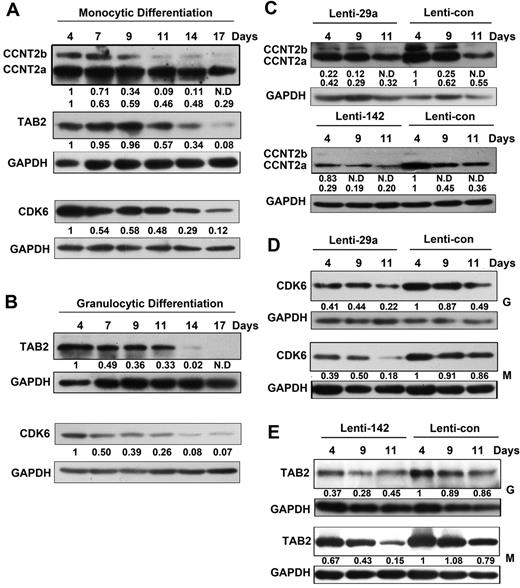
Western blot analysis showed the levels of the 3 miRNA target proteins decrease beginning on day 7 after differentiation induction (Figure 6A-B). To prove the interaction of the miRNAs and their targets in the differentiating cells, the target protein levels in the induction cultures were analyzed. Decreased CCNT2 levels were detected in the monocytic induction cultures of CD34+ cells infected with either Lenti-29a or Lenti-142 compared with the cells infected with Lenti-control (Figure 6C). Decreased CDK6 and TAB2 levels were also detected in the monocytic and granulocytic induction cultures of CD34+ cells infected with Lenti-29a and Lenti-142, respectively, compared with control cells (Figure 6D-E).
CDK6, TAB2, and CCNT2 expression levels in monocytic and granulocytic induction cultures of CD34+ cells. (A-B) Western blot analysis of CDK6, TAB2, and CCNT2 in extracts from monocytic (A) and granulocytic (B) induction cultures at the indicated times. Whole cell extracts were incubated with anti-CDK6, TAB2, or CCNT2 antibodies. (C) CCNT2 protein expression was detected by Western blot analysis at day 4, 9, and 11 of monocytic induction cultures of CD34+ cells infected with Lenti-29a, Lenti-142, or Lenti-control. (D) CDK6 protein expression was detected by Western blot analysis at day 4, 9, and 11 of monocytic and granulocytic induction cultures of CD34+ cells infected with Lenti-29a or Lenti-control. (E) TAB2 protein expression was detected by Western blot analysis at day 4, 9, and 11 of monocytic and granulocytic induction cultures of CD34+ cells infected with Lenti-142 or Lenti-control. For the Western blots shown in this figure, GAPDH antibody was used to assess equal protein loading. The signal in each lane was quantified using Gelpro software, and the ratio of CCNT2, CDK6, and TAB2 to GAPDH was determined. Because the CD34+ cells are a mixture containing various HPCs and the sample amount was limited, the 0-day samples was not included in the Western blot analysis.
CDK6, TAB2, and CCNT2 expression levels in monocytic and granulocytic induction cultures of CD34+ cells. (A-B) Western blot analysis of CDK6, TAB2, and CCNT2 in extracts from monocytic (A) and granulocytic (B) induction cultures at the indicated times. Whole cell extracts were incubated with anti-CDK6, TAB2, or CCNT2 antibodies. (C) CCNT2 protein expression was detected by Western blot analysis at day 4, 9, and 11 of monocytic induction cultures of CD34+ cells infected with Lenti-29a, Lenti-142, or Lenti-control. (D) CDK6 protein expression was detected by Western blot analysis at day 4, 9, and 11 of monocytic and granulocytic induction cultures of CD34+ cells infected with Lenti-29a or Lenti-control. (E) TAB2 protein expression was detected by Western blot analysis at day 4, 9, and 11 of monocytic and granulocytic induction cultures of CD34+ cells infected with Lenti-142 or Lenti-control. For the Western blots shown in this figure, GAPDH antibody was used to assess equal protein loading. The signal in each lane was quantified using Gelpro software, and the ratio of CCNT2, CDK6, and TAB2 to GAPDH was determined. Because the CD34+ cells are a mixture containing various HPCs and the sample amount was limited, the 0-day samples was not included in the Western blot analysis.
