Abstract
HSC fate decisions are regulated by cell-intrinsic and cell-extrinsic cues. The latter cues are derived from the BM niche. Membrane-type 1 matrix metalloproteinase (MT1-MMP), which is best known for its proteolytic role in pericellular matrix remodeling, is highly expressed in HSCs and stromal/niche cells. We found that, in MT1-MMP−/− mice, in addition to a stem cell defect, the transcription and release of kit ligand (KitL), stromal cell–derived factor-1 (SDF-1/CXCL12), erythropoietin (Epo), and IL-7 was impaired, resulting in a trilineage hematopoietic differentiation block, while addition of exogenous KitL and SDF-1 restored hematopoiesis. Further mechanistic studies revealed that MT1-MMP activates the hypoxia-inducible factor-1 (HIF-1) pathway via factor inhibiting HIF-1 (FIH-1) within niche cells, thereby inducing the transcription of HIF-responsive genes, which induce terminal hematopoietic differentiation. Thus, MT1-MMP in niche cells regulates postnatal hematopoiesis, by modulating hematopoietic HIF-dependent niche factors that are critical for terminal differentiation and migration.
Introduction
The adult hematopoietic system is maintained by a small number of HSCs that reside in the BM in a specialized microenvironment (the niche).1,2 Here, HSCs undertake fate decisions including differentiation to progenitor cells and self-renewal, which ensures a lifelong supply of terminally differentiated blood cells. Intrinsic cellular programming and external stimuli such as adhesive interactions with the microenvironmental stroma and cytokine activities regulate HSC fate. However, it is unclear how niche factor production is controlled to adjust to external demand with a fine-tuned response.
Hypoxia-inducible factors (HIFs) consist of an α (HIF-α) and a β (HIF-β, or ARNT) subunit and activate the expression of genes encoding proteins that regulate cell metabolism, motility, angiogenesis, hematopoiesis, and other functions. HSCs maintain cell-cycle quiescence by regulating HIF-1α levels.3,4 Mice with mutations in the heterodimeric transcription factor HIF develop extensive hematopoietic pathologies: embryos lacking Arnt have defects in primitive hematopoiesis.5 Mice lacking endothelial PAS domain protein 1 (EPAS1, also known as HIF-2alpha/HRF/HLF/MOP3), a second HIF family member, exhibited pancytopenia, and it was shown that EPAS1 is necessary to maintain a functional microenvironment in the BM for effective hematopoiesis.6 HIFs bind to canonical DNA sequences in the promoters or enhancers of target genes such as erythropoietin (Epo), vascular endothelial growth factor-A, SDF-1α/CXCL12, angiopoietin-2, platelet-derived growth factor-B and Kit Ligand (KitL)/stem cell factor, which are involved in HSC maintenance within the BM niche.7-10 The chemokine SDF-1α/CXCL12 (SDF-1α) is expressed by perivascular, endosteal, mesenchymal stem and progenitor cells as well as by osteoblasts.11,12 SDF-1α deficiency leads to a reduction in HSCs and impaired B-cell development in mice.13,14 IL-7 is another stromal cell–derived niche factor, which, in cooperation with CXCL12, functions at sequential stages of B-cell development.15,16 IL-7 or IL-7R deficiency results in impaired B-cell development.17,18
Proteases such as matrix metalloproteinase-9 (MMP-9) and the serine proteinase plasmin(ogen) regulate HSC fate through KitL release in the BM.10,19 Membrane type-1 MMP (MT1-MMP, also known as MMP-14) can proteolytically degrade extracellular matrix (ECM) components and cleave membrane receptors and ligands.20 MT1-MMP is expressed within mesenchymal stem and immature hematopoietic cells.21,22 MT1-MMP can control the migration of hematopoietic stem/progenitor cells and monocytes.21,23,24 MT1-MMP function is essential for angiogenesis, wound healing, connective tissue remodeling, arthritis, tumor growth, and metastasis.25-27 MT1-MMP–deficient (MT1-MMP−/−) mice showed skeletal dysplasia, arthritis, and osteopenia.28 However, the role of MT1-MMP in hematopoiesis is unclear.
We found that MT1-MMP inactivation in mice resulted in severe pancytopenia, characterized by an impaired stem cell pool and a block in hematopoietic differentiation. MT1-MMP deletion from hematopoietic cells generated normal hematopoiesis in recipient mice, thereby demonstrating that MT1-MMP is an essential regulator of the BM microenvironment. Mechanistically, we demonstrate that MT1-MMP deficiency blocked the transcription of typical HIF-1–dependent niche factors including Epo, SDF-1, KitL, and IL-7 by modulating the HIF-1 pathway through factor inhibiting HIF-1 (FIH1), resulting in a 3-lineage terminal differentiation block. Thus, MT1-MMP controls HSC fate by regulating the BM niche.
