Abstract
5-Azacytidine (5-azaC) is an azanucleoside approved for myelodysplastic syndrome. Approximately 80%-90% of 5-azaC is believed to be incorporated into RNA, which disrupts nucleic acid and protein metabolism leading to apoptosis. A smaller fraction (10%-20%) of 5-azaC inhibits DNA methylation and synthesis through conversion to decitabine triphosphate and subsequent DNA incorporation. However, its precise mechanism of action remains unclear. Ribonucleotide reductase (RR) is a highly regulated enzyme comprising 2 subunits, RRM1 and RRM2, that provides the deoxyribonucleotides required for DNA synthesis/repair. In the present study, we found for the first time that 5-azaC is a potent inhibitor of RRM2 in leukemia cell lines, in a mouse model, and in BM mononuclear cells from acute myeloid leukemia (AML) patients. 5-azaC–induced RRM2 gene expression inhibition involves its direct RNA incorporation and an attenuated RRM2 mRNA stability. Therefore, 5-azaC causes a major perturbation of deoxyribonucleotide pools. We also demonstrate herein that the initial RR-mediated 5-azaC conversion to decitabine is terminated through its own inhibition. In conclusion, we identify RRM2 as a novel molecular target of 5-azaC in AML. Our findings provide a basis for its more widespread clinical use either alone or in combination.
Introduction
5-Azacytidine (5-azaC; Vidaza) is a pyrimidine analog that was first synthesized 45 years ago and later found to be a natural product from Streptoverticillium ladakanus.1–3 Currently, 5-azaC is approved by the US Food and Drug Administration (FDA) for the treatment of myelodysplastic syndrome (MDS), but it has also been widely used for the treatment of acute myeloid leukemia (AML).4 Despite decades of efforts made to delineate the mechanisms of the antileukemia activity for this compound in terms of its interference with RNA and DNA metabolism,5 the precise basis of its clinical efficacy remains to be elucidated fully.6
After cell uptake, 5-azaC is first anabolized to its nucleoside monophosphate, 5-aza–CMP, by uridine-cytidine kinase (UCK) and is eventually phosphorylated by diphosphate kinase to its triphosphate metabolite, 5-aza–CTP, which is thought to be incorporated into RNA.7,8 Approximately 80%-90% of 5-azaC is processed through the entire sequence and incorporated into RNA.7 This ultimately disrupts mRNA and protein metabolism, leading to apoptosis.4,6 A small fraction (10%-20%) of the 5-azaC disphosphate intermediate product 5-aza–CDP is instead converted to 5-aza–dCDP by ribonucleotide reductase (RR), followed by further phosphorylation to 5-azaC–dCTP (5-aza-2'-deoxycyridine triphosphate; DAC-TP).4,7 DAC-TP is incorporated into DNA and covalently binds DNA methyltransferases (DNMTs), thereby inhibiting the activity of these proteins. It has been recognized that DNMTs add methyl groups to the cytosine in the context of the CpG dinucleotides in promoter region thereby promoting gene hypermethylation and epigenetic silencing. Thus 5-azaC has been classified as a hypomethylating agent. The deoxy-analog, decitabine (5-aza-2′-deoxycytidine, DAC), differs structurally from 5-azaC by having a deoxyribose and therefore does not require RR-mediated reduction before DNA incorporation. At higher doses (50-100 mg/m2/d), DAC is incorporated into DNA, leading to inhibition of DNA synthesis and cell death,9,10 whereas at lower doses (5-20 mg/m2/d), it induces DNA demethylation and reactivates hypermethylation-associated silenced tumor suppressor genes.11–13 DAC has also been approved for the treatment of MDS.
Whereas recent attention has focused on the hypomethylating activity of 5-azaC via its DNA pathway through the converted DAC,4 the antileukemia activity of this compound via its RNA pathway is less understood. In the present study, we report a previously unidentified activity of 5-azaC, showing it to be a potent inhibitor of M2 subunits of RR (RRM2) via underlying molecular mechanisms that involve 5-azaC direct RNA incorporation and an attenuated RRM2 mRNA stability. Inhibition of RR leads to a reduced deoxyribonucleotide pool, which is crucial for DNA synthesis and repair. Our present findings support a profoundly different mode of action between 5-azaC and DAC, which may ultimately explain some of the differences in their clinical activities.
Methods
Chemicals
5-azaC, DAC, cytidine, uridine, and 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) were purchased from Sigma-Aldrich. Triapine and GTI-2040 (LOR-2040) were obtained from the National Cancer Institute (Bethesda, MD). A 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) cell viability kit was purchased from Promega.
Cell lines, cell culture, and transfection
MV4-11 and K562 were obtained from ATCC and cultured in RPMI 1640 medium supplemented with 10% FBS, l-glutamine, and penicillin-streptomycin antibiotics (Invitrogen). Mononuclear cells (MNCs) from BM samples of 6 untreated AML patients were obtained from The Ohio State University Leukemia Tissue Bank. MNCs were prepared and cultured as described previously.14 All experiments with these cells were performed in accordance with the protocols approved by The Ohio State University institutional review board. GTI-2040 or siRRM2 (LOR-1284; Invitrogen) were transfected into cells by nucleoporation (Lonza).
Western blotting
Tissues or cell samples were lysed and the proteins were separated, transferred, and analyzed by immunoblotting. Abs to RRM2 (E-16), DNMT3a, and GAPDH were purchased from Santa Cruz Biotechnology; Abs to DNMT1 were from New England Biolabs; and Abs to β-actin were from Sigma-Aldrich. Protein expression was quantified by densitometry and normalized to GAPDH.
Intracellular deoxyribonucleotide/ribonucleotide, DAC, and DAC-TP Levels
Cells were lysed and intracellular deoxyribonucleotides/ribonucleotides, DAC, and DAC-TP were extracted and quantified by our previously described methods.15,16 Briefly, cell pellets were washed, deprotonized with 70% methanol, and sonicated in an ice bath. Cell extracts were centrifuged and the supernatant was dried under a stream of nitrogen. The residues were reconstituted with water and centrifuged again. A 50-μL aliquot of the supernatants was injected into an ion-trap liquid chromatography–tandem mass spectrometry (LC-MS/MS) system for deoxyribonucleotide and DAC-TP measurements.15 The DAC level was measured by the triple-quadruple LC-MS/MS system.16
Quantitative RT-PCR
Total RNA was isolated and cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Invitrogen). The cDNA templates and primers were then mixed with reagents from a SYBR Green PCR Master Mix (Applied Biosystems). The primers used were as follows: RRM2+: 5′-GCCTGGCCTCACATTTTCTAAT-3′; RRM2−: 5′-GAACATCAGGCAAGCAAAATCA-3′; abl+: 5′-TGGAGATTAACACTCTAAGCATAACTAAAGGT-3′; abl−: 5′-GATGTAGTTGCTTGGGACCCA-3′; GAPDH+: 5′-TCCACCCATGGCAAATTCC-3′; GAPDH−: 5′-TCGCCCCACTTGATTTTGG-3′. Reactions were carried out in triplicate in the StepOne Plus Real-time PCR system (Applied Biosystems), and data were analyzed with the comparative CT method. The amount of RRM2 mRNA was normalized by an internal control, abl. The relative changes in the treated groups were expressed as a percentage of the untreated control (arbitrarily set at 100).
mRNA stability
MV4-11 cells were treated with 1μM 5-azaC. At the same time, 1μM actinomycin D (Sigma-Aldrich) was used to block mRNA synthesis. Cells were harvested at various time points after actinomycin D treatment. RRM2, abl, and GAPDH mRNA levels were quantified by quantitative RT-PCR. Their half-lives were calculated from the plots of their mRNA levels as a function of time. We also performed the experiment in the presence of the potent protein-synthesis inhibitor cycloheximide (Sigma-Aldrich). MV4-11 cells were pretreated with 10 μg/mL of cycloheximide for 2 hours. Actinomycin D was added after the cells were treated with 5-azaC for 2 hours. The cells continued to be incubated with drugs for additional 4 hours and were then harvested for the measurement of RRM2 and abl mRNAs.
RNA reduction and hydrolysis
MV4-11 cells were treated with 5-azaC and the total RNA was isolated and incubated with NaBH4 or NaBD4. The solution was then neutralized to pH 7.0 by HCl. RNA hydrolysis was performed as described previously.17 Briefly, a 1/10 volume of 0.1M ammonium acetate (pH 5.3) and 2 units of nuclease P1 were added to 1 μg of total RNA. The mixture was incubated at 45°C for 2 hours, followed by the addition of 1/10 volume of 1M ammonium bicarbonate and 0.002 units of venom phosphodiesterase I. The resulting mixture was incubated at 37°C for 1 hour. After adding 0.5 units of alkaline phosphatase, the mixture was incubated at 37°C for another 1 hour. Fifty microliters of the resulting mixture was injected into LC-MS for quantification.
Xenograft mouse model
Male nu/nu athymic mice (4-6 weeks of age) were obtained from Charles River Laboratory. All experiments were conducted in accordance with the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care. MV4-11 cells were suspended with Matrigel (BD Biosciences) and subcutaneously inoculated into mice flanks. Tumor diameters were measured and body weights were monitored weekly after implantation. Treatments were initiated when the average tumor size reached 100-150 mm3. 5-azaC was injected IP at a dose of 20 mg/kg twice a week for 4 weeks. At the end of the experiment, the tumor tissues were collected for further analysis.
Results
5-azaC down-regulates RRM2 expression in vitro and in vivo
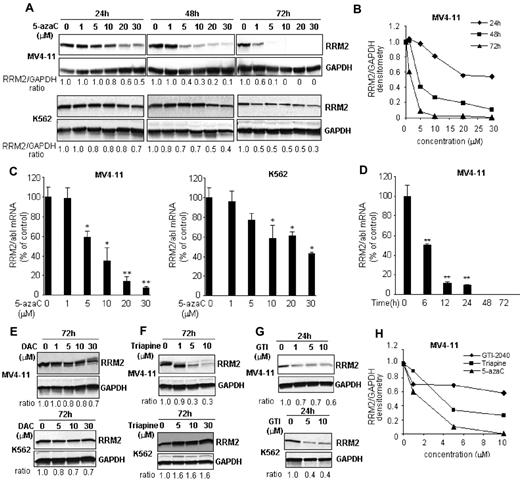
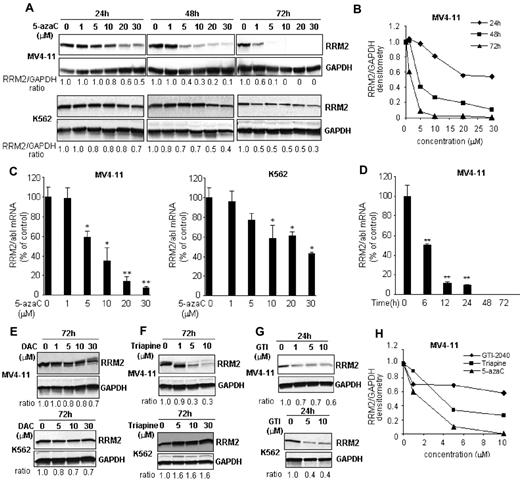
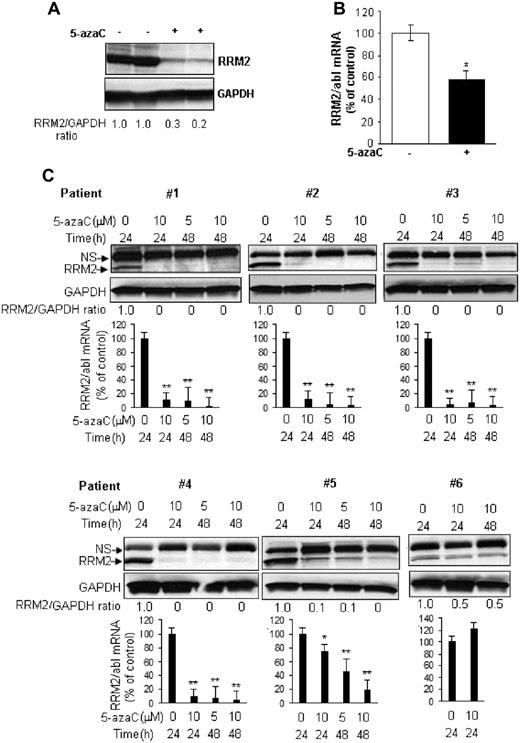
We initially intended to investigate the maximal RNA effects and the associated cytotoxicity of 5-azaC using a RRM2 antisense GTI-2040 to block its conversion to DAC. However, we discovered that the RRM2 protein was completely depleted in samples treated with only 5-azaC (control) during the course of the study. We therefore hypothesized that 5-azaC itself may interfere with the synthesis of RRM2, 1 of the 2 subunits of RR, somewhat limiting its own conversion to DAC and in turn DNA incorporation. To validate this hypothesis, we first characterized the activity of 5-azaC on RRM2 expression in leukemia cell lines. Cells were exposed to 5-azaC at clinically relevant concentrations (ie, 0.7-4.8μM) based on previous pharmacokinetics studies,18 and a robust dose- and time-dependent decrease of RRM2 protein was observed (Figure 1A-D). In MV4-11 cells, RRM2 protein was reduced by 60% at 48 hours and completely depleted at 72 hours with 5μM 5-azaC treatment (Figure 1A-B). RRM2 mRNA was also down-regulated by approximately 40% after 24 hours of treatment with 5μM 5-azaC (Figure 1C). K562 cells showed similar results, although they appeared to be less sensitive than MV4-11 cells (Figure 1A,C). At a 5μM concentration, 5-azaC only decreased RRM2 protein by 30% and 50% after 48 and 72 hours of incubation, respectively (Figure 1A).
5-azaC down-regulates RRM2 expression in vitro. (A) The RRM2 protein level was reduced by 5-azaC in a dose- and time-dependent manner in MV4-11 and K562 cells. Densitometry was performed to quantify each lane and the ratio of RRM2 over the loading control GAPDH is presented under each blot. (B) Representative densitometry plots. (C) 5-azaC down-regulates RRM2 mRNA in a dose-dependent manner. Cells were treated with serial concentrations of 5-azaC for 24 hours and collected for analysis. (D) 5-azaC down-regulates RRM2 mRNA in a time-dependent manner. MV4-11 cells were treated with 5μM 5-azaC for the indicated times and the RRM2 mRNA level was quantified. The RRM2 mRNA levels were normalized by an internal control, abl. Data are presented as the percentage of the untreated controls. *P < .05 and **P < .01 compared with the controls. (E) Decitabine (DAC) has minimal effects on RRM2 protein expressions. (F-G) Effects of 2 RRM2 inhibitors, Triapine (F) and GTI-2040 (G), on the RRM2 protein level. (H) Comparative plot of RRM2 protein levels in MV4-11 cells after 5-azaC, Triapine, or GTI-2040 treatment.
5-azaC down-regulates RRM2 expression in vitro. (A) The RRM2 protein level was reduced by 5-azaC in a dose- and time-dependent manner in MV4-11 and K562 cells. Densitometry was performed to quantify each lane and the ratio of RRM2 over the loading control GAPDH is presented under each blot. (B) Representative densitometry plots. (C) 5-azaC down-regulates RRM2 mRNA in a dose-dependent manner. Cells were treated with serial concentrations of 5-azaC for 24 hours and collected for analysis. (D) 5-azaC down-regulates RRM2 mRNA in a time-dependent manner. MV4-11 cells were treated with 5μM 5-azaC for the indicated times and the RRM2 mRNA level was quantified. The RRM2 mRNA levels were normalized by an internal control, abl. Data are presented as the percentage of the untreated controls. *P < .05 and **P < .01 compared with the controls. (E) Decitabine (DAC) has minimal effects on RRM2 protein expressions. (F-G) Effects of 2 RRM2 inhibitors, Triapine (F) and GTI-2040 (G), on the RRM2 protein level. (H) Comparative plot of RRM2 protein levels in MV4-11 cells after 5-azaC, Triapine, or GTI-2040 treatment.
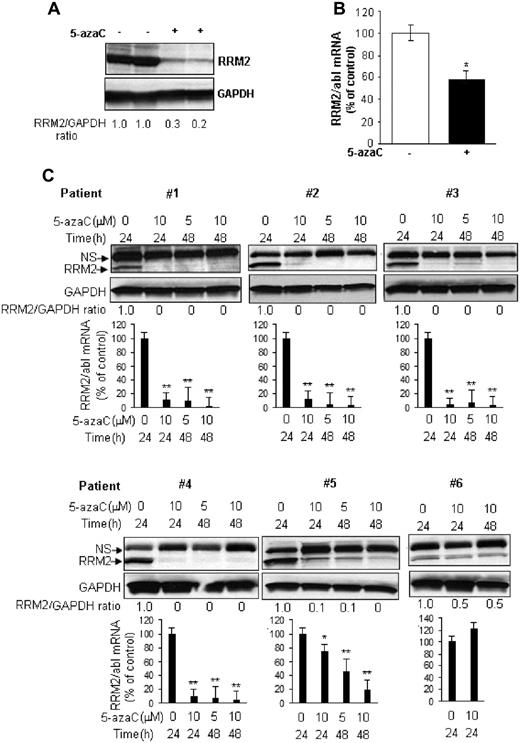
RRM2 down-regulation was specific to 5-azaC. Its azanucleoside analog, DAC, did not appreciably reduce RRM2 protein levels in MV4-11 and K562 cells (Figure 1E). In addition, we compared the potency of 5-azaC with 2 other established RRM2 inhibitors, Triapine and GTI-2040. Consistent with previous results,19–21 Triapine dose-dependently decreased the RRM2 protein level in MV4-11 but not in K562 cells (Figure 1F), whereas GTI-2040 down-regulated RRM2 in both MV4-11 and K562 cells (Figure 1G). 5-azaC was more potent than Triapine and GTI-2040, as shown by a leftward shift of the dose-response curve (Figure 1H). We also confirmed 5-azaC–mediated RRM2 down-regulation in vivo. MV4-11 cells were subcutaneously inoculated in athymic nu/nu mouse. When the tumor volume reached 100-150 mm3 in about 2 weeks, mice were intraperitoneally injected with 20 mg/kg of 5-azaC twice a weak. This 5-azaC dose for the treatment of mice is equivalent to the 60 mg/m2 dose used in humans.22 After 4 weeks of treatment, mice were killed and RRM2 levels were tested in the engrafted tissues. 5-azaC treatment decreased RRM2 by 40% and 70%-80% of mRNA and protein levels, respectively, compared with vehicle-treated controls (Figure 2A-B). These data demonstrate that 5-azaC is an in vitro and in vivo RRM2 inhibitor.
5-azaC down-regulates RRM2 expression ex vivo and in vivo. (A-B) 5-azaC inhibits RRM2 protein (A) and mRNA (B) expression in MV4-11 tumor xenografts in mice. Athymic nu/nu mice subcutaneously inoculated with MV4-11 cells were treated with 5-azaC and the engrafted tumor tissues were subjected to analysis. (C) 5-azaC reduces the RRM2 level in primary BM cells from AML patients. Primary cells from 6 patients were cultured in vitro and treated with the indicated concentrations of 5-azaC for 24 or 48 hours. Cells were then collected for Western blot and quantitative RT-PCR analyses. For Western blot analysis, densitometry was performed to quantify each lane, and the ratio of RRM2 over the loading control GAPDH is presented under each blot. For quantitative RT-PCR analysis, the data are presented as a percentage of the untreated controls. The results are means ± SD of the representative experiment performed in triplicate. *P < .05 and **P < .01 versus untreated controls.
5-azaC down-regulates RRM2 expression ex vivo and in vivo. (A-B) 5-azaC inhibits RRM2 protein (A) and mRNA (B) expression in MV4-11 tumor xenografts in mice. Athymic nu/nu mice subcutaneously inoculated with MV4-11 cells were treated with 5-azaC and the engrafted tumor tissues were subjected to analysis. (C) 5-azaC reduces the RRM2 level in primary BM cells from AML patients. Primary cells from 6 patients were cultured in vitro and treated with the indicated concentrations of 5-azaC for 24 or 48 hours. Cells were then collected for Western blot and quantitative RT-PCR analyses. For Western blot analysis, densitometry was performed to quantify each lane, and the ratio of RRM2 over the loading control GAPDH is presented under each blot. For quantitative RT-PCR analysis, the data are presented as a percentage of the untreated controls. The results are means ± SD of the representative experiment performed in triplicate. *P < .05 and **P < .01 versus untreated controls.
To further demonstrate the relevance of our findings to AML, we evaluated the effects of 5-azaC on RRM2 expression in primary MNCs from the BM from 6 AML patients (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Cells were treated in vitro with 5 or 10μM 5-azaC for 24 or 48 hours. As shown in Figure 2C, 5-azaC depleted both the mRNA and protein of RRM2 in samples from 4 patients (66%), induced a dose- and time-dependent RRM2 decrease in 1 patient (17%), and repressed only the RRM2 protein but not the mRNA in another patient (17%).
5-azaC perturbs the intracellular deoxyribonucleotide/ribonucleotide pool in leukemia cells
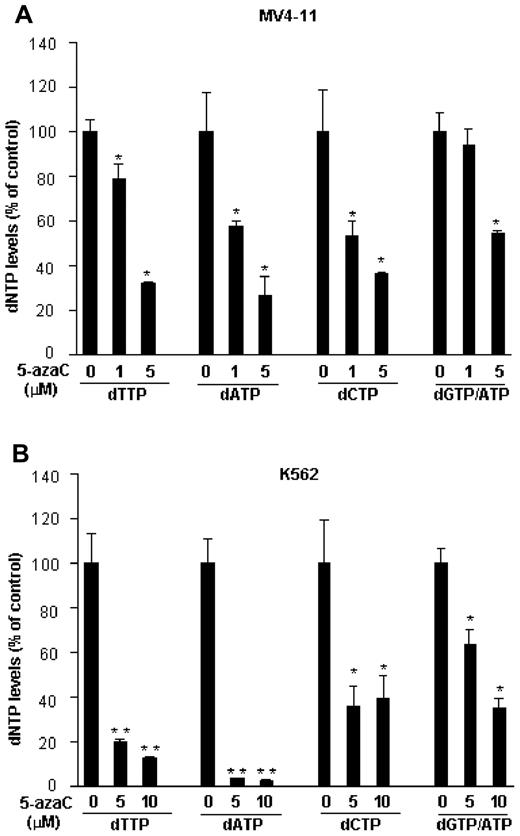
Because RRM2 catalyzes the rate-limiting step for DNA synthesis and repair by reducing ribonucleotides to their corresponding deoxyribonucleotides23 and because its level is dramatically decreased by 5-azaC, we investigated whether 5-azaC decreases the intracellular deoxyribonucleotide levels. After exposure of cells to 1-10μM 5-azaC for 72 hours, the deoxyribonucleotides were extracted and measured by the nonradioactive LC-MS/MS method.15 A significant dose-dependent decrease in the intracellular levels of dTTP, dATP, dCTP, and dGTP/ATP was observed. All 4 deoxyribonucleotide levels were reduced by approximately 70% or 60%-90% by 5μM 5-azaC in MV4-11 and K562 cells, respectively (Figure 3A-B).
5-azaC decreases intracellular deoxyribonucleotide pools. MV4-11 or K562 cells were treated with the indicated concentrations of 5-azaC for 72 hours and subjected to LC-MS/MS analysis for deoxyribonucleotide levels. 5-azaC dose-dependently reduces the dTTP, dATP, dCTP, and dGTP/ATP levels in MV4-11 (A) and K562 (B) cells. Data are presented as a percentage of the untreated controls. The results are means ± SD of a representative experiment performed in triplicate. *P < .05 and **P < .01 versus untreated controls.
5-azaC decreases intracellular deoxyribonucleotide pools. MV4-11 or K562 cells were treated with the indicated concentrations of 5-azaC for 72 hours and subjected to LC-MS/MS analysis for deoxyribonucleotide levels. 5-azaC dose-dependently reduces the dTTP, dATP, dCTP, and dGTP/ATP levels in MV4-11 (A) and K562 (B) cells. Data are presented as a percentage of the untreated controls. The results are means ± SD of a representative experiment performed in triplicate. *P < .05 and **P < .01 versus untreated controls.
Conversion of 5-azaC to DAC is RRM2 mediated and self-limited
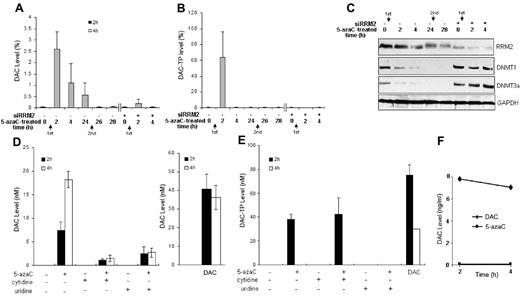
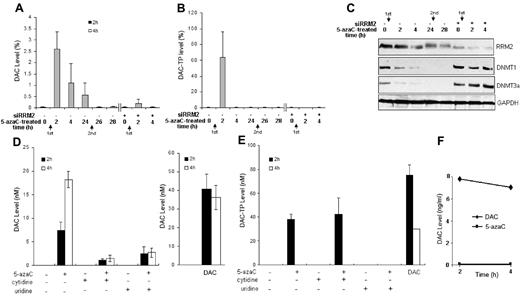
RR is believed to catalyze the conversion of 5-azaC to DAC through the intermediate product 5-aza–CDP to 5-aza–dCDP, which is eventually phosphorylated to 5-aza–dCTP (DAC-TP) and incorporated into DNA4,7 or dephosphorylated to DAC. We therefore monitored the dynamics of this conversion process by measuring the intracellular DAC and DAC-TP level after 5-azaC exposure. We found that the peak levels of DAC and DAC-TP were reached at approximately 2 hours and declined over time after 5-azaC treatment (Figure 4A-B). A second exposure to 5-azaC at 24 hours could not restore DAC/DAC-TP levels in leukemia cells, suggesting that the initial 5-azaC treatment had induced a sustained RRM2 down-regulation that inhibited further conversion of 5-azaC into DAC/DAC-TP (Figure 4A-B). The role of RR in the conversion of 5-azaC into DAC/DAC-TP was validated by showing that RNAi silencing of RRM2 expression followed by 5-azaC treatment led to a near depletion of DAC/DAC-TP levels (Figure 4A-B).
RRM2 is involved in the conversion of 5-azaC to DAC in MV4-11 cells. (A-C) 5-azaC rapidly converts to DAC and DAC-TP, which diminishes with time. Knock-down of RRM2 expression prevents 5-azaC conversion to DAC (A) and DAC-TP (B) and associated events (C). MV4-11 cells were transfected with siRRM2 or mock-transfected for 24 hours, followed by treatment with 10μM 5-azaC for 2-28 hours. The arrows indicate sequences of fresh addition of 5-azaC. The intracellular DAC and DAC-TP were extracted and quantified by LC-MS/MS. Cells treated with 10μM DAC for 2 hours serve as a positive control. Data are presented as a percentage of the positive controls. Knock-down of RRM2 expression inhibits 5-azaC–induced down-regulation of DNMT1 and DNMT3a (C). Western blot analysis shows that 5-azaC alone time-dependently decreases the protein levels of DNMT1 and DNMT3a, whereas RRM2 depletion by siRRM2 abolishes its effects. Experiments were repeated and the representative results are presented. (D-E) Conversion of 5-azaC to DAC is reduced by UCK inhibitors. MV4-11 cells were treated with 10μM 5-azaC for 2 or 4 hours. UCK was inhibited competitively by its natural substrates, cytidine or uridine. Intracellular DAC (D) and DAC-TP (E) levels were measured. Cells treated with 10μM DAC for 2 or 4 hours serve as positive controls. (F) Stability of DAC. MV4-11 cells in PBS were heat-inactivated at 100°C for 6 minutes and then exposed to 10μM 5-azaC or DAC for 2 or 4 hours at 37°C.
RRM2 is involved in the conversion of 5-azaC to DAC in MV4-11 cells. (A-C) 5-azaC rapidly converts to DAC and DAC-TP, which diminishes with time. Knock-down of RRM2 expression prevents 5-azaC conversion to DAC (A) and DAC-TP (B) and associated events (C). MV4-11 cells were transfected with siRRM2 or mock-transfected for 24 hours, followed by treatment with 10μM 5-azaC for 2-28 hours. The arrows indicate sequences of fresh addition of 5-azaC. The intracellular DAC and DAC-TP were extracted and quantified by LC-MS/MS. Cells treated with 10μM DAC for 2 hours serve as a positive control. Data are presented as a percentage of the positive controls. Knock-down of RRM2 expression inhibits 5-azaC–induced down-regulation of DNMT1 and DNMT3a (C). Western blot analysis shows that 5-azaC alone time-dependently decreases the protein levels of DNMT1 and DNMT3a, whereas RRM2 depletion by siRRM2 abolishes its effects. Experiments were repeated and the representative results are presented. (D-E) Conversion of 5-azaC to DAC is reduced by UCK inhibitors. MV4-11 cells were treated with 10μM 5-azaC for 2 or 4 hours. UCK was inhibited competitively by its natural substrates, cytidine or uridine. Intracellular DAC (D) and DAC-TP (E) levels were measured. Cells treated with 10μM DAC for 2 or 4 hours serve as positive controls. (F) Stability of DAC. MV4-11 cells in PBS were heat-inactivated at 100°C for 6 minutes and then exposed to 10μM 5-azaC or DAC for 2 or 4 hours at 37°C.
It was thought previously that the conversion of 5-azaC to DAC involved multiple processes, first the reduction of 5-azaCDP to DAC-DP, followed by reversible dephosphorylation to DAC. However, the dephosphorylation processes have not been well defined; in particular, the enzyme that dephosphorylates the intermediate product 5-aza–dCMP to DAC has not been identified. Therefore, a relatively slow or an inefficient formation of DAC from 5-azaC would be expected. In fact, the low conversion (∼ 15%-20%) of 5-azaC to DAC as a whole would be consistent with this scenario. We further determined the DAC/DAC-TP level in 5-azaC–treated MV4-11 cells in the presence of cytidine or uridine,24,25 the competitive inhibitors of UCK. After this inhibition, the DAC/DAC-TP levels were reduced significantly (Figure 4D-E). These results demonstrate that the UCK-RRM2–mediated pathway is a major pathway for 5-azaC conversion.
As control experiments, we incubated the heat-inactivated cell lysates with 5-azaC or DAC. As expected, there was no detectable DAC in 5-azaC–treated cell samples because the heat inactivates the enzymes required for conversion. In addition, the DAC level was found to be stable in DAC-treated cell samples during the time course of the experiment, supporting the idea that the lack of detectable DAC was not because of its own degradation (Figure 4F).
Lack of RRM2-mediated conversion of 5-azaC to DAC limits the activity of 5-azaC on DNMTs
It has been shown previously that DAC/DAC-TP down-regulates DNMT1 by forming a DNA-drug-enzyme complex.26 Therefore, one should expect that knockout of RRM2 expression should abolish this effect by 5-azaC because its conversion to DAC/DAC-TP is blocked. Indeed, we observed that the DNMT1 and DNMT3a proteins were not down-regulated in siRRM2-treated MV4-11 cells after 5-azaC incubation (Figure 4C). The DNMT1 and DNMT3a protein levels remained depleted even though there was no detectable (free) DAC/DAC-TP 24 hours after a second exposure of 5-azaC, indicating that other mechanisms may be involved (Figure 4C).
5-azaC destabilizes RNA
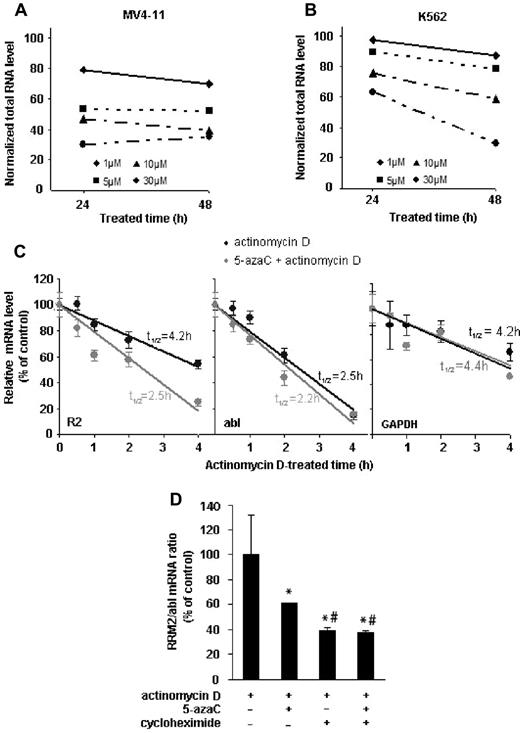
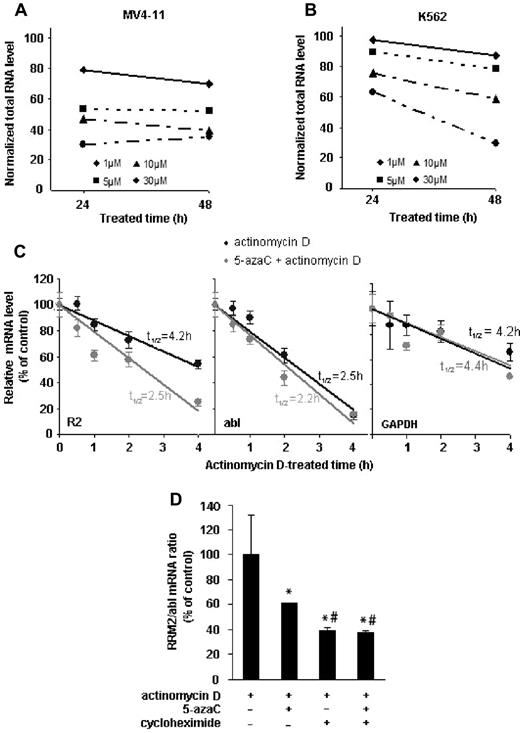
To investigate the underlying mechanisms for RRM2 mRNA inhibition by 5-azaC, we first examined the effects of 5-azaC on RNA stability. We observed that 5-azaC induces a dose- and time-dependent decline in total RNA levels in both MV4-11 and the K562 cell lines (Figure 5A-B). However, this decrease is not because of 5-azaC cytotoxicity, because at this dose 5-azaC did not cause a significant cell death after 24 or 48 hours of exposure (supplemental Figure 1). Our data thus suggest that 5-azaC destabilizes RNA and promotes its degradation. We further addressed possible mechanisms by which 5-azaC specifically down-regulates RRM2 expression. We determined the effects of 5-azaC on RRM2 mRNA stability by comparing the half-life of RRM2 mRNA (t1/2) in the presence or absence of 5-azaC. The mRNA turnover rates were estimated by inhibiting de novo transcription with actinomycin D and monitoring the decline in mRNA level with time.27 In the absence of 5-azaC, the RRM2 mRNA turnover is rather rapid, with a half-life of 4.2 hours in MV4-11 cells. After 5-azaC treatment, RRM2 mRNA was remarkably destabilized, with a much shortened half-life of 2.5 hours (Figure 5C). The half-lives of the housekeeping controls abl and GAPDH mRNA (t1/2 = 2.5 hours for abl and 4.2 hours for GAPDH in control samples), however, were not changed significantly by 5-azaC (t1/2 = 2.2 hours for abl and 4.4 hours for GAPDH in 5-azaC–treated samples, Figure 5C). Similar results were obtained when we used another transcription inhibitor, DRB, to block mRNA synthesis28 (supplemental Figure 2). These data suggest that the inhibition of 5-azaC on the RRM2 mRNA level was due at least in part to its effects on mRNA stability. Furthermore, we found that protein synthesis is involved in stabilizing RRM2 mRNA, because addition of the protein synthesis inhibitor cycloheximide further decreased 5-azaC–mediated destabilization of RRM2 mRNA (although cycloheximide itself also has similar destabilizing effect; Figure 5D). These results suggest that 5-azaC and cycloheximide exert different mechanisms to decrease RRM2 mRNA, and this requires further investigation.
5-azaC reduces the stability of total RNA and RRM2 mRNA. (A-B) 5-azaC causes a dose- and time-dependent decline in the total RNA level in MV4-11 (A) and K562 cells (B). (C) 5-azaC shortens the half-life (t1/2) of RRM2 but not abl or GAPDH mRNA. MV4-11 cells were treated with 1μM actinomycin D to block the de novo transcription in the presence or absence of 5-azaC. mRNA levels of RRM2 and 2 internal controls, abl and GAPDH, were measured by quantitative RT-PCR at the indicated time points after actinomycin D treatment. The data are presented as a percentage of the mRNA level measured at time 0 (without actinomycin D). mRNA half-lives were calculated and are indicated in the plots. (D) Blockade of protein synthesis facilitates 5-azaC–induced destabilization of RRM2 mRNA. MV4-11 cells were pretreated with cycloheximide to block protein synthesis, followed by treatment with 5-azaC and actinomycin D. Normalized mRNA levels are presented as the percentage of actinomycin D only, and are shown as means ± SD from triplicate experiments. *P < .05 compared with actinomycin D only; #P < .05 compared with treatment of both 5-azaC and actinomycin D.
5-azaC reduces the stability of total RNA and RRM2 mRNA. (A-B) 5-azaC causes a dose- and time-dependent decline in the total RNA level in MV4-11 (A) and K562 cells (B). (C) 5-azaC shortens the half-life (t1/2) of RRM2 but not abl or GAPDH mRNA. MV4-11 cells were treated with 1μM actinomycin D to block the de novo transcription in the presence or absence of 5-azaC. mRNA levels of RRM2 and 2 internal controls, abl and GAPDH, were measured by quantitative RT-PCR at the indicated time points after actinomycin D treatment. The data are presented as a percentage of the mRNA level measured at time 0 (without actinomycin D). mRNA half-lives were calculated and are indicated in the plots. (D) Blockade of protein synthesis facilitates 5-azaC–induced destabilization of RRM2 mRNA. MV4-11 cells were pretreated with cycloheximide to block protein synthesis, followed by treatment with 5-azaC and actinomycin D. Normalized mRNA levels are presented as the percentage of actinomycin D only, and are shown as means ± SD from triplicate experiments. *P < .05 compared with actinomycin D only; #P < .05 compared with treatment of both 5-azaC and actinomycin D.
5-azaC incorporates into RNA
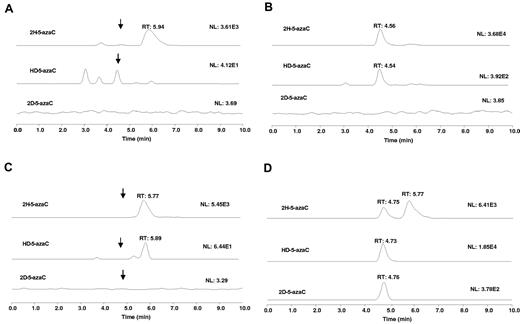
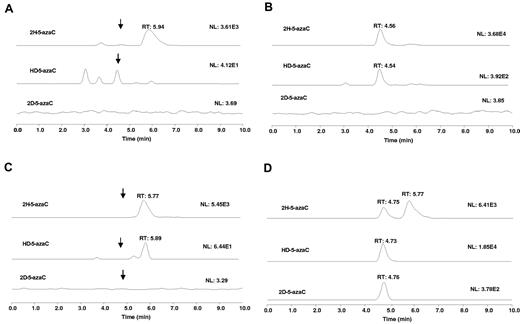
The RNA destabilization effect of 5-azaC may in part because of RNA incorporation. Previous studies have suggested that 5-azaC is incorporated into RNA,7,29,30 a potential mechanism contributing to 5-azaC's RNA destabilizing effects. However, the provided evidence was rather indirect by measuring the incorporation of radiolabeled 5-azaC.7,31 To investigate this possibility directly, we used a chemical method coupled to the LC-MS/MS assay. Because 5-azaC is chemically labile,32–34 it would not be expected to be detectable in the total RNA acidic hydrolysate. However, 5-azaC can be readily converted to stable dihydro-5-azaC35 and dideuterio-5-azaC with NaBH4 and NaBD4, respectively, in aqueous solution under mild conditions. These products can be monitored by LC-MS/MS as a means to probe 5-azaC RNA incorporation. Therefore, we first examined the LC-MS characteristics of standard dihydro-5-azaC, HD-5-azaC, and dideuterio-5-azaC in the mobile phase. The total RNA isolated from the control or the 5-azaC–treated MV-411 cells was treated with NaBH4 followed by acid hydrolysis. No peak of significant intensity was observed at the retention time (∼ 4.5 minutes) and the ion transitions for 5-azaC (data not shown) or dihydro-5-azaC in the control RNA samples (Figure 6A). However, a peak with significant intensity was observed at 4.56 minutes, corresponding to dihydro-5-azaC with the correct ion transition in the treated samples, indicating the presence of 5-azaC in the total RNA (Figure 6B). To further substantiate these data, we also used NaBD4 to reduce the blank and 5-azaC–treated total RNA. Because each of the deuterium atom increases the mass by 1 relative to the hydrogen atom, we would expect that the observed new peaks, if they relate to 5-azaC, would have a proportional increase in the mass unit corresponding to dihydro-5-azaC, HD-5-azaC, and dideuterio-5-azaC. Similar to the NaBH4 experiment, after NaBD4 treatment, no significant peak was observed at 4.7 minutes for any of the reduced products in the controls (Figure 6C). However, a significant peak was observed at 4.7 minutes with ion transitions corresponding to dihydro-5-azaC, HD-5-azaC, and dideuterio-5-azaC (Figure 6D). These data indicate the presence of 5-azaC in the total RNA of the 5-azaC–treated samples and confirm the expected 5-azaC incorporation into total RNA in a human AML cell line.
5-azaC incorporates into RNA. Extracted ion chromatograms for: (A) RNA hydrolysate from a blank cell sample treated with NaBH4 (scale is enhanced). Arrows show the approximate locations of the ion peaks. (B) RNA hydrolysate from 5-azaC–treated cell sample followed by treatment with NaBH4. (C) RNA hydrolysate from blank cell sample treated with NaBD4 (scale is enhanced). Arrows show the approximate location of the ion peaks. (D) RNA hydrolysate from a 5-azaC–treated cell sample followed by treatment with NaBD4. To simplify the figure, the 5-azaC channel is not shown.
5-azaC incorporates into RNA. Extracted ion chromatograms for: (A) RNA hydrolysate from a blank cell sample treated with NaBH4 (scale is enhanced). Arrows show the approximate locations of the ion peaks. (B) RNA hydrolysate from 5-azaC–treated cell sample followed by treatment with NaBH4. (C) RNA hydrolysate from blank cell sample treated with NaBD4 (scale is enhanced). Arrows show the approximate location of the ion peaks. (D) RNA hydrolysate from a 5-azaC–treated cell sample followed by treatment with NaBD4. To simplify the figure, the 5-azaC channel is not shown.
Discussion
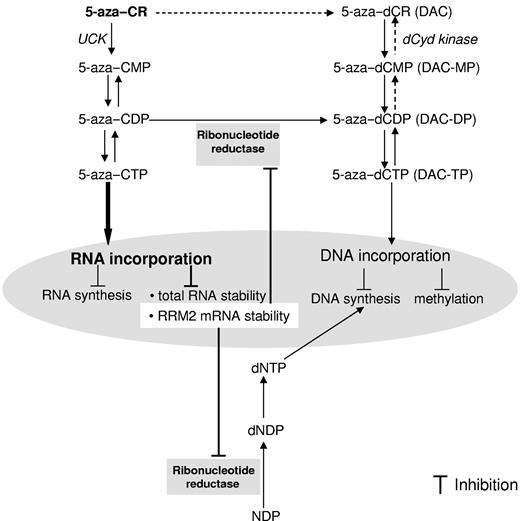
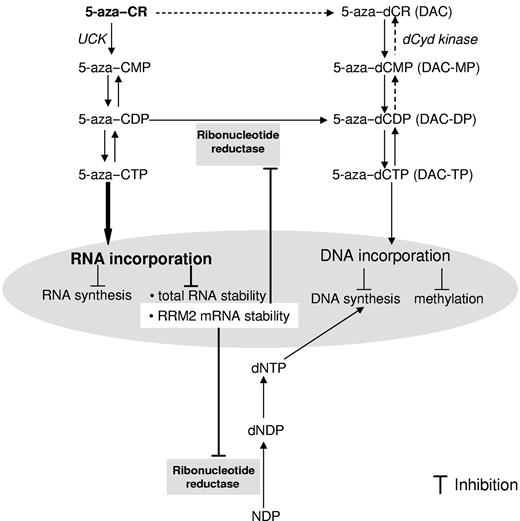
5-azaC and DAC have long been considered as similar antitumor agents promoting DNA synthesis inhibition and hypomethylation, even though they differed chemically in the ribose moiety. Clinically, these agents are mainly used for the treatment of hematopoietic malignancies and both have been approved recently by the FDA for the treatment of MDS. It has long been recognized that DAC has a primarily modulating effect on DNA, whereas 5-azaC has effects on both the DNA and RNA pathways, and it is believed that 5-azaC's DNA effect was because of its conversion to DAC. More importantly, only 20% of 5-azaC is converted to DAC and the major portion of 5-azaC (∼ 80%) acts on the RNA pathway.7 Despite this, little attention has been focused on the RNA mechanism, which was therefore the focus of the present investigation. We used RRM2 antisense to inhibit 5-azaC's conversion and its subsequent DNA pathway and studied its RNA effects. Unexpectedly, we discovered that RRM2 is a novel target for 5-azaC itself (Figure 7).
Proposed mechanism of actions for 5-azaC in AML. 5-azaC is majorly phosphorylated to 5-aza–CTP and incorporated into RNA. A fraction of the 5-azaC intermediate product 5-aza–CDP is converted to 5-aza–dCDP by RR, followed by further phosphorylation to 5-azaC–dCTP, which is believed to incorporate into DNA. The results of the present study identify RRM2 as a novel target of 5-azaC. The underlying molecular mechanisms for the inhibitory effects of 5-azaC on RRM2 involve its RNA incorporation and attenuated RRM2 mRNA stability.
Proposed mechanism of actions for 5-azaC in AML. 5-azaC is majorly phosphorylated to 5-aza–CTP and incorporated into RNA. A fraction of the 5-azaC intermediate product 5-aza–CDP is converted to 5-aza–dCDP by RR, followed by further phosphorylation to 5-azaC–dCTP, which is believed to incorporate into DNA. The results of the present study identify RRM2 as a novel target of 5-azaC. The underlying molecular mechanisms for the inhibitory effects of 5-azaC on RRM2 involve its RNA incorporation and attenuated RRM2 mRNA stability.
RR is a highly regulated enzyme comprising 2 nonidentical dimeric subunits, RRM1 and RRM2. The RRM1/RRM2 complex serves as a major provider of deoxyribonucleotides for DNA replication during the S phase.20,36 Overexpression of RR is found commonly in malignant cells36–38 and increases the endogenous pool of deoxyribonucleotides, thereby increasing the DNA synthesis rate to serve the proliferative nature of malignant cells. RR overexpression is also a potential mechanism for chemoresistance to nucleoside analogs competing for DNA incorporation.39 These important roles have made RRM2 an attractive target for chemotherapeutic development.40,41 Studies have demonstrated that RRM2 inhibition provides antiproliferative and antineoplastic benefits.20,36 However, several established RR inhibitors have considerable limitations. For example, hydroxyurea, one of the most commonly used RRM2 inhibitors, is limited by its low affinity for RR, high hydrophilicity, short half-life, and early development of resistance.20,23,42 Newer classes of RRM2 inhibitors, such as antisense GTI-2040 (LOR-2040) and ion chelators (eg, Triapine) are under development, and their effectiveness is still being evaluated. We have demonstrated in the present study that 5-azaC induces a dose- and time-dependent decrease of RRM2 protein in leukemia cells, and this effect was specific to 5-azaC and not to its reduced analog, DAC. We further compared 5-azaC with other established RRM2 inhibitors, antisense GTI-2040, and Triapine, in down-regulating RRM2. 5-azaC is more potent than Triapine, as indicated by a leftward shift of the dose-response curve. In addition, the in vitro findings were confirmed in mice-engrafted tumor tissues and BM cells from AML patients. However, because these are ex vivo studies and the number of patient samples (n = 6) was relatively small, it was not possible for us to correlate response to 5-azaC treatment with patients' World Health Organization subtype or clinical, cytogenetic, or molecular characteristics (supplemental Table 1). With regard to the mechanisms for 5-azaC down-regulation of the RRM2 mRNA level, we found that 5-azaC attenuates total RNA and RRM2 mRNA stability. The decrease in RNA stability is likely because of the incorporation of 5-azaC, as confirmed by our LC-MS/MS data. We also observed a discrepancy between the decrease in RRM2 mRNA levels and protein levels, suggesting that 5-azaC may have additional, transcription-independent effects on RRM2 protein, the nature of which warrants further studies.
Because the formation of deoxyribonucleotides from the corresponding ribonucleotides is controlled by RR,43 the enzyme activity of RR relates directly to the production of deoxyribonucleotides in cells.44 A substantial number of deoxyribonucleotides is required by cells in the S phase and is linked closely to the cellular growth control mechanisms. Therefore, altered deoxyribonucleotide pools by 5-azaC–mediated RRM2 inhibition is likely to cause perturbations in DNA synthesis and replication processes, resulting in chromosomal instability.36 Our present data indeed showed that after 5-azaC treatment, the dTTP, dATP, and dCTP pools were reduced in a dose-dependent manner.
In addition to its major effects on DNA synthesis, RRM2 is also involved in the intracellular metabolism of 5-azaC. We have shown that RRM2 expression knockout decreases the intracellular DAC and DAC-TP level dramatically and abolishes 5-azaC–mediated down-regulation of DNMTs, strongly supporting the idea that the hypomethylation effect of 5-azaC is through its conversion to DAC. However, because 5-azaC induces strong inhibition of RRM2 expression, it is likely that 5-azaC inhibits its own further conversion to DAC over time. Indeed, we have shown herein that after the initial conversion, both the DAC and DAC-TP levels diminished to nondetectable levels in approximately 4 hours. Because there was no detectable DAC or DAC-TP even after a second dose of 5-azaC at 24 hours, we concluded that RRM2 was completely blocked by the initial 5-azaC dosing. However, the downstream DNMT1 and DNMT3a protein expression remained down-regulated, suggesting that a rather stable covalent complex of DNA/DNMT/DAC-TP formed after the initial 5-azaC exposure. We further extended the time course of 5-azaC treatment to evaluate its effects on DNMT protein expression. Our data suggested that 5-azaC has similar DNMT–down-regulating effects as DAC. Although the conversion of 5-azaC to DAC is self-limiting, the 5-azaC–derived DAC could elicit a similar DNMT down-regulation as DAC that persisted over time for at least 72 hours (supplemental Figure 3). In addition, the kinetics of RRM2 down-regulation and DNA methylation caused by 5-azaC are likely different. Our present results suggest that the down-regulation of RRM2 by 5-azaC is dose and time dependent and may require higher drug concentrations and longer exposure times than what is required for DNMTs.
We also found that RRM2 participates in a direct conversion of 5-azaC to DAC, although it only accounts for a small fraction of conversion (Figure 4D). Previously, 5-azaC was believed to be converted to DAC only through the diphosphate stage and presumably by subsequent dephosphorylations. A molecular mechanism of reduction of nucleotides has been proposed,45–47 in which binding of the nucleotide to the reductase active site is required before the reduction of the 2-hydroxy on the ribose moiety. The 5′-phosphate group(s) helps to anchor the nucleotide to the binding pocket. Our present data indicate that the nucleoside itself (5-azaC) could also be reduced directly, which implies that 5-azaC itself could anchor the binding site sufficiently before the reduction.
In conclusion, in the present study, we found that 5-azaC is a potent and specific RRM2 inhibitor. This represents a major advance in the pharmacology of 5-azaC and will enable efficient translation of preclinical studies to clinical effectiveness. For example, our discovery expands the therapeutic use of 5-azaC to combination regiments containing cytarabine (Ara-C) or other nucleoside analogs by increasing the Ara-CTP/deoxyribonucleotide ratio in favor of Ara-CTP, ultimately resulting in increased DNA incorporation of Ara-CTP and enhanced antileukemia activity. Our proposed combination study is being evaluated presently in a clinical trial at our institution. Furthermore, direct comparisons between 5-azaC and DAC activities are also presented herein, and our data support the distinction between these 2 compounds. 5-azaC can be converted to DAC in cells and this process is rapid but transient and self-limited. The results of the present study may have relevance for the design and dosing of 5-azaC in regimens pursuing gene hypomethylation and RRM2 inhibition in MDS or AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Joy Scott for editorial and administrative assistance.
This work was supported by the National Cancer Institute, National Institutes of Health (grants UO1-CA76576 and R21 CA 133879 ARRA to K.K.C., G.M., R.B.K., and W.B. and R01 CA102031 to G.M. and K.K.C.).
National Institutes of Health
Authorship
Contribution: J.A. and H.W. designed the study, contributed experimental data, and wrote the manuscript; P.C. developed the LC-MS/MS methods, contributed experimental data, and wrote the manuscript; Z.X. and J.W. contributed experimental data; S.L. provided technical support and helpful discussions; R.K., A.M., and W.B. provided patient samples and information; G.M. designed the study and wrote the manuscript; and K.K.C. conceived and designed the study, wrote the manuscript, and provided administrative and financial support.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kenneth K. Chan, PhD, College of Pharmacy, Ohio State University, 500 W 12th Ave, Columbus, OH 43210; e-mail: chan.56@osu.edu.
References
Author notes
J.A. and H.W. contributed equally to this work.