It should be noted that the precursor of miR-142 can be processed into 2 mature miRNAs: miR-142-3p and miR-142-5p.29 By real-time PCR analysis, we found that the expression level of miR-142-5p was always much lower than that of miR-142-3p during the monocytic and granulocytic differentiation of CD34+ cells (supplemental Figure 9). An increased expression of miR-142-3p, but not of miR-142-5p, was detected in the induction cultures infected with Lenti-142 compared with the controls. These suggested that the promotion of myeloid differentiation effected by the lentivirus-mediated gene transfer primarily reflected the action of miR-142-3p.
Validation that miR-29a and miR-142-3p negatively regulate the target genes in AML PBMNCs
We previously demonstrated significantly decreased expression of miR-29a and miR-142-3p in PBMNCs by comparing 100 normal subjects and 52 primary diagnosed AML patients.9 To validate that these miRNAs regulate their identified targets in AML, we recently collected PBMNCs from 30 healthy donors and 20 primary diagnosed AML patients (supplemental Table 1). Again, we observed that expression of the 2 miRNAs significantly decreased in AML patients (supplemental Figure 10A). Western blot analysis revealed that levels of the target proteins CCNT2, CDK6, and TAB2 were, to a great extent, inversely correlated with miR-29a and miR-142-3p expression in the samples (supplemental Figure 10B).
Enforced expression of miR-29a or miR-142-3p in AML BM blasts improved myeloid differentiation
We expanded our analysis to BM CD34+ cells derived from partial projects and observed significantly decreased miR-29a and miR-142-3p expression in the AML patients compared with healthy donors (Figure 7A). We also stochastically selected BM CD34+ cells derived from an AML M5 patient (patient 12) and an M3b patient (patient 4; supplemental Table 1) to examine their expression levels after myeloid differentiation induction. Up-regulation of the 2 miRNAs during granulocytic and monocytic induction cultures was detected (supplemental Figure 11), but the ratios of expression increase were much less than that in the normal CD34+ cells (Figure 5A).
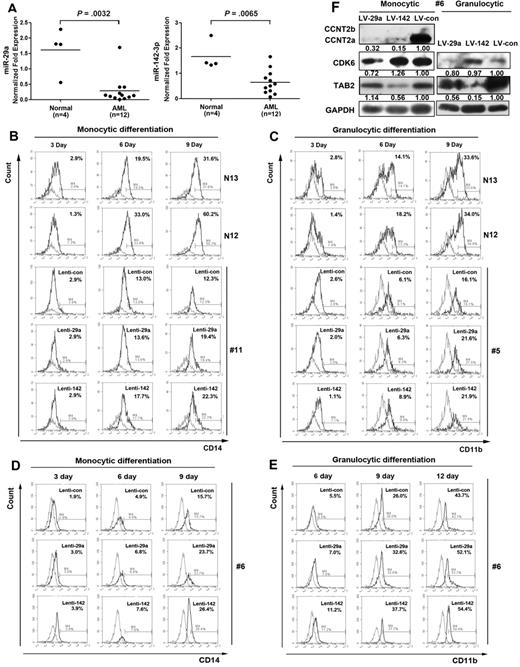
Expression of miR-29a and miR-142-3p in AML BM blasts and their function in promoting myelopoiesis. (A) Expression levels of miR-29a and miR-142-3p were determined by real-time PCR analysis of BM CD34+ cells from AML patients and healthy donors using TaqMan probes. The differences were shown to be significant by one-tailed unpaired Mann-Whitney U nonparametric t test analysis. (B-E) BM CD34+ cells from 3 AML patients (patients 11, 5, and 6 were diagnosed as French-American-British M5, M2, and M4, respectively; all demonstrated low expression levels of miR-29a and miR-142-3p; supplemental Table 1) were infected with Lenti-miR-29a, Lenti-miR-142-3p, or Lenti-control. After infection for 24 hours, the cells were cultured in monocytic or granulocytic induction medium for the indicated number of days, after which the cells were collected and the GFP-positive cells were analyzed for myeloid marker expression. The expression levels of CD14 (B) and CD11b (C) in lentivirus-infected blast cells (#11 and #5 of supplemental Table 1) were analyzed by FACS analysis compared with their expression levels in CD34+ BM cells from normal donors (N13 and N12). Red histogram represents unstained CD34+ cells; and black histogram, cells induced for the indicated number of days. The monocytic (D) and granulocytic (E) differentiation of CD34+ cells from AML patient 6 were determined by FACS analysis. (F) Western blot analysis of the target proteins, CCNT2, CDK6, and TAB2. The protein extracts were obtained from the cells at day 6 of granulocytic and monocytic induction cultures of BM CD34+ cells derived from AML 6, and infected with Lenti-29a, Lenti-142-3p, or Lenti-control. GAPDH was detected as the loading control.