Methods
Animals
Age-matched (14-day) MT1-MMP+/+ and MT1-MMP−/− mice were obtained by heterozygous breeding.24 Animal procedures were approved by the Animal Care Committee of The Institute of Medical Science (University of Tokyo). C57BL/6 and C57BL/6-Tg (CAG-EGFP) mice were purchased from Japan SLC Inc, and Ly5.1 mice were purchased from Sankyo Lab Service.
In vivo assays
Competitive transplantation experiments.
Lethally irradiated Ly-5.1 mice were injected with BM cells (10 recipient mice per cell concentration) from MT1-MMP+/+ or MT1-MMP−/− mice together with 2 × 105 CD45.1 BM competitive cells. Peripheral blood (PB) cells of the recipient mice were analyzed 4 months after transplantation. Cells were stained with PE-conjugated anti-CD4 and anti-CD8, FITC-conjugated anti-CD45.2, allophycocyanin-conjugated anti-CD11b and anti-Gr-1, PE-cy7–conjugated anti-B220, and biotinylated anti-CD45.1 Abs. The biotinylated Ab was developed using streptavidin-PE–Cy5. The percentage of donor-derived lineage contributions in PBMCs was assessed using Abs against CD45.2, Gr-1/CD11b, or B220. Total chimerism of > 1% for all Abs tested using PBMCs was considered as long-term reconstitution. The frequencies were determined using L-Calc software (StemCell Technologies).
Growth factor rescue experiments.
Recombinant mouse KitL (PeproTech) was administered IP into MT1-MMP+/+ and MT1-MMP−/− mice at a concentration of 150 μg/kg body weight, daily from postnatal day 7 to day 10. Recombinant mouse SDF-1α (PeproTech; 100 ng/mice) was injected twice intraperitoneally on postnatal day 10. Blood was collected and blood cells counted on day 12.
CFU-S assay.
Mobilized PBMCs were obtained and subjected to a CFU-S assay as previously described.29 Mice were killed on day 12. The number of visible splenic colonies was counted.
In vitro assays
Peripheral blood analysis.
Blood was collected from mice by retro-orbital bleeding using heparinized capillaries. White blood cell (WBC), RBC, and platelet (PLT) counts were determined. Plasma samples were stored at −80°C until further analysis.
Hematopoietic progenitor assay.
BM mononuclear cells (BMMCs; 104 cells/plate) were plated in triplicate in 1 mL of a commercially available methylcellulose-based assay solution (Methocult; StemCell Technologies).
Lineage-negative cell separation.
Murine BM cells were obtained after flushing mouse femur and tibiae. Cells were stained using a lineage cell separation kit (StemCell Technologies). After MACS cell separation (Miltenyi Biotec), cells were stained with c-Kit, Sca-1, and lineage Abs (BD Pharmingen), and were then analyzed by FACS.
B-cell colony-forming assay (CFU–IL-7).
The CFU–IL-7 assay was carried out in medium (Invitrogen) containing 1.2% methylcellulose (StemCell Technologies), 30% FCS (HyClone), 1% BSA, 0.1mM 2-ME, and mouse IL-7 (PeproTech). On day 7 of culture, aggregates consisting of > 50 cells were scored as a colony.
Cell culture.
Mouse stromal cells (MS-5) were maintained in IMDM supplemented with 10% FBS. Human BM endothelial cells (BMEC-1) were maintained in Medium 199 supplemented with 10% FBS, 0.146 mg/mL l-glutamine, and 2.2 mg/mL sodium bicarbonate. Mouse embryonic fibroblast cells (NIH3T3) were maintained in DMEM supplemented with 10% FBS. These cells were cultured at 37°C, in a 5% CO2 incubator. Human osteoblastic cells (FOB) were maintained in a 1:1 mixture of Ham F12 medium DMEM supplemented with 2.5mM l-glutamine and 10% FBS, and the cells were cultured at 34°C, in a 5% CO2 incubator. MT1-MMP+/+ and MT1-MMP−/− mouse embryonic fibroblasts (MEF) cells (kindly provided by M.S.) were maintained in DMEM (Invitrogen) supplemented with 10% FBS at 37°C, in a 5% CO2 incubator.30
Knockdown experiment using shRNA.
The shRNA sequences used for knockdown of mouse MT1-MMP and FIH-1 were: 5′-caccgctgtggtgttccggataagtcgaaacttatccggaacaccacagc-3′ and 5′-caccggacctcgaatacctgcaagacgaatcttgcaggtattcgaggtcctttt-3′, respectively. These sequences were subcloned into pENTR/U6 TOPO (Invitrogen) and then transferred via recombination into the lentivirus vector pLenti6 BLOCKiT (Invitrogen). shRNA-expressing lentiviral vectors were generated and used according to the manufacturer's instructions.
FACS analysis.
Cells were flushed out from mouse BM. For cell-surface analysis, cells were stained with the following Abs: PE-conjugated anti-CD19, -Sca-1, -B220, CD44, c-kit, and -Gr1, FITC-conjugated anti-CD43, -CD34, -NK1.1, and -CD8, allophycocyanin-conjugated anti-B220, -CD4, CD11b, and a lineage cocktail, PerCP-Cy5.1–conjugated anti-c-kit. Cells were analyzed using FACS Aria (BD Biosciences). Common myeloid progenitor (CMP), granulocyte/macrophage progenitor (GMP), and megakaryocyte/erythroid progenitor (MEP) were determined in Lin− BM cells of 14-day-old MT1-MMP+/+ and MT1-MMP−/− mice. The following Abs were used: c-kit–allophycocyanin, Sca1-PE/Cy7, CD34-FITC, FcgRIII-PE all from BD Pharmingen. For the detection of Lin− cells, biotinylated Abs (B220-bio, Gr-1-bio, CD11b-bio, CD5-bio, Ter119-bio, 7-4-bio, from Miltenyi Biotec) were costained with allophycocyanin/Cy7-conjugated (BD Biosciences/BD Pharmingen) streptavidin. For mesenchymal stem cells (MSCs) detection, cells were stained with the following Abs: CD45-PE, Sca1-FITC, PDGFRα-allophycocyanin (eBioscience), TER119-PE (BD Bioscience), CD45–Pacific Blue (BioLegend), and Alexa 488–conjugated nestin Ab (Abcam).
RNA extraction, RT-PCR, and quantitative real-time PCR analysis.
Total RNA was extracted using RNA TRIzol (Invitrogen), and cDNA was generated according to the manufacturer's protocols. This cDNA (10 ng) was used as a template for each PCR amplification using the following specific forward and reverse primers, respectively: mouse β-actin (5′-tggaatcctgtggcatccatgaaac-3′) and (5′-taaaacgcagctcagtaacagtccg-3′); mouse MT1-MMP (5′-tccggataagtttgggactg-3′) and (5′-ccttcaccatcaaagggtgt-3′); human GAPDH (5′-gagtcaacggatttggtcgt-3′) and (5′-ttgattttggagggatctcg-3′); human MT1-MMP (5′-caagcattgggtgtttgatgatg-3′) and (5′-cttggggtactcgctatcca-3′). For quantitative real-time PCR, PCR mixtures were prepared using SYBR Premix Ex TaqII (Takara) containing 0.2mM of each primer, and amplification reactions were performed. Specific forward and reverse primers, respectively, were designed as follows: β-actin (5′-gctggaaggtggacagtgag-3′) and (5′-tgacaggatgcagaaggaga-3′); KitL (5′-gcctagtcattgttggctac-3′) and (5′-cccaagtttgtctatgatgg-3′); SDF-1 (5′-agaaaccttccaccagagca-3′) and (5′-aacggctaggaaagggtctc-3′); IL-7 (5′-tgcaatcatgtcaactgcaa-3′) and (5′-tgcaatcatgtcaactgcaa-3′); EPO (5′-catctgcgacagtcgagttctg-3′) and (5′-cacaacccatcgtgacattttc-3′); G-CSF (5′-cctgcttagagcagagagag-3′) and (5′-cagcagcaggaatcaatact-3′). Gene expression levels were measured using the ABI Prism 7500 sequence detection system (Applied Biosystems). PCR product levels were estimated by measurement of the intensity of SYBR Green fluorescence. Gene expression levels were normalized to β-actin mRNA.
Knockdown experiment using siRNA.
Target sequences for siRNA were commercially designed and synthesized (B-bridge). siRNAs were provided as a mixture containing 3 different siRNA target sequences. The RNAi transfection solution was prepared by preincubating a mixture of 5nM siRNAs dissolved in 1 mL of serum-free and antibiotic-free medium (OptiMEM; Invitrogen) and 10 μL of RNAiMAX for 20 minutes at room temperature. Cells (3-4.5 × 105 cell/4 mL growth medium without antibiotics) suspended by trypsinization were added to the mixture and cultured overnight, and the medium was replaced with fresh growth medium.
Overexpression of MT1-MMP.
The transfection solution was prepared by preincubating a mixture of 1 μg of plasmid DNA dissolved in 250 μL of OptiMEM and 3 μL of Lipofectamine 2000 (Invitrogen) dissolved in 250 μL of OptiMEM for 20 minutes at room temperature. The transfection mixture (500 μL in total) was then added to the cells. After 48 hours of culture, the culture media were replaced with fresh growth medium.
Immunoprecipitation.
Cells were lysed with cell lysis buffer (Cell Signaling Technology) and the supernatants were collected. Total protein content was measured using the Bradford assay (Bio-Rad). Lysates were incubated with protein-G–Sepharose beads (Santa Cruz Biotechnology) and anti–HIF-1α Ab (BD Biosciences) overnight and were then spun down for 1 minute at 5800g. The supernatant was removed and the pellets were solubilized with 6 × SDS sample buffer (0.35M Tris-HCl, pH 6.8, 10% SDS, 30% glycerol, 9.3% DTT) and analyzed by immunoblot analysis.
Immunoblotting.
Cells were lysed with lysis buffer (Cell Signaling Technology) according to the manufacturer's instructions. The supernatants were collected, and total protein content was measured using the Bradford assay (Bio-Rad). The subcellular proteome extraction kit (Merck) was used to fractionate subcellular proteins. Lysates were separated by SDS-PAGE, transferred to a PVDF membrane, subjected to immunoblotting using anti–FIH-1 (1:200; Santa Cruz Biotechnology), anti-integrinβ1 (1/1000; Chemicon), and anti-α/β tubulin (1:1000; Cell Signaling Technology), washed with TBS-T, and immunoprobed for 1 hour at room temperature with HRP-conjugated or alkaline phosphatase–conjugated Ab, then washed with PBS-T. Finally, membranes were incubated with ECL-Plus (Amersham), and the chemiluminescent signal was detected using LAS4000 (Fujifilm) according to the manufacturer's instructions. The alkaline phosphatase signal was then detected using the Histofine Kit (Nichirei).
Immunoassay.
After cell transfection with siRNA, the growth medium was replaced and the cells were cultured for 48 hours. Supernatants were then collected and analyzed for murine KitL, SDF-1α, and IL-7 using commercially available ELISAs (R&D Systems).
Stromal-based expansion cultures.
A total of 1 × 104 Lin− cells from the BM of GFP mice were cocultured with a confluent layer of stromal cells (MS-5). Recombinant mouse KitL (20 ng/mL, every other day) or recombinant mouse SDF-1α (100 ng/mL, every other day) was added to MT1-MMP KD MS-5 and control MS-5 cell cultures. Twelve days later, adherent cells were retrieved using trypsin (Sigma-Aldrich) and pooled with the nonadherent cells. Hematopoietic cells (GFP positive) were counted.
Migration assay.
Migration assays were performed in 24-well plates using 5-μm polycarbonate Transwell inserts (Costar). BM Lin− cells were collected and isolated from normal adult mice. The cells were resuspended in X-vivo 15 (Lonza). MT1-MMP+/+ and MT1-MMP−/− MEF culture supernatant was aliquoted (600-μL aliquots) into 24-well plates, which formed the bottom chamber, in the presence or absence of neutralizing Abs against mouse SDF-1 (R&D Systems). Lin− cells derived from the BM cells of GFP mice (2 × 105 cells in 100 μL) were added into the Transwell insert (top chamber), and the cells were allowed to migrate through the porous bottom for 4 hours at 37°C. The number of cells that migrated into the lower chamber was determined using flow cytometry. Cells had been stained with Abs against CD11b and Gr1 (BD Biosciences). The medium from the lower chamber was passed through a FACSCalibur for 60 seconds, gating on forward (FSC) and side scatter to exclude cell debris. The number of live cells was compared with a 100% migration control in which 2 × 105 cells had been added directly into the lower chamber and then counted on the FACScalibur for 60 seconds.
Immunohistochemistry.
BM sections were fixed with 4% paraformaldehyde and permeabilized using 0.2% Triton X-100 for 20 minutes. After blocking in PBS containing 5% goat serum and 0.2% BSA, the sections were incubated with anti–FIH-1 Ab (Novus Biologicals) for 16 hours, washed 3 times with PBS and incubated for 1 hour with anti–rabbit Alexa 568 (Invitrogen). After washing, the sections were incubated with Alexa 488–conjugated nestin Ab (Abcam) for 2 hours. For HIF-1 staining, fixed and blocked sections were incubated with Alexa 488–conjugated HIF-1 Ab (BD Pharmingen) for 2 hours. The sections were mounted using Vectashield Mounting Medium with DAPI (Vector Laboratories). Images were acquired with an Olympus DP71 camera.
Micro-CT analysis.
We evaluated the BM shaft surface area by using micro-CT (Rigaku). We measured the shaft area at the center of the femur by NIH ImageJ software.
Proliferation assay.
A total of 2 × 104 Lin− cells isolated by MACS (Miltenyi Biotec) from BM cells of GFP mice were cocultured with a confluent layer of MEF cells. The cells were maintained in X-vivo 15 supplemented recombinant mouse IL-3 (20 ng/mL, every other day) with or without neutralizing Abs against mouse KitL (R&D Systems). One week later, all cells were retrieved using trypsin (Sigma-Aldrich). GFP-positive were counted using FACS.
Statistical analysis
Data were analyzed using the unpaired 2-tailed Student t test and are expressed as means ± SEM. P values of < .05 were considered significant.
Results
MT1-MMP deletion leads to severe pancytopenia
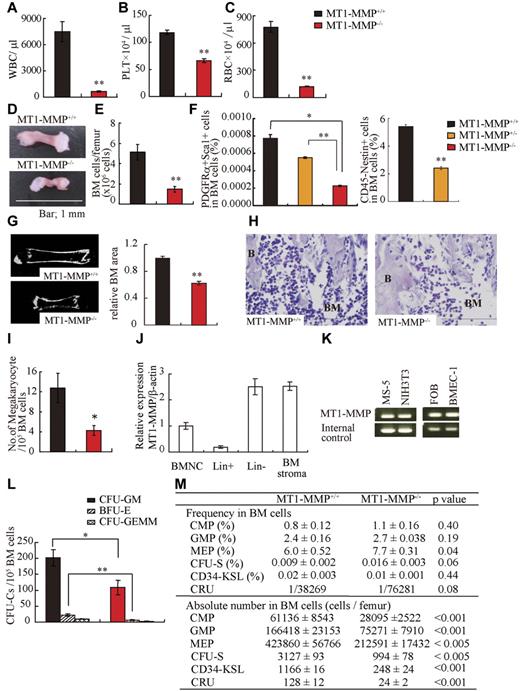
We used 14-day-old MT1-MMP−/− mice to explore the role of MT1-MMP in the regulation of postnatal hematopoiesis, as mortality of older MT1-MMP−/− mice is very high.24,28 Compared with MT1-MMP+/+ mice, the body weight of MT1-MMP−/− mice was decreased (7.22 ± 0.48 and 2.99 ± 0.05 g/mouse, respectively; n = 5, P < .001) and there was a decline in all hematopoietic lineages examined in MT1-MMP−/− animals, including WBCs (Figure 1A), PLTs (Figure 1B), and RBCs (Figure 1C) in the PB. MT1-MMP−/− hematopoietic femurs were smaller than MT1-MMP+/+ femurs (Figure 1D). Under steady-state conditions, BM cellularity in MT1-MMP mice was decreased compared with MT1-MMP+/+ mice (Figure 1E). We next analyzed the number of MSCs by flow cytometry. The frequency of PDGFRα+/Sca1+/CD45−/Ter119− BM-derived MSCs and nestin+ niche cells was lower in MT1-MMP−/− BM cells than in MT1-MMP+/+ BM cells (Figure 1F).31 Because the skeletal malformation described in MT1-MMP−/− mice could affect the intrafemural space where hematopoiesis occurs, the BM shaft surface area was determined by microcomputer tomography (microCT). The BM shaft surface area was reduced in MT1-MMP−/− femurs compared with MT1-MMP+/+ mice (Figure 1G).
MT1-MMP gene deletion causes myelosuppression. (A-C) The number of (A) WBCs, (B) platelets (PLT), and (C) RBCs in the PB of 14-day-old mice was counted (n = 8). (D) Images of mouse femurs (bars, 1 mm). (E) Total BM cell number per femur. (F) The percentage of PDGFRα+Sca1+ cells (MSCs) was determined by flow cytometry (n = 2). (G) (left panel) Representative MicroCT scan images, and (right panel) quantification of the relative BM area within the BM femur shaft are presented (n = 3). (H) H&E-stained femur sections. Right panel shows the quantification of the BM area per femur. B indicates bone, (bars, 200 μm). (I) Megakaryocyte number (n = 5). (J-K) Real-time PCR analysis of MT1-MMP expression in (J) BM nuclear cells (BMNCs), Lin+, Lin−, and BM stroma cells, and (K) in MS-5, NIH3T3, FOB, and BMEC-1 cells. (L-M) The (L) absolute number of CFU-C per 105 BM cells and (M) frequency and absolute number/femur of CFU-S, CMP, GMP, MEP, CD34-KSL cells, and competitive repopulating units (CRU) in wild-type and knockout BM cells (n = 3). For CRU determination, freshly isolated BM cells of different cell concentration were transplanted into recipients (n = 10). Four months posttransplantation, the repopulating unit (RU) with trilineage engraftment (> 1% of donor-derived cells) was calculated. The absolute number of CRU was calculated based on the observed number of BM cells per femur. Errors in bar graphs are SEM; *P < .05, **P < .01. CFU-C indicates CFU cells; CFU-S, CFU-spleen; CMP, common myeloid progenitor; GMP, granulocyte/macrophage lineage-restricted progenitor; and MEP, megakaryocyte/erythrocyte lineage-restricted progenitor.
MT1-MMP gene deletion causes myelosuppression. (A-C) The number of (A) WBCs, (B) platelets (PLT), and (C) RBCs in the PB of 14-day-old mice was counted (n = 8). (D) Images of mouse femurs (bars, 1 mm). (E) Total BM cell number per femur. (F) The percentage of PDGFRα+Sca1+ cells (MSCs) was determined by flow cytometry (n = 2). (G) (left panel) Representative MicroCT scan images, and (right panel) quantification of the relative BM area within the BM femur shaft are presented (n = 3). (H) H&E-stained femur sections. Right panel shows the quantification of the BM area per femur. B indicates bone, (bars, 200 μm). (I) Megakaryocyte number (n = 5). (J-K) Real-time PCR analysis of MT1-MMP expression in (J) BM nuclear cells (BMNCs), Lin+, Lin−, and BM stroma cells, and (K) in MS-5, NIH3T3, FOB, and BMEC-1 cells. (L-M) The (L) absolute number of CFU-C per 105 BM cells and (M) frequency and absolute number/femur of CFU-S, CMP, GMP, MEP, CD34-KSL cells, and competitive repopulating units (CRU) in wild-type and knockout BM cells (n = 3). For CRU determination, freshly isolated BM cells of different cell concentration were transplanted into recipients (n = 10). Four months posttransplantation, the repopulating unit (RU) with trilineage engraftment (> 1% of donor-derived cells) was calculated. The absolute number of CRU was calculated based on the observed number of BM cells per femur. Errors in bar graphs are SEM; *P < .05, **P < .01. CFU-C indicates CFU cells; CFU-S, CFU-spleen; CMP, common myeloid progenitor; GMP, granulocyte/macrophage lineage-restricted progenitor; and MEP, megakaryocyte/erythrocyte lineage-restricted progenitor.
Histologic analysis of BM sections showed that MT1-MMP−/− mice exhibited a striking paucity of hematopoietic cells within the BM shaft and a reduction in the number of megakaryocytes, which are responsible for platelet production (Figure 1H-I).
We next examined HSCs and hematopoietic progenitor populations. In wild-type mice, MT1-MMP is expressed in primary BMMCs within the lineage-negative (Lin−) cell fraction, which contains mostly hematopoietic progenitor cells and a small fraction of HSCs, and in BM stromal cells, including BM endothelial cells (BMEC-1), fetal osteoblasts (FOB), and fibroblast-like stromal cells (MS-5; Figure 1J-K). The absolute number of CFU cells (CFU-C; Figure 1L) and of more primitive progenitor populations within BM cells, including day-8 CFU-spleen (CFU-S; Figure 1M), CMPs, GMPs, and MEPs and HSC-enriched CD34−c-Kit+, Sca-1+, Lin− (KSL) cells were diminished in MT1-MMP−/− BMMCs. To determine the HSC content of the BM, we performed limiting-dilution competitive repopulation analysis using MT1-MMP+/+ or MT1-MMP−/− BMMCs. There was a lower frequency, and a significantly reduced absolute number of competitive repopulation units (CRUs) per femur in MT1-MMP−/− BMMCs than in MT1-MMP+/+ BMMCs. These data indicate that MT1-MMP is required for HSC maintenance and for normal hematopoietic differentiation.
MT1-MMP−/− mice show a T- and B-cell differentiation defect
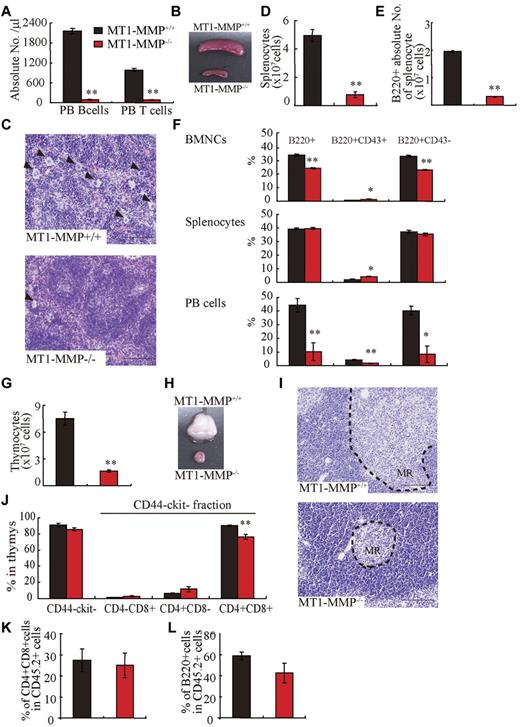
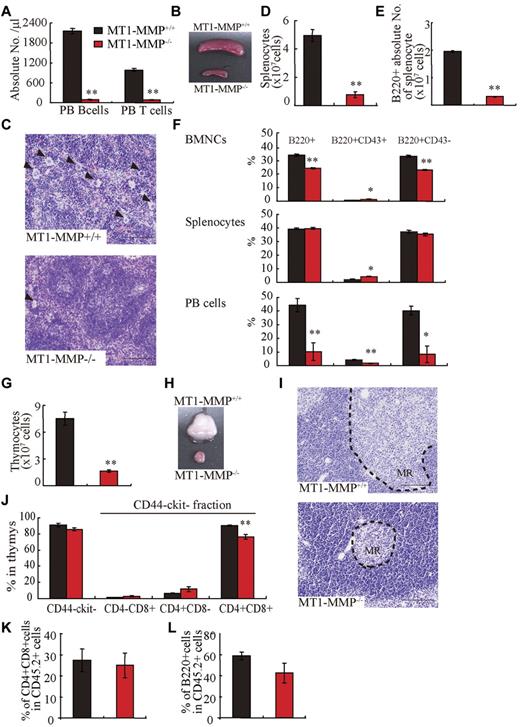
Compared with MT1-MMP+/+ mice, the absolute numbers of PB CD3+ T and B220+ B lymphocytes were reduced (Figure 2A) and the spleen was much smaller (Figure 2B) in MT1-MMP−/− mice. Similar to the BM, histologic examination revealed less megakaryocytes in the MT1-MMP−/− spleen (Figure 2C). The white pulp appeared smaller in MT1-MMP−/− spleens (Figure 2C), which correlated with a reduction in the absolute numbers of lymphoid and total B (B220+) spleen cells (Figure 2D-E). The percentage of B220+, early- (B220+CD43+), and late-stage (B220+CD43−) B cells was reduced in MT1-MMP−/− BM, spleen, and PBMCs (Figure 2F). Although the absolute number of early- and late-stage B cells in splenocytes was reduced in MT1-MMP−/− mice (data not shown), the relative percentage of early (B220+CD43+) B cells was augmented in MT1-MMP−/− splenocytes. These data demonstrate that MT1-MMP is required for normal terminal B-cell differentiation within the BM with impaired release of B cells into the circulation.
T and B lymphopoiesis are impaired in MT1-MMP−/− mice. (A) PB B- and T-cell numbers (FACS analysis). (B) Images of mouse spleens. (C) H&E-stained spleen sections. Arrowheads indicate megakaryocytes (bars, 100 μm). (D-E) The (D) total number of splenocytes and (E) B220+ cells in splenocytes (n ≥ 6). (F) The percentage of B220+ populations in BMNCs (n = 6), splenocytes (n = 3), and PB cells (n = 2). (G) Thymocyte number (n = 4). (H) Images of thymi. (I) H&E-stained thymus sections. MR indicates medullar region (bars, 100 μm). (J) The percentage of thymic T-lineage subpopulations. (K-L) BM cells from MT1-MMP+/+ and MT1-MMP−/− mice were transplanted into wild-type animals (CD45.2). (K) The percentage of donor CD4+/CD8+ T and (L) B220+B cell lineage contribution of donor-derived cells in the PB 4 months after transplantation (n = 10/group). Errors in bar graphs are SEM; *P < .05, **P < .01.
T and B lymphopoiesis are impaired in MT1-MMP−/− mice. (A) PB B- and T-cell numbers (FACS analysis). (B) Images of mouse spleens. (C) H&E-stained spleen sections. Arrowheads indicate megakaryocytes (bars, 100 μm). (D-E) The (D) total number of splenocytes and (E) B220+ cells in splenocytes (n ≥ 6). (F) The percentage of B220+ populations in BMNCs (n = 6), splenocytes (n = 3), and PB cells (n = 2). (G) Thymocyte number (n = 4). (H) Images of thymi. (I) H&E-stained thymus sections. MR indicates medullar region (bars, 100 μm). (J) The percentage of thymic T-lineage subpopulations. (K-L) BM cells from MT1-MMP+/+ and MT1-MMP−/− mice were transplanted into wild-type animals (CD45.2). (K) The percentage of donor CD4+/CD8+ T and (L) B220+B cell lineage contribution of donor-derived cells in the PB 4 months after transplantation (n = 10/group). Errors in bar graphs are SEM; *P < .05, **P < .01.
The earliest T-cell progenitors are produced in the BM. We found an increase in the percentage of immature CD4−CD8−c-Kit−CD44− cells T cells in MT1-MMP−/− BM cells (4.1 × 105 ± 0.2 × 105 cells in MT1-MMP−/− and 2.6 × 105 ± 0.3 × 105 cells in MT1-MMP+/+ BM, respectively; n = 4, P < .05). The MT1-MMP−/− thymus (Figure 2G) was small with a low number of thymocytes (Figure 2H). Thymic lobes, and specifically the medullar region, appeared smaller than in MT1-MMP+/+ mice (Figure 2I). Flow cytometric analysis of thymocytes revealed no difference in the percentage of immature CD4− or CD8− double-negative (DN) thymocytes or of CD4−CD8−c-Kit−CD44− thymocytes between the MT1-MMP+/+ and the MT1-MMP−/− mice. However, a relative decrease in the percentage of the more mature CD4+CD8+ double-positive (DP) thymocyte population and in the CD4+ single-positive (SP) cell population was revealed in thymocytes from the MT1-MMP−/− mice (Figure 2J). In absolute terms, CD4 SP numbers were reduced 2.5-fold, CD8 SP 2.1-fold, and DP thymocytes 5.3-fold, whereas the DN population remained unaffected. These data indicate that T-cell progenitors developed within the BM even in the absence of MT1-MMP, but that their further differentiation in the thymus was blocked at or before development of the immature DN thymocyte population, causing default differentiation into CD4 cell SP and NK-cell lineages.
To assess whether the abnormal thymic colonization observed in MT1-MMP−/− mice was because of the loss of MT1-MMP in hematopoietic cells or in stromal cells, MT1-MMP+/+ and MT1-MMP−/− BM donor cells were transplanted into lethally irradiated wild-type recipients. Chimeras were analyzed 4 months after transplantation. T- and B-cell differentiation of transplanted MT1-MMP−/− cells (expressing the Ag Ly5.2 (CD45.2) on leukocytes) within the BM were normal (Figure 2K-L). These data indicated that MT1-MMP−/− mice showed impaired T- and B-cell development. Even though MT1-MMP−/− mice showed a hematopoietic cell defect (see Figure 1), MT1-MMP−/− BM cells retained the potential to differentiate into T- and B-cell lineages in an MT1-MMP+/+ environment. These data indicate that, aside from its influence on stem cells, MT1-MMP also plays an additional role in regulating the BM niche.
MT1-MMP ablation impairs erythroid and myeloid differentiation
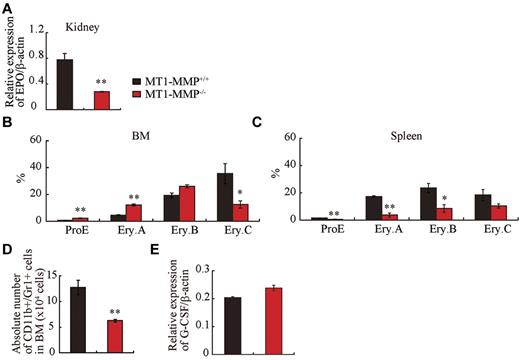
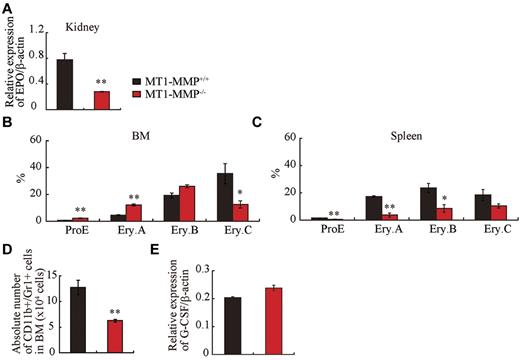
Anemia, as seen in 14-day-old MT1-MMP mice (see Figure 1C), is expected to trigger a compensatory response that is mediated principally through increased serum levels of erythropoietin (Epo), a master cytokine of erythropoiesis. Epo controls growth, survival, and differentiation of erythroid progenitors, either cooperatively with, or independently of KitL.32-34 Despite severe anemia, Epo mRNA expression in kidney tissues of MT1-MMP−/− mice was low (Figure 3A) and the expected compensatory increase in spleen size did not occur (see Figure 2B).
Erythroid and myeloid differentiation block in MT1-MMP−/− mice. (A) Real-time PCR of Epo in kidney (n = 3). (B-C) Percentage of different erythroblast populations during erythroid development (proE-EryA-EryB-EryC) of (B) BM cells or of (C) dissociated mouse spleen cells from MT1-MMP+/+ (n = 5) and MT1-MMP−/− mice (n = 3). (D) Percentage of CD11b+/Gr-1+ cells per femur as determined by FACS (n = 5). Errors in bar graphs are SEM; **P < .01. (E) Real-time PCR of G-CSF in BM cells (n = 3).
Erythroid and myeloid differentiation block in MT1-MMP−/− mice. (A) Real-time PCR of Epo in kidney (n = 3). (B-C) Percentage of different erythroblast populations during erythroid development (proE-EryA-EryB-EryC) of (B) BM cells or of (C) dissociated mouse spleen cells from MT1-MMP+/+ (n = 5) and MT1-MMP−/− mice (n = 3). (D) Percentage of CD11b+/Gr-1+ cells per femur as determined by FACS (n = 5). Errors in bar graphs are SEM; **P < .01. (E) Real-time PCR of G-CSF in BM cells (n = 3).
To assess erythroid differentiation, we examined CD71 and TER119 expression in the BM and splenic erythroblasts, and classified erythroid cells into 4 populations at progressive levels of differentiation: proerythroblasts TER119medCD71highFSChigh (ProE), TER119highCD71highFSChigh (Ery.A), TER119highCD71highFSClow (Ery.B), and TER119highCD71lowFSClow (Ery.C).35
MT1-MMP−/− BM erythroid lineage cells accumulated at the ProE, Ery.A, and Ery.B stage, and cells at the late erythroblast differentiation stages (Ery.C) were reduced (Figure 3B). In spleen, all stages of erythroid lineage cells were reduced in MT1-MMP−/− mice (Figure 3C). These data suggest that MT1-MMP is involved in both BM and splenic erythropoