Abstract
The transcription factor C/EBPα is a critical mediator of myeloid differentiation and is often functionally impaired in acute myeloid leukemia. Recent studies have suggested that oncogenic FLT3 activity disrupts wild-type C/EBPα function via phosphorylation on serine 21 (S21). Despite the apparent role of pS21 as a negative regulator of C/EBPα transcription activity, the mechanism by which phosphorylation tips the balance between transcriptionally competent and inhibited forms remains unresolved. In the present study, we used immuno-affinity purification combined with quantitative mass spectrometry to delineate the proteins associated with C/EBPα on chromatin. We identified DEK, a protein with genetic links to leukemia, as a member of the C/EBPα complexes, and demonstrate that this association is disrupted by S21 phosphorylation. We confirmed that DEK is recruited specifically to chromatin with C/EBPα to enhance GCSFR3 promoter activation. In addition, we demonstrated that genetic depletion of DEK reduces the ability of C/EBPα to drive the expression of granulocytic target genes in vitro and disrupts G-CSF–mediated granulocytic differentiation of fresh human BM-derived CD34+ cells. Our data suggest that C/EBPα and DEK coordinately activate myeloid gene expression and that S21 phosphorylation on wild-type C/EBPα mediates protein interactions that regulate the differentiation capacity of hematopoietic progenitors.
Introduction
C/EBPα is the founding member of the C/EBP family of transcription factors that share a conserved leucine-zipper dimerization domain.1 Although C/EBPα participates in the development of many tissues, the phenotype of knock-out mice best illustrates the basic requirement of C/EBPα for life,2 along with its central role in hematopoiesis in general3 and granulopoiesis in particular.4 Fetal livers of C/EBPα–null mice are hyperproliferative and exhibit limited capacity for the development of bipotent granulocyte/monocyte progeny and terminally differentiated granulocytes.5 Similarly, conditional disruption of C/EBPα expression disrupts the formation of granulocytes and leads to a concomitant increase in self-renewal of hematopoietic stem cells. In addition, studies using ectopic expression illustrate that C/EBPα is a key molecular determinant in myeloid lineage commitment.4,6,7
C/EBPα drives myeloid differentiation through distinct roles (reviewed by Friedman et al8 ): activation of myeloid target genes (including CEBPE, GCSFR3, MPO, ELA, LF, mim-1, and miR34a) and inhibition of the cell cycle through interactions with regulatory proteins (including CDK2/4, p21, E2F, and pRb family members). In the context of granulopoiesis, the antiproliferative role of C/EBPα has been well described,9 whereas recent studies demonstrated that various transcription coregulatory proteins, including CBP/p300,10 c-Jun,11 histone acetyltransferase TIP60,12 and Max,13 participate with C/EBPα to mediate myeloid differentiation. Although recent studies have focused on C/EBPα–mediated gene activation in hematopoietic cells, along with its role in chromatin structure,14 the underlying molecular mechanisms remain largely unresolved.
The observation that C/EBPα is required for proper regulation of the myeloid compartment suggests that loss-of-function may result in differentiation arrest. Consistent with this hypothesis, the BM of adult C/EBPα−/− mice contains myeloblasts, which is similar to the phenotype observed in human acute myeloid leukemia (AML).15 Further, an increasing body of evidence suggests that impaired function of C/EBPα leads to the accumulation of myeloid progenitors and eventually leukemic blasts.16,17 In hematopoietic malignancies, genetic lesions or dominant-negative interactions represent the most common mechanisms for disruption of C/EBPα activity. For example, somatic mutations result in expression of C/EBPα proteins that exhibit diminished DNA binding, whereas mutations at the 5′ end of the gene result in translation from an alternative start site and the expression of an N-terminally truncated protein (C/EBPα p30) product that functions as a dominant negative with respect to the wild-type protein.18 In a subset of AML patients who do not harbor C/EBPα mutations, overexpression of TRIB2 is correlated with hypermethylation of the CEBPα promoter region19 ; moreover, TRIB2 inactivates C/EBPα and induces leukemia in mice.20 Finally, translational and transcriptional repression in the context of oncogenic BCR-ABL and AML1-ETO translocation products (reviewed by Pabst et al17 ), respectfully, lead to diminished expression of C/EBPα and inhibition of granulopoiesis.
Interestingly, oncogenic FLT3-ITD mediates the phosphorylation of serine 21 (pS21) on C/EBPα in model cell lines and AML blasts and disrupts granulocytic differentiation in vitro.21 The link between FLT3 and C/EBPα is particularly intriguing given that multiple oncogenic events are required for a highly penetrant AML phenotype and that aberrant expression of FLT3 or genetic mutations that confer constitutive activity to this kinase represent the most frequent molecular abnormalities in AML.22 In addition, S21 is differentially phosphorylated in lineage-depleted mouse progenitor cells by downstream mediators of the monocyte (M-CSF) and granulocyte (G-CSF) pathways, respectively; moreover, targeted inhibition of effectors in these pathways (MEK and SHP2, respectively) diminishes pS21 significantly.23 These results, combined with evidence that pS21 does not alter C/EBPα affinity for DNA appreciably,24 led us to hypothesize that protein-protein interactions mediated by phosphorylation may play a critical role in the regulation of myeloid gene expression and, ultimately, granulocytic differentiation.
The protein DEK was originally identified as a fusion with the CAN/NUP214 nucleoporin in a subset of AML patients who harbored the t(6;9)(p23;q64) translocation.25 DEK-CAN induces leukemia in the hematopoietic stem cell compartment of mice, specifically from long-term multipotent progenitors.26 Overexpression of DEK mRNA has been reported in AML and in a variety of solid tumors, including melanoma, glioblastoma, and retinoblastoma, along with small-cell lung, cervical, and bladder cancers (reviewed by Riveiro-Falkenbach and Soengas27 ). Inactivation of DEK in mouse models results in reduced papilloma formation. The coordinate activity of DEK with HPV-E6/7 and HRAS in the formation and proliferation of keratinocyte tumors28 further illustrates the oncogenic capacity of this gene. Interestingly, nearly 50% of patients with a DEK-CAN translocation are also heterozygous for FLT3-ITD, a mutation that confers constitutive kinase activity and is correlated with poor clinical outcomes in AML.29 The role of DEK in transcription varies based on cell type, gene target, and developmental context. For example, DEK localizes preferentially at sites proximal to the promoters of expressed genes,30 enhances the transcription capacity of AP-2 in human malignant glioblastoma (U-87MG) and canine osteosarcoma (D17) cell lines,31 and acts as a coactivator of the nuclear splicing factor U2AF in HeLa cells.32 Conversely, DEK's membership in complexes such as those associated with DAXX and PCAF/p300 or, similarly, its negative regulation of p65/NFκB, suggests that it has a repressive role (reviewed by Riveiro-Falkenbach and Soengas27 ).
In the present study, we report that DEK is a novel protein partner of C/EBPα and demonstrate that this protein-protein interaction is mediated by reversible phosphorylation of S21. In addition, we observed that DEK is an essential component of transcriptionally competent C/EBPα complexes at the promoters of GCSFR3 and CEBPE, two early myeloid-specific target genes. We also found that depletion of DEK diminishes the C/EBPα–induced expression of GCSFR3 and CEBPE and reduces the differentiation capacity of primary CD34+ hematopoietic progenitors. Our data demonstrate that the interaction between the C/EBPα and DEK, which is mediated in-part by pS21, plays a role in gene activation and ultimately granulocytic differentiation.