Expression of miR-29a and miR-142-3p in AML BM blasts and their function in promoting myelopoiesis. (A) Expression levels of miR-29a and miR-142-3p were determined by real-time PCR analysis of BM CD34+ cells from AML patients and healthy donors using TaqMan probes. The differences were shown to be significant by one-tailed unpaired Mann-Whitney U nonparametric t test analysis. (B-E) BM CD34+ cells from 3 AML patients (patients 11, 5, and 6 were diagnosed as French-American-British M5, M2, and M4, respectively; all demonstrated low expression levels of miR-29a and miR-142-3p; supplemental Table 1) were infected with Lenti-miR-29a, Lenti-miR-142-3p, or Lenti-control. After infection for 24 hours, the cells were cultured in monocytic or granulocytic induction medium for the indicated number of days, after which the cells were collected and the GFP-positive cells were analyzed for myeloid marker expression. The expression levels of CD14 (B) and CD11b (C) in lentivirus-infected blast cells (#11 and #5 of supplemental Table 1) were analyzed by FACS analysis compared with their expression levels in CD34+ BM cells from normal donors (N13 and N12). Red histogram represents unstained CD34+ cells; and black histogram, cells induced for the indicated number of days. The monocytic (D) and granulocytic (E) differentiation of CD34+ cells from AML patient 6 were determined by FACS analysis. (F) Western blot analysis of the target proteins, CCNT2, CDK6, and TAB2. The protein extracts were obtained from the cells at day 6 of granulocytic and monocytic induction cultures of BM CD34+ cells derived from AML 6, and infected with Lenti-29a, Lenti-142-3p, or Lenti-control. GAPDH was detected as the loading control.
To test whether ectopic expression of the miRNAs could promote myeloid maturation of AML BM blasts, we infected BM CD34+ cells derived from 3 AML patients with the recombination viruses. Using flow cytometry, we observed that the ectopic expression of miR-29a or miR-142-3p partially overcame differentiation arrest in AML blasts (Figure 7B-E). Western blot analysis further confirmed that the ectopic expression of miR-29a or miR-142-3p decreased their target protein levels (Figure 7F).
Discussion
In this study, we demonstrated a positive regulation of miR-29a and miR-142-3p on the monocytic and granulocytic differentiations and validated their 3 targets. In addition, we showed that abnormal expression of these 2 miRNAs and the 3 identified targets is involved in AML development.
Expression signature of miR-142-3p had been previously implicated in hematopoietic differentiation of AML cells, but some contradict results were reported, which may be resulted from the differences in cell induction and RNA isolation methods.13,29-31 The miR-142-3p expression was reported to increase by 2.5-fold after 2 weeks of granulocytic differentiation in UCB-derived CD34+ cells.13 A study compared miRNA expression among HSPCs and peripheral blood leukocytes also identified miR-142-3p as the most up-regulated miRNA in peripheral blood leukocytes.30 Meanwhile, previous reports regarding miR-29a did not demonstrate its involvement in myeloid differentiation but focused on its abnormal expression in many kinds of leukemia.32 Moreover, we previously detected a significantly decreased expression of miR-29a and miR-142-3p in the PBMNCs of AML patients,9 and confirmed the result in AML BM blast cells in the current study.
We confirmed CCNT2 as a common target of miR-29a and miR-142-3p. CCNT2 is a component of the positive transcription elongation factor b (P-TEFb).33 In mammals, P-TEFb is essential for the elongation of transcription and cotranscriptional processing by RNA polymerase II, and different P-TEFb complexes can regulate subsets of distinct genes that are important for embryonic development.34
We observed inhibition of proliferation in THP-1 cells and a decreased level of phosphorylated Rb by siRNA-mediated knockdown of CCNT2, which is consistent with previous findings.27 We also showed that the ectopic expression of miR-29a or miR-142-3p could decrease pRb level, which might be a consequence of the reduction of CCNT2 expression.
Previous studies showed that the monocytic differentiation of BM cells is correlated to high levels of pRb, whereas during neutrophilic differentiation, pRb levels are low.35 It has been demonstrated that the importance of Rb phosphorylation in different pathways other than the cell cycle.36 In contrary, mechanistic studies of the response of THP-1 cells to PMA treatment showed that high levels of pRb are indispensible, and growth arrest at the G1 phase of the cell cycle is involved.37 Thus, reduced levels of pRb are indispensible for the monocytic differentiation of both promyeloid cell lines and BM HSPCs, whether G1 phase arrest is induced or not. This also accounts for our observation that the CCNT2 protein levels decreased only during monocytic induction cultures but remained almost unchanged during granulocytic induction cultures.
We validated CDK6 as another target of miR-29a during myeloid differentiation. In vitro, CDK6 levels rapidly decrease after the onset of terminal granulocytic differentiation in 32Dcl3 cells.38 In vivo, loss of CDK6 in the mouse affected the production of terminally differentiated myeloid and erythroid cells.39 It is clear that CDK6 blocks myeloid differentiation by interfering with Runx1′ ability to bind DNA and with Runx1-C/EBPα interaction in immature proliferating cells.40 Down-regulation of CDK6 therefore provides a molecular switch that allows the differentiation-promoting activity of Runx proteins to be selectively activated in terminally differentiating cells.
We also identified TAB2 as another target of miR-142-3p. TAB2 has been shown to play a role in vitro as an adapter protein in the receptor proximal signaling events of the proinflammatory IL-1 and of several TNF-family members, including RANKL, to activate the NF-κB pathway via binding to ubiquitinated TRAF2 and TRAF6 in the cytoplasm.41-43 In this way, RANKL stimulates osteoclast differentiation from hematopoietic precursor cells. We propose that the up-regulation of miR-142-3p blocks TAB2 expression, which results in inhibition of osteoclast development, and as a consequence, drives monocytic precursors to differentiate into mature macrophages. This miRNA-based mechanism underlying the commitment of monocyte progenitors to a single-cell fate was also observed by Mann et al.44 They demonstrated that miR-155 drives the commitment of monocyte progenitors toward activated macrophages by interfering with the expression of MITF, which propels the same progenitors toward osteoclast differentiation.44 In the current study, we found that knockdown of TAB2 also induced granulocytic differentiation in NB4 cells on ATRA treatment. Future studies are needed to explain the mechanism underlying this regulation of TAB2.
Because miR-29a and miR-142-3p have many proven and potential targets, we could have expected to find that their function in promoting myeloid differentiation is implemented by various targets. However, given the pro-myelopoiesis effects of CCNT2, CDK6, and TAB2 siRNA transfection in the current study, and that the expression of these proteins in the CCNT2, CDK6, and TAB2 knockdown assays can be rescued by anti–miR-29a or anti–miR-142-3p transfection, it is therefore conceivable that miR-29a and miR-142-3p promote myeloid differentiation primarily by affecting these 3 targets. Moreover, significantly increased levels of the target proteins were detected in the AML blasts with abnormally reduced expression of miR-29a and miR-142-3p, which further suggested that the 2 miRNAs regulate myeloid differentiation and AML development via the 3 target genes.
Collectively, our data demonstrated that miR-29a and miR-142-3p play a key role in promoting granulopoiesis and monopoiesis, and the abnormally decreased expression of these 2 miRNAs is responsible for the differentiation block of blast cells in some AML patients, whereas enforced expression of either miR-29a or miR-142-3p could improve the differentiation status. Because we have observed the universal down-regulation of these 2 miRNAs in AML M1 to M5 subtypes, increasing endogenous expression or ectopic implantation of miR-29a and miR-142-3p may be a potential strategy for AML treatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Shao-Wei Wang from Beijing Hospital for assistance in UCB collection.
This work was supported by the National Natural Science Foundation of China (30970616 and 31171311; J.-W.Z.) and the Specific Fund of National Laboratory of China (3060204; J.-W.Z.).
Authorship
Contribution: X.-S.W. designed and performed experiments, analyzed data, and wrote the paper; J.-N.G. performed experiments and analyzed data; J.Y., F.W., G.-H.Y., and C.S. performed experiments; X.-H. Z., X.-L.Y., Z.-Q.T., and Z.-M.L. provided the experimental materials of AML patients and healthy donors; and J.-W.Z. designed the research, analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jun-Wu Zhang, National Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and Peking Union Medical College, 5 Dong Dan San Tiao, Beijing 100005, China; e-mail: junwu_zhang@pumc.edu.cn.
References
Author notes
X.-S.W. and J.-N.G. contributed equally to this study.