Methods
Cell lines
293T cells were obtained from the ATCC and cultured according the manufacturer's recommendations. K562 ER mutant cells were cultured as described previously.24 MOLM-14 cells were obtained from the laboratory of Dr J. Griffin (Dana-Farber Cancer Institute, Boston, MA). The generation of the tetracycline-inducible C/EBPα–FLAG/HA and control MOCK MOLM-14 cell lines; cellular fractionation and immunoprecipitation; digestion and on column iTRAQ labeling; 2-dimensional chromatography, mass spectrometry data processing, immunodetection, electrophoretic mobility shift analysis, luciferase reporter assay, chromatin immunoprecipitation, and gene-expression analysis by RT-PCR are all described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Patient AML samples
The study protocols were in accordance with the Declaration of Helsinki and approved by the institutional review board at The Ohio State University. All patients provided written informed consent. Sample lyses conditions are described in supplemental Methods.
RNA knockdown
Lentiviral transduction of 300 000 ER- and C/EBPα–mutant K562 cell lines was performed by spinoculation in the presence of protamine sulfate (5 μg/mL; Sigma-Aldrich) at a multiplicity of infection of 5 for 1.5 hours in 24-well plates coated with Retronectin (Takara Bio). Sorting of green fluorescent protein (GFP) populations was carried out with a FACSAria II sorter (BD Biosciences). To induce C/EBPα–ER nuclear translocation, β-estradiol was added to a final concentration of 1μM 24 hours after sorting. After an additional 16 or 24 hours, cells were harvested for total RNA purification.
Proximity ligation assay
Anti–mouse and anti–rabbit proximity ligation assay (PLA) probes (“plus” and “minus” PLA forms) along with Duolink detection kit 563 were purchased from Olink Bioscience. The PLA assay was performed with primary Abs (anti-C/EBPα, SC-61, and anti-DEK; 610948) and PLA probes according to the manufacturer's recommendations. The detailed protocol for this assay is provided in supplemental Methods.
Propagation and manipulation of CD34+ cells
Fresh BM-derived CD34+ cells were obtained from Lonza and cultured in 24-well plates at a density of 100 000 cells/mL. Cells were maintained in an undifferentiated state by passage every 3 days in StemSpan SFEM supplemented with StemSpan CC100 (StemCell Technologies) containing the following recombinant human cytokines: rhFlt3 (rhFlt3; 100 ng/mL), rhSCF (100 ng/mL), rhIL-3 (20 ng/mL), and rhIL-6 (20 ng/mL). Two different lentiviral depletion systems were used and were based on: KH1 vector (KH1 control, shDEK1, and shDEK2) and Mission shRNA vector (nontargeting sequence, shDEK#13104, and shDEK#04; Sigma-Aldrich). Lentiviral transduction was performed as described in “RNA knockdown” at a multiplicity of infection of 20. Three days after transduction, an aliquot of each culture was washed with HBSS/1% FBS/1% goat serum, and GFPhigh populations were selected by FACS for subsequent expansion for 12 hours and Western blot analysis. Separate aliquots of sorted cells were used in differentiation, proliferation, and CFU assays. Granulocytic differentiation assay of transduced cells was conducted in the presence of rhG-CSF (20 ng/mL) for up to 10 days, with fresh medium added every 3 days.
FACS analysis
For staining of surface markers, cells were prepared in HBSS/1% FBS/1% goat serum (The Jackson Laboratory) and labeled with multicolor mAb combinations. For intracellular staining, cells were fixed and permeabilized using Cytofix/Cytoperm (BD Biosciences). Samples were analyzed using an LSRII flow cytometer (BD Biosciences), and the resulting data were processed with FlowJo Version 7.6.4 software (TreeStar). Specific Ab conjugates and corresponding IgG controls used in this study included: PerCP/Cy5.5 CD11c (Bu15), PE CD11b (CBRM1/5), APC/Cy7 CD14 (M5E2), and Alexa Fluor 647 CD34 (4H11), all from BioLegend; Alexa Fluor 647 CD11b (CBRM1/5) and Pacific blue CD15 (HI98) from eBiosciences; PE-Cy7 CD16 (GO22) from BD Biosciences; and PE lactoferrin (CLB13.17) from Beckman Coulter. Cell-cycle analysis was conducted as described previously.33
Hematopoietic CFU assay
Hematopoietic clonogenic assays were performed using MethoCultSF (4536) complete methylcellulose medium with the following cytokines: rhSCF, rhGM-CSF, rhG-CSF, rhIL-3, and rhIL-6 (StemCell Technologies). Cell-plating densities were optimized and 1000 cells were seeded in triplicate. Colonies were scored after 14 days of incubation based on morphologic criteria such as granulocyte/macrophage (CFU-GM), mixed granulocyte/monocyte/megakaryocyte (CFU-GMM), granulocyte (CFU-G), and macrophage (CFU-M). Large colonies containing multiple centers of myeloid (granulocytes and macrophages) and erythromegakaryocytic origin were identified as multilineage (CFU-GMM) colonies. Colonies were counted on an Optipat 2 microscope (Nikon).
Results
DEK interacts with C/EBPα on chromatin as a function of S21 phosphorylation
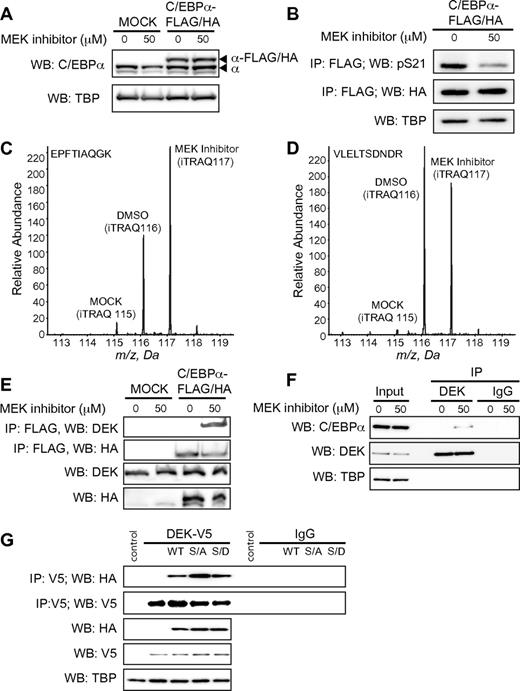
Given the role of C/EBPα in cell-cycle arrest, we reasoned that a successful proteomics screen of C/EBPα protein complexes would require an inducible and affinity-tagged form of C/EBPα. We generated a tetracycline-inducible, C-terminally tagged form of C/EBPα (C/EBPα–FLAG/HA) in MOLM-14 cells, a line originally derived from the peripheral blood of a patient with AML and characterized by the mutation FLT3-ITD. We chose MOLM-14 cells for our model system because they recapitulate the reported molecular links between oncogenic FLT3 and C/EBPα.21 Figure 1A shows that C/EBPα–FLAG/HA expression was comparable to the level of endogenous protein in the chromatin fraction. Furthermore, treatment with a small-molecule MEK inhibitor did not affect the expression of C/EBPα but did reduce S21 phosphorylation (Figure 1B). We also verified the DNA-binding activity of C/EBPα–FLAG/HA (supplemental Figure 1). These data demonstrate that the Tet-inducible C/EBPα–FLAG/HA MOLM-14 cell line is a suitable model with which to study C/EBPα protein interactions as a function of S21 phosphorylation.
DEK interacts preferentially with dephosphorylated C/EBPα. (A) Expression of C/EBPα–FLAG/HA and endogenous C/EBPα proteins in the chromatin fraction of MOLM-14 cells after doxycycline withdrawal for 14 hours and subsequent treatment with MEK inhibitor PD 98059 for 6 hours. TATA-binding protein (TBP) served as the loading control. (B) Detection of S21 phosphorylation on C/EBPα–FLAG/HA immunoprecipitated from the chromatin compartment 6 hours after treatment with MEK inhibitor. Representative quantification data (iTRAQ reporter ion signals) corresponding to DEK peptide (C; EPFTIAQGK) and C/EBPα peptide (D; VLELTSDNDR) after treatment with MEK inhibitor, immunoprecipitation of C/EBPα, and proteomics analysis, as described in supplemental Methods and supplemental Figure 2. (E) Detection of DEK protein after immunoprecipitation of C/EBPα–FLAG/HA from the chromatin fraction of MOLM-14 cells subjected to doxycycline induction and treatment with MEK inhibitor. Western blots against DEK and C/EBPα–FLAG/HA in total cell extracts served as a control for each immunoprecipitation. (F) Western blot for C/EBPα after immunoprecipitation of endogenous DEK from MOLM-14 cells treated with MEK inhibitor. Immunoblot against TBP served as a loading control. (G) Coimmunoprecipitation of C/EBPα–FLAG/HA WT and mutants S21A and S21D (designated as WT, S/A and S/D) by DEK-V5 in 293T cells. Western blots were performed against V5 and HA, with TBP probed as a loading control.
DEK interacts preferentially with dephosphorylated C/EBPα. (A) Expression of C/EBPα–FLAG/HA and endogenous C/EBPα proteins in the chromatin fraction of MOLM-14 cells after doxycycline withdrawal for 14 hours and subsequent treatment with MEK inhibitor PD 98059 for 6 hours. TATA-binding protein (TBP) served as the loading control. (B) Detection of S21 phosphorylation on C/EBPα–FLAG/HA immunoprecipitated from the chromatin compartment 6 hours after treatment with MEK inhibitor. Representative quantification data (iTRAQ reporter ion signals) corresponding to DEK peptide (C; EPFTIAQGK) and C/EBPα peptide (D; VLELTSDNDR) after treatment with MEK inhibitor, immunoprecipitation of C/EBPα, and proteomics analysis, as described in supplemental Methods and supplemental Figure 2. (E) Detection of DEK protein after immunoprecipitation of C/EBPα–FLAG/HA from the chromatin fraction of MOLM-14 cells subjected to doxycycline induction and treatment with MEK inhibitor. Western blots against DEK and C/EBPα–FLAG/HA in total cell extracts served as a control for each immunoprecipitation. (F) Western blot for C/EBPα after immunoprecipitation of endogenous DEK from MOLM-14 cells treated with MEK inhibitor. Immunoblot against TBP served as a loading control. (G) Coimmunoprecipitation of C/EBPα–FLAG/HA WT and mutants S21A and S21D (designated as WT, S/A and S/D) by DEK-V5 in 293T cells. Western blots were performed against V5 and HA, with TBP probed as a loading control.
We also used quantitative proteomics to analyze multicomponent C/EBPα protein complexes (supplemental Methods and supplemental Figure 2). We identified more than 65 proteins specifically associated with C/EBPα–FLAG/HA complexes, spanning diverse functional classes such as chromatin organization (DEK, SET, CTCF, CBX3, and CBX5), transcription initiation (ILF2, ILF3, TPX2, N-PAC, and others), and elongation (SSRP1, SPT16, ruvB-like1, and others; supplemental Table 1). In addition, we detected many proteins that had been reported previously to interact with C/EBPα, including: PARP1,34 SMARCC1 and SMARCC2,35 Jun-D,11 CREB1,36 UBF-1,37 and C/EBPβ.38 We also observed that participation of several proteins in the C/EBPα complex was dependent on the phosphorylation status of S21 (Table 1), supporting our hypothesis that C/EBPα protein complexes are remodeled in the context of oncogenic FLT3 signaling. We next queried the Online Mendelian Inheritance in Man (OMIM) database and identified DEK, NUMA1, CBFB, and SET as C/EBPα partners with genetic links to hematopoietic malignancy. From these candidates, we selected DEK for further study because it was detected reproducibly across 3 independent bioreplicate IPs and had not been reported previously in association with C/EBPα. Figure 1C-D shows the iTRAQ reporter ions from representative DEK and C/EBPα peptides, respectively; the observed ratios indicate that (1) DEK and C/EBPα–FLAG/HA were not purified in appreciable quantities from the negative control, (2) C/EBPα–FLAG/HA was purified in equal quantities from cultures ± MEK inhibitor, and (3) participation of DEK in the C/EBPα complex was correlated inversely with S21 phosphorylation, suggesting that reversible phosphorylation at this site mediates protein-protein interactions on chromatin.
Proteins detected with higher relative abundance in association with dephosphorylated S21 C/EBPα–FLAG/HA multicomponent complexes
| Protein description . | Accession no. . | Gene name . | Experiments detected, n . | Ratio, Inh/dmso . | Log ratio, Inh/dmso . |
|---|---|---|---|---|---|
| Protein SET isoform 2 | gi 170763498 | SET | 1 | 3.0 | 0.477 |
| Antigen KI-67 isoform 1 | gi 103472005 | MKI67 | 1 | 2.0 | 0.309 |
| Transformer-2 protein homolog beta | gi 4759098 | SFRS10 | 3 | 2.0 | 0.292 |
| Transcription elongation factor SPT6 | gi 27597090 | SUPT6H | 2 | 1.8 | 0.261 |
| Peptidyl-prolyl cis-trans isomerase B precursor | gi 4758950 | PPIB | 2 | 1.7 | 0.241 |
| Histone H2A type 1 | gi 4504239 | HIST1H2AI | 3 | 1.7 | 0.240 |
| Poly (ADP-ribose) polymerase 1 | gi 156523968 | PARP1 | 3 | 1.7 | 0.239 |
| Serine/arginine-rich splicing factor 1 isoform 1 | gi 5902076 | SFRS1 | 3 | 1.7 | 0.221 |
| Chromobox protein homolog 5 | gi 6912292 | CBX5 | 1 | 1.7 | 0.219 |
| Protein DEK isoform 1 | gi 4503249 | DEK | 3 | 1.6 | 0.212 |
| Nucleolin | gi 55956788 | NCL | 3 | 1.6 | 0.210 |
| Nucleolar RNA helicase 2 | gi 50659095 | DDX21 | 2 | 1.6 | 0.207 |
| Chromobox protein homolog 1 | gi 5803076 | CBX1 | 1 | 1.6 | 0.203 |
| Protein ELYS | gi 262359929 | AHCTF1 | 3 | 1.6 | 0.202 |
| Peptidyl-prolyl cis-trans isomerase A | gi 10863927 | PPIA | 2 | 1.6 | 0.198 |
| CCAAT/enhancer binding protein alpha | gi 28872794 | CEBPA | 3 | 0.9 | −0.051 |
| Protein description . | Accession no. . | Gene name . | Experiments detected, n . | Ratio, Inh/dmso . | Log ratio, Inh/dmso . |
|---|---|---|---|---|---|
| Protein SET isoform 2 | gi 170763498 | SET | 1 | 3.0 | 0.477 |
| Antigen KI-67 isoform 1 | gi 103472005 | MKI67 | 1 | 2.0 | 0.309 |
| Transformer-2 protein homolog beta | gi 4759098 | SFRS10 | 3 | 2.0 | 0.292 |
| Transcription elongation factor SPT6 | gi 27597090 | SUPT6H | 2 | 1.8 | 0.261 |
| Peptidyl-prolyl cis-trans isomerase B precursor | gi 4758950 | PPIB | 2 | 1.7 | 0.241 |
| Histone H2A type 1 | gi 4504239 | HIST1H2AI | 3 | 1.7 | 0.240 |
| Poly (ADP-ribose) polymerase 1 | gi 156523968 | PARP1 | 3 | 1.7 | 0.239 |
| Serine/arginine-rich splicing factor 1 isoform 1 | gi 5902076 | SFRS1 | 3 | 1.7 | 0.221 |
| Chromobox protein homolog 5 | gi 6912292 | CBX5 | 1 | 1.7 | 0.219 |
| Protein DEK isoform 1 | gi 4503249 | DEK | 3 | 1.6 | 0.212 |
| Nucleolin | gi 55956788 | NCL | 3 | 1.6 | 0.210 |
| Nucleolar RNA helicase 2 | gi 50659095 | DDX21 | 2 | 1.6 | 0.207 |
| Chromobox protein homolog 1 | gi 5803076 | CBX1 | 1 | 1.6 | 0.203 |
| Protein ELYS | gi 262359929 | AHCTF1 | 3 | 1.6 | 0.202 |
| Peptidyl-prolyl cis-trans isomerase A | gi 10863927 | PPIA | 2 | 1.6 | 0.198 |
| CCAAT/enhancer binding protein alpha | gi 28872794 | CEBPA | 3 | 0.9 | −0.051 |
For reference, data for C/EBPα are listed in the last row. Detected protein ratio in inhibitor-treated cells versus control is denoted as Inh/dmso.
We verified the pS21-dependent association of DEK in C/EBPα protein complexes by coimmunoprecipitation followed by Western blot in C/EBPα–FLAG/HA MOLM-14 cells (Figure 1E). For the reverse coimmunoprecipitation, endogenous C/EBPα was only detected when DEK was immunoprecipitated in the presence of the MEK inhibitor (Figure 1F). To further explore the pS21-dependent nature of the C/EBPα–DEK interaction, we expressed wild-type (WT), phospho-null (S21A), and phospho-mimetic (S21D) C/EBPα, along with V5-tagged DEK in HEK 293T cells (this line does not express detectable levels of endogenous C/EBPα nor do they harbor activated forms of FLT3). Immunoprecipitation directed against V5, followed by Western blot for C/EBPα, revealed that DEK interacted preferentially with the dephosphorylated form of C/EBPα (Figure 1G).
C/EBPα and DEK colocalize in the nucleus
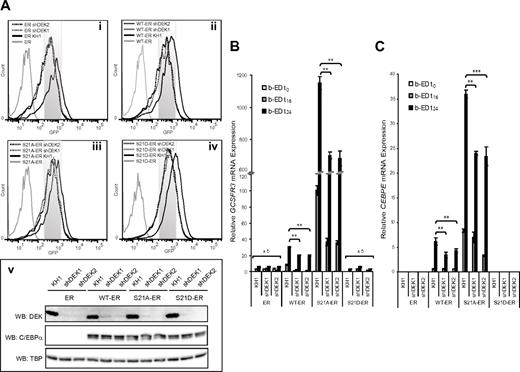
We used a PLA to provide further evidence for colocalization of C/EBPα and DEK. First we carried out shRNA-mediated protein depletion with lentiviral particles containing either a control vector (KH1) or a sequence targeting DEK (shDEK1)39 in myeloid K562 cells that expressed either WT C/EBPα fused to the estrogen receptor ligand–binding domain (WT-ER)24 or a control (ER domain alone). The WT-ER cells provide a useful model system with which to study the dynamics of C/EBPα–dependent granulocytic differentiation24,40 because they respond to β-estradiol treatment by nuclear translocation of C/EBPα–ER protein, followed by up-regulation of the C/EBPα target genes GCSFR3 and CEBPE. The nuclear localization of C/EBPα–ER quickly reaches its maximum and is completely eliminated by 24 hours (data not shown). Western blot analysis demonstrated that C/EBPα is expressed only in WT-ER cells and that DEK protein expression was efficiently abrogated in ER/shDEK1 and ER-WT/shDEK1 cells (Figure 2A). PLA assays confirmed these results and also demonstrated that both proteins are localized individually in the nucleus (supplemental Figure 3A). Moreover, probing with PLA reagents in WT-ER/KH1 cells generated readily detectable foci (Figure 2Bi), suggesting that C/EBPα and DEK were closely colocalized in the nuclei. The reaction specificity was confirmed in cells lacking either DEK or C/EBPα (Figure 2Bii-iii) and in cells lacking both proteins (Figure 2Biv). Our proteomics, coimmunoprecipitation, and PLA data confirm that C/EBPα and DEK are closely associated within a multicomponent complex on chromatin and that phosphorylation of S21 diminishes this interaction.
C/EBPα and DEK colocalize in K562 cells. K562 lines expressing control (ER alone) and C/EBPα–ER (WT-ER) were transduced with lentiviral particles encoding GFP alone (KH1) or GFP along with shRNA targeting DEK (shDEK1). After 48 hours, GFP+ populations were enriched by FACS. (A) Aliquots of sorted populations were probed by immunoblot for C/EBPα, DEK, and TATA-binding protein protein expression 24 hours after FACS enrichment. (B) Separate aliquots of sorted populations were treated with β-estradiol for 2 hours to induce C/EBPα nuclear translocation. Fixed cells were incubated with Abs against DEK and C/EBPα, followed by incubation with matched OLINK in situ PLA probes and imaged with fluorescence microscopy using blue (DAPI) and red (TxRed) filters, respectively (see supplemental Methods and supplemental Figure 3A-B for protocol, control images, and single-channel images, respectively). The scale bar represents 10 μm.
C/EBPα and DEK colocalize in K562 cells. K562 lines expressing control (ER alone) and C/EBPα–ER (WT-ER) were transduced with lentiviral particles encoding GFP alone (KH1) or GFP along with shRNA targeting DEK (shDEK1). After 48 hours, GFP+ populations were enriched by FACS. (A) Aliquots of sorted populations were probed by immunoblot for C/EBPα, DEK, and TATA-binding protein protein expression 24 hours after FACS enrichment. (B) Separate aliquots of sorted populations were treated with β-estradiol for 2 hours to induce C/EBPα nuclear translocation. Fixed cells were incubated with Abs against DEK and C/EBPα, followed by incubation with matched OLINK in situ PLA probes and imaged with fluorescence microscopy using blue (DAPI) and red (TxRed) filters, respectively (see supplemental Methods and supplemental Figure 3A-B for protocol, control images, and single-channel images, respectively). The scale bar represents 10 μm.
The DEK-C/EBPα interaction regulates activation of GCSFR3
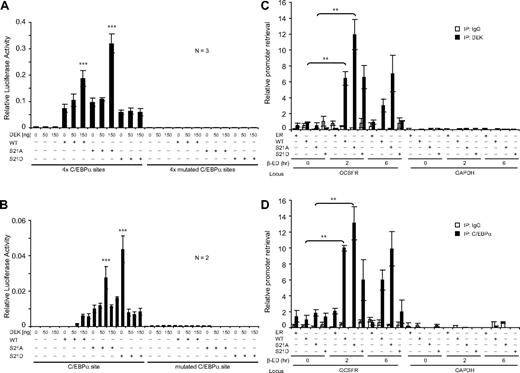
Having established that a bona fide and phosphorylation-dependent interaction existed between DEK-C/EBPα, we next investigated whether DEK played a functional role in the activation of GCSFR3, a myeloid target gene of C/EBPα. We used a luciferase reporter construct containing the TK minimal promoter with tetramers of either a WT or mutated C/EBPα–binding site from the human GCSFR3 promoter.18 These experiments were performed in 293T cells that express endogenous DEK protein but have no detectable C/EBPα. Figure 3A shows that DEK activated the WT C/EBPα–dependent reporter in a dose-dependent manner only when expressed with either the C/EBPα WT or S21A phospho-null mutant. Conversely, neither the individual nor combined expression of C/EBPα and DEK was sufficient to drive luciferase activity of the mutant reporter construct. To explore the possibility that DEK has affinity to regions of the GCSFR3 promoter that neighbor the C/EBPα–binding site, we next used luciferase constructs that contained a short sequence (+74/−67) derived from the native GCSFR3 promoter region or the corresponding sequence with a mutated C/EBPα–binding site.41 Again, we observed a dose-dependent response to DEK only in the presence of the WT and S21A C/EBPα expression vectors (Figure 3B), suggesting that DEK cooperates with dephosphorylated C/EBPα to augment transactivation capacity.
DEK binds in vivo to the GCSFR3 promoter region in a C/EBPα pS21–dependent manner to enhance C/EBPα transactivation capacity. (A) Cells (293T) were cotransfected in triplicate with C/EBPα–WT, S21A, and S21D (0 or 50 ng); increasing quantities of DEK (0, 50, or 150 ng); and a luciferase reporter driven by the TK minimal promoter along with a repeat of 4 C/EBPα–binding sites corresponding to those on the GCSFR3 promoter (left) or a vector containing 4 mutated sequences of the C/EBPα binding site as a negative control (right). (B) Cells (293T) were cotransfected in duplicate with C/EBPα (0 or 50 ng), increasing quantities of DEK (0, 50, or 150 ng), and a luciferase reporter driven by a short sequence (+74/−67) corresponding to the native GCSFR3 promoter region (left) or the same GCSFR3 promoter sequence but with a mutated C/EBPα binding site as a negative control (right). Transfection efficiency was normalized by cotransfection with Renilla TK (7 ng; Promega). Error bars indicate the SD resulting from independent experiments (n = 3 or 2). (C-D) Cross-linked chromatin was prepared from C/EBPα mutants (WT-ER, S21A-ER, and S21D-ER) and control (ER) K562 cells treated with 1μM β-estradiol for 0, 2, and 6 hours, and then immunoprecipitated using either anti-DEK (C) or C/EBPα (D) Abs. The precipitated chromatin was subjected to quantitative PCR using primer pairs that spanned the promoters of human GCSFR3 and GAPDH, respectively. Immunoprecipitation with rabbit IgG served as a negative control. Quantitative PCR signals of the immunoprecipitated chromatin were normalized against that from the input chromatin (1:10 dilution). Error bars indicate the SD resulting from 2 independent experiments performed in triplicate. ***P < .001; **P < .01.
DEK binds in vivo to the GCSFR3 promoter region in a C/EBPα pS21–dependent manner to enhance C/EBPα transactivation capacity. (A) Cells (293T) were cotransfected in triplicate with C/EBPα–WT, S21A, and S21D (0 or 50 ng); increasing quantities of DEK (0, 50, or 150 ng); and a luciferase reporter driven by the TK minimal promoter along with a repeat of 4 C/EBPα–binding sites corresponding to those on the GCSFR3 promoter (left) or a vector containing 4 mutated sequences of the C/EBPα binding site as a negative control (right). (B) Cells (293T) were cotransfected in duplicate with C/EBPα (0 or 50 ng), increasing quantities of DEK (0, 50, or 150 ng), and a luciferase reporter driven by a short sequence (+74/−67) corresponding to the native GCSFR3 promoter region (left) or the same GCSFR3 promoter sequence but with a mutated C/EBPα binding site as a negative control (right). Transfection efficiency was normalized by cotransfection with Renilla TK (7 ng; Promega). Error bars indicate the SD resulting from independent experiments (n = 3 or 2). (C-D) Cross-linked chromatin was prepared from C/EBPα mutants (WT-ER, S21A-ER, and S21D-ER) and control (ER) K562 cells treated with 1μM β-estradiol for 0, 2, and 6 hours, and then immunoprecipitated using either anti-DEK (C) or C/EBPα (D) Abs. The precipitated chromatin was subjected to quantitative PCR using primer pairs that spanned the promoters of human GCSFR3 and GAPDH, respectively. Immunoprecipitation with rabbit IgG served as a negative control. Quantitative PCR signals of the immunoprecipitated chromatin were normalized against that from the input chromatin (1:10 dilution). Error bars indicate the SD resulting from 2 independent experiments performed in triplicate. ***P < .001; **P < .01.
We next used the ChIP assay in K562 cells expressing C/EBPα mutants (WT-ER, S21A-ER, and S21D-ER)24 or a negative control (ER) to determine whether DEK participated in C/EBPα transcription complexes at the endogenous GCSFR3 promoter. DEK immunoprecipitation followed by PCR with primers to the GCSFR3 promoter region12 revealed that this protein bound to DNA specifically and in close proximity to the canonical C/EBPα–binding site on GCSFR3 (Figure 3C). Binding of DEK to the GCSFR3 promoter region was in direct response to C/EBPα nuclear localization (via β-estradiol stimulation), as evidenced by the lack of signal before β-estradiol stimulation. We also observed increased binding of DEK to the GCSFR3 promoter region in the presence of C/EBPα S21A-ER. Conversely, there was no binding of DEK to the GAPDH promoter region under any of the conditions tested. We observed similar results for ChIP analysis performed with an Ab to C/EBPα (Figure 3D). The kinetic profiles shown in Figure 3A-B are consistent with colocalization of DEK and C/EBPα at the GCSFR3 promoter region (prolonged incubation with β-estradiol leads to the elimination of C/EBPα–ER from chromatin, data not shown). These data demonstrate that localization of C/EBPα at the GCSFR3 promoter enhances DEK binding to the same regulatory region.
Based on previous reports suggesting that dysregulation of C/EBPα and DEK are implicated independently in the pathogenesis of AML,29,42 and our evidence that DEK augmented C/EBPα transactivation capacity in vitro, we speculated that these proteins coordinately regulate myeloid gene expression. To test this hypothesis, we used K562 cells expressing C/EBPα mutants (WT-ER, S21A-ER, and S21D-ER)24 or a negative control (ER), along with lentiviral vectors carrying GFP and an insertless vector control, a single DEK shRNA sequence, or two different shRNA sequences (in same vector)39 targeting DEK. Each shRNA construct abolished DEK protein expression in these cell lines (Figure 4A). Depletion of DEK in K562 ER cells did not affect GCSFR3 mRNA levels (Figure 4B). Consistent with our luciferase data, we observed that endogenous DEK (KH1) in the presence of WT (WT-ER) or dephosphorylated (S21A-ER) C/EBPα drove robust, time-dependent expression of GCSFR3 mRNA upon C/EBPα nuclear translocation. Depletion of DEK reduced C/EBPα–mediated GCSFR3 expression in WT-ER and S21A-ER cells. In contrast, we observed that GCSFR3 mRNA expression was low and largely independent of both DEK and the phosphomimetic form (S21D-ER) of C/EBPα. CEBPE mRNA levels were similarly dependent on the coexpression of DEK with either WT or S21A-ER C/EBPα, albeit with somewhat different kinetics (Figure 4C). These data provide further support for the hypothesis that DEK is a coactivator of C/EBPα–mediated transcription. The affinity of DEK to dephosphorylated C/EBPα, along with its capacity to positively regulate C/EBPα–induced transcription, suggests that DEK is required for the assembly of a functionally active complex at the promoter of GCSFR3.
DEK is required for C/EBPα–induced expression of GCSFR3 and CEBPE in vitro. K562 cells expressing control (ER) and C/EBPα mutants (WT-ER, S21A-ER, and S21D-ER) were transduced (72 hours) with lentiviral particles carrying either shRNA sequences that targeted DEK (shDEK1 or shDEK2) or an insertless construct as a negative control (KH1). (A) GFP+ cells (i-iv) were assessed by immunoblot for expression of DEK and C/EBPα, along with TATA-binding protein as a loading control (v). Expression of GCSFR3 (B) and CEBPE (C) mRNA in sorted populations after β-estradiol treatment for 0, 16, and 24 hours. Expression levels are normalized against mRNA expression of 36B4. Error bars indicate the SD resulting from 2 independent experiments performed in triplicate. ***P < .001; **P < .01.
DEK is required for C/EBPα–induced expression of GCSFR3 and CEBPE in vitro. K562 cells expressing control (ER) and C/EBPα mutants (WT-ER, S21A-ER, and S21D-ER) were transduced (72 hours) with lentiviral particles carrying either shRNA sequences that targeted DEK (shDEK1 or shDEK2) or an insertless construct as a negative control (KH1). (A) GFP+ cells (i-iv) were assessed by immunoblot for expression of DEK and C/EBPα, along with TATA-binding protein as a loading control (v). Expression of GCSFR3 (B) and CEBPE (C) mRNA in sorted populations after β-estradiol treatment for 0, 16, and 24 hours. Expression levels are normalized against mRNA expression of 36B4. Error bars indicate the SD resulting from 2 independent experiments performed in triplicate. ***P < .001; **P < .01.
DEK is necessary for normal granulocytic differentiation of CD34+ cells
C/EBPα regulates granulocytic differentiation via transcriptional activation of myeloid genes (eg, CEBPE, GCSFR3, MPO, ELANE, ITAX, and LTF), along with concomitant repression of proliferation through direct interactions with cell-cycle machinery proteins.8 Independent reports have demonstrated that DEK participates in the transcription regulation of CD21/CR2, APOE, cIAP2, and IL-8 in vitro and that the translocation product DEK-CAN has the potential to initiate a leukemic phenotype in hematopoietic stem cells.26 However, a functional link for DEK in myeloid differentiation has not been established. Multipotent hematopoietic CD34+ progenitor cells can undergo granulocytic differentiation after stimulation with the G-CSF ligand. To verify that human BM CD34+ cells are a viable model with which to study the association of C/EBPα and DEK, we first evaluated C/EBPα and DEK protein expression after G-CSF treatment for up to 10 days. Supplemental Figure 4A shows that C/EBPα and DEK were coexpressed after G-CSF stimulation. In parallel, we verified that the expression of the myeloid markers CD11b, CD15, CD11c, and lactoferrin exhibited a robust response to G-CSF (supplemental Figure 4B), whereas expression of the intermediate myeloid marker CD38 increased modestly and then declined (data not shown), which is consistent with the molecular profile of myelocytes or metamyelocytes.43,44 These data confirmed the expression of C/EBPα and DEK during differentiation and suggest that our ex vivo model recapitulates granulopoiesis as defined by coordinate expression of specific surface markers. To assess whether DEK is functionally required for terminal myeloid differentiation, we used shRNA-based depletion of DEK in human BM-derived CD34+ cells and observed a significant reduction in DEK protein levels using 2 different lentiviral constructs (Figure 5A-B). We next assessed the role of DEK in myeloid colony formation. Figure 5C and D show that depletion of DEK reduced the capacity of CD34+ cells to form granulocytic colonies (CFU-G), whereas bipotent granulocytic-monocytic colonies (CFU-GM) increased more than 2-fold. Finally, DEK-depleted populations and corresponding controls were subjected to a differentiation assay and analyzed separately for expression of surface markers to establish their relative capacity to undergo myeloid differentiation. Figure 5E through H shows that depletion of DEK inhibited granulocytic differentiation, as evidenced by reduced levels of CD11b (10% and 8%, respectively) and CD15 (9% and 19%, respectively) compared with control cells. DEK depletion did not affect the expression of the granulocytic markers MPO or LF. Likewise, both CD16, a marker for activated granulocytes, and the monocytic marker CD14 were unaffected under these conditions (data not shown). Interestingly, we observed a reduction in CEBPE and GCSFR3 mRNA levels in G-CSF–stimulated CD34+ cells that were depleted of DEK (Figure 5I-J). Because C/EBPα inhibits proliferation by inducing cell-cycle arrest, we evaluated the effect of DEK depletion on cell-cycle progression by staining CD34+ cells with propidium iodide 7 days after stimulation with G-CSF (supplemental Table 4). These data illustrate that DEK does not play a major role in CD34+ cell-cycle regulation. In summary, our findings suggest that C/EBPα–DEK complexes couple GCSFR3-mediated signaling to the expression of specific genes within the myeloid differentiation program.
DEK is necessary for normal granulocytic differentiation of CD34+ cells. BM-derived CD34+ cells were transduced with lentiviral particles carrying shRNA sequences that targeted either DEK (shDEK1 or shDEK2) or an insertless construct as a negative control (KH1), or shRNA sequences targeting DEK #13104 and 04 or a nontargeting sequence (NT) as a negative control (Mission pLKO.1-puro UbC-TurboGFP vector, SHC014; Sigma-Aldrich; supplemental Table 3). After 3 days, separate aliquots of transduced cell cultures were assayed for either DEK protein expression or differentiation capacity. Cells were enriched for a GFPhigh population and then probed by Western blot using 2 different anti-DEK Abs to determine the efficiency of DEK depletion through use of KH1 (A) or Mission (B) constructs. TATA-binding protein was used as a loading control. GFPhigh enriched populations (1000 cells) from KH1 (C) and Mission (D) transduction constructs were plated in triplicate in MethoCult SF complete methylcellulose medium supplemented with cytokines the rhSCF, rhGM-CSF, rhG-CSF, rhIL-3, and rhIL-6. Colonies were scored after 14 days of incubation according to morphologic criteria: CFU-GM, CFU-GMM, CFU-G, or CFU-M. Each bar represents the mean of triplicate measurements ± SEM. (E-H) Transduced cells were incubated in the presence of rhG-CSF (20 ng/mL) for up to 10 days to induce granulocytic differentiation. Expression of the myeloid surface markers CD11b (E-F) and CD15 (G-H) were assessed after 7 or 10 days as indicated. Each bar represents the mean of 2 independent experiments ± SEM. Expression of CEBPE (I) and GCSFR3 (J) mRNA in GFPhigh populations carrying KH1 transduction constructs after G-CSF treatment for 0 and 4 days. Expression levels are normalized against mRNA expression of 36B4. Error bars indicate the SD resulting from 2 independent experiments performed in triplicate. ***P < .001; **P < .01; *P < .05.
DEK is necessary for normal granulocytic differentiation of CD34+ cells. BM-derived CD34+ cells were transduced with lentiviral particles carrying shRNA sequences that targeted either DEK (shDEK1 or shDEK2) or an insertless construct as a negative control (KH1), or shRNA sequences targeting DEK #13104 and 04 or a nontargeting sequence (NT) as a negative control (Mission pLKO.1-puro UbC-TurboGFP vector, SHC014; Sigma-Aldrich; supplemental Table 3). After 3 days, separate aliquots of transduced cell cultures were assayed for either DEK protein expression or differentiation capacity. Cells were enriched for a GFPhigh population and then probed by Western blot using 2 different anti-DEK Abs to determine the efficiency of DEK depletion through use of KH1 (A) or Mission (B) constructs. TATA-binding protein was used as a loading control. GFPhigh enriched populations (1000 cells) from KH1 (C) and Mission (D) transduction constructs were plated in triplicate in MethoCult SF complete methylcellulose medium supplemented with cytokines the rhSCF, rhGM-CSF, rhG-CSF, rhIL-3, and rhIL-6. Colonies were scored after 14 days of incubation according to morphologic criteria: CFU-GM, CFU-GMM, CFU-G, or CFU-M. Each bar represents the mean of triplicate measurements ± SEM. (E-H) Transduced cells were incubated in the presence of rhG-CSF (20 ng/mL) for up to 10 days to induce granulocytic differentiation. Expression of the myeloid surface markers CD11b (E-F) and CD15 (G-H) were assessed after 7 or 10 days as indicated. Each bar represents the mean of 2 independent experiments ± SEM. Expression of CEBPE (I) and GCSFR3 (J) mRNA in GFPhigh populations carrying KH1 transduction constructs after G-CSF treatment for 0 and 4 days. Expression levels are normalized against mRNA expression of 36B4. Error bars indicate the SD resulting from 2 independent experiments performed in triplicate. ***P < .001; **P < .01; *P < .05.
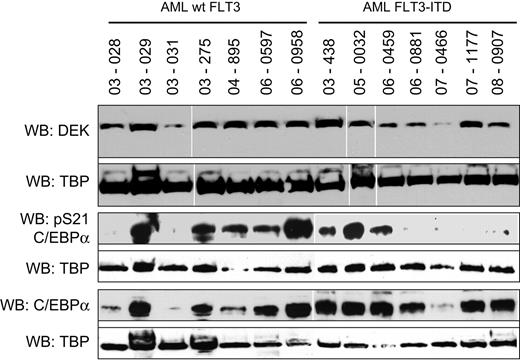
Cancer cell lines and primary CD34+ myeloid progenitors are valuable tools for the discovery and interrogation of functionally relevant protein interactions; however, biochemical validation of these molecular pathways in primary tumors is complicated by cellular and genomic heterogeneity. With these caveats in mind, we probed primary patient AML blasts genotyped as FLT3-WT or FLT3-ITD for their expression of DEK and C/EBPα and for their S21 phosphorylation status (Figure 6). We detected DEK protein in all of the AML samples, suggesting that the expression of DEK in primary blasts is independent of FLT3 activity. Expression of C/EBPα was detectable in 13 of 14 AML samples, whereas phosphorylation on S21 was observed in 8 of 14 patient samples. These data are consistent with our hypothesis that FLT3-mediated phosphorylation on S21 of C/EBPα disrupts protein interactions required for competent gene activation and contributes to differentiation arrest in AML.
DEK and C/EBPα protein expression in primary AML blasts. AML blasts were isolated from cryopreserved patient samples and lysed in RIPA buffer. Expression of DEK, pS21 C/EBPα, and C/EBPα was assessed by Western blot. TATA-binding protein was used as a loading control.
DEK and C/EBPα protein expression in primary AML blasts. AML blasts were isolated from cryopreserved patient samples and lysed in RIPA buffer. Expression of DEK, pS21 C/EBPα, and C/EBPα was assessed by Western blot. TATA-binding protein was used as a loading control.
Discussion
Myeloid cell fate is coordinately regulated by several transcription factors, including C/EBPα, PARP1, GATA1, PU.1, C-MYB, and AML1/CBFβ. C/EBPα functions as a primary molecular determinant of granulopoiesis and mediates the differentiation of granulocyte-monocyte progenitors through interactions with a diverse set of chromatin modifiers and transcription factors that collectively activate (CBP/p300,10 c-Jun,11 histone acetyltransferase TIP60,12 and Max13 ) or repress (CDP/cut45 and CA15046 ) specific target genes. Posttranslational modifications represent an additional regulatory mechanism for C/EBPα function.17 In particular, multiple studies have demonstrated that reversible phosphorylation on C/EBPα S21 attenuates transcription capacity and inhibits granulocytic21,24,47 or erythrocytic differentiation.48
We reasoned that reversible S21 phosphorylation may mediate the assembly and maintenance of competent transcription complexes at the promoters of C/EBPα target genes. To explore this hypothesis, in the present study, we used quantitative proteomics to profile C/EBPα–associated proteins and their response to S21 phosphorylation. Our results revealed several proteins not previously known to be associated with C/EBPα complexes. We chose DEK for further study because: (1) it was detected reproducibly across multiple proteomic analyses, (2) it was associated with C/EBPα in a pS21-dependent manner, and (3) it has well-established genetic links to leukemia. We confirmed that the interaction of DEK and C/EBPα is S21 phosphorylation dependent. Our quantitative analysis of multicomponent complexes formed in vivo complements previous studies21,24,40 and provides further evidence that the N-terminal region of C/EBPα mediates interactions that are critical for transcription regulation.
The transcription activity of C/EBPα is connected intrinsically with GCSFR-mediated signaling. For example, whereas GCSFR3 is a direct C/EBPα target gene,41 the coordinate activity of both pathways is required for the regulated expression of MPO and ELANE in 32Dcl3 cells.49 Furthermore, G-CSF stimulation of myeloid FDN1.1 cells activates the expression of MPO and CCR2 through recruitment of C/EBPα to the respective promoters and activating modifications on associated histones.14 The results of the present study provide multiple lines of evidence to suggest that DEK is an important coactivator of C/EBPα–induced expression of GCSFR3. First, we demonstrated that DEK enhances C/EBPα transactivation capacity on both synthetic and native GCSFR3 promoters, and that recruitment of DEK to the C/EBPα–binding region at GCSFR3 promoters occurs in a S21 phosphorylation–dependent manner. In addition, we demonstrated that C/EBPα–mediated activation of GCSFR3 and CEBPE was diminished in the absence of DEK in the context of either C/EBPα–WT or the dephosphorylated (S21A) mutant. It is important to note that, although multiple transcription factors (eg, PU.1 and SP1) cooperate with C/EBPα to promote GCSFR3 and CEBPE expression, we found in the present study that depletion of DEK inhibited the activation of these target genes, suggesting a functional role for DEK in C/EBPα–mediated gene expression. Consistent with previous results describing a positive regulatory role for DEK in AP2-dependent transcription in human malignant glioblastoma D17 cells, our present data demonstrate that DEK facilitates C/EBPα activity at the GCSFR3 promoter in myeloid cells. Interestingly, the phosphomimetic form (S21D-ER) of C/EBPα exhibited no apparent capacity to drive GCSFR3 or CEBPE expression in K562 cells despite binding to GCSFR3 promoters in vivo and activating luciferase constructs in vitro. These data provide compelling evidence that S21 phosphorylation plays a significant role in the assembly of transcriptionally competent C/EBPα protein complexes on chromatin. Given the multitude of C/EBPα target genes and, more generally, the complexity of transcription regulation, it is plausible that coactivators are required to ensure maximal activation of specific target genes. Our results suggest that DEK is a coregulator that is required for fully competent C/EBPα–mediated GCSFR3 transcription.
To define the role of DEK in granulocyte lineage commitment, we performed biochemical assays in a model system that encompassed endogenous protein levels, a relevant cellular context (ie, immature normal BM CD34+ cells), and an appropriate response to G-CSF, a cytokine that regulates proliferation of myeloid cells and their differentiation into mature neutrophils. Important limitations of our approach include the use of asynchronously differentiating cells and shRNA sequences that targeted only 1 of the 2 known DEK isoforms, both of which may partially obscure the role of DEK in the commitment of early progenitors. Our depletion experiments demonstrated that DEK is necessary for terminal differentiation of CD34+ cells, as evidenced by reduced expression of terminal neutrophil markers CD11b and CD15 in the context of G-CSF stimulation. We found that the expression levels of the monocytic marker CD14 were independent of DEK, as was the expression of CD16, a marker of activated granulocytes. Loss of DEK expression from myeloid precursors led to a reduced number of mature granulocyte colonies with a concomitant accumulation of granulocyte-monocyte colonies. This is consistent with the reduced expression of CD11b and CD15 after stimulation with G-CSF. We found no significant difference in monocyte colony formation when DEK was depleted in progenitor cells. Furthermore, neither the cell cycle nor colony size was influenced significantly as a result of DEK depletion for cells cultured in liquid or semisolid media, respectively. These results support a role for DEK specifically in granulocytic, rather than monocytic, lineage commitment.
Previous studies demonstrated that the expression of WT C/EBPα or the S21A mutant induced CD15 in CD34+ cells.7,48 Our present data demonstrate that depletion of DEK disrupts G-CSF–mediated expression of CD15 and terminal differentiation in CD34+ progenitors. These results suggest that DEK participates in a common pathway, including C/EBPα and GCSFR3, which drives CD15 expression and ultimately granulocytic differentiation. We observed reduced GCSFR3 and CEBPE transcription levels in the context of DEK loss-of-function in both C/EBPα-ER–induced K562 and G-CSF–stimulated CD34+ cells. The 2-fold reduction observed for expression of these target genes in our ex vivo differentiation assay suggests that DEK augments the GCSFR3- C/EBPα signaling axis in myeloid progenitors. Given the central role that C/EBPα plays in the expression of primary and secondary granulocytic genes, it is likely that a diverse set of protein partners, many of which are not recognized as canonical transcription factors, coregulate C/EBPα transcription activity in a gene-specific manner. Our results provide compelling evidence that proteins with disparate genetic links to AML can coordinately regulate normal hematopoiesis in the absence of somatic or other mutations. It will be of interest to probe the C/EBPα–DEK interaction in cells expressing a DEK-CAN translocation product given the fact that the latter has been implicated in the pathogenesis of AML.25
In the present study using a small panel of primary AML patient samples, we observed that DEK protein expression was independent of FLT3 status (WT or ITD). Moreover, several samples also exhibited robust phosphorylation on S21. These data are consistent with the notion that disruption of C/EBPα–DEK associations via pS21 may contribute to differentiation arrest in AML. In summary, our results demonstrate that DEK is a constituent of transcriptionally competent C/EBPα multicomponent protein complexes and that it participates in normal granulopoiesis. Based on the well-established roles of FLT3 and C/EBPα in AML, it is interesting to speculate that oncogenic signaling from FLT3-ITD commandeers downstream mediators (MEK and ERK) of endogenous pathways (eg, M-CSF and G-CSF) to disrupt key regulatory interactions (eg, DEK and C/EBPα on chromatin via pS21) at specific stages of lineage commitment (monocyte/granulocyte), ultimately coupling aberrant proliferation with differentiation arrest to support leukemogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff of the flow cytometry, confocal/light microscopy, and cell manipulation core facilities at the Dana-Farber Cancer Institute; the staff of The Ohio State University Leukemia Tissue Bank; Brian Bergeron and members of the Marto laboratory for technical assistance and useful discussions; Dr D. Markovitz (University of Michigan, Ann Arbor) for kindly providing KH1 DEK depletion constructs; Dr F. Kappes (University of Michigan, Ann Arbor) for valuable discussions; Dr S. Schwind (The Ohio State University, Columbus) for assistance with the identification and procurement of AML samples; and Drs C. Stiles and S. Blacklow (Dana-Farber Cancer Institute, Boston) for critical reading of the manuscript.
This work was supported by the Dana-Farber Cancer Institute, and the National Institutes of Health (P01NS047572 to J.M., P30 CA016058 to G.M., and CA118316 and DK080665 to D.T.).
National Institutes of Health
Authorship
Contribution: R.I.K. designed the experiments, performed the research, and analyzed the data; S.B.F. designed and implemented the RP-RP LC-MS/MS platform; H.S.R. and J.A.A. performed the research; M.J.C.-A. and C.J.L. provided reagents and guidance for the FACS analysis and differentiation assays; J.T.W. assisted with the proteomic data analysis; G.M. and D.G.T. provided reagents and guidance for the biochemical and differentiation assays; and J.A.M. and R.I.K. conceived of the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for H.S.R. is Comprehensive Cancer Center, The Ohio State University, Columbus, OH.
Correspondence: Jarrod A. Marto, Department of Cancer Biology, Dana-Farber Cancer Institute, 450 Brookline Ave, Smith 1158A, Boston, MA 02215-5450; e-mail: jarrod_marto@dfci.harvard.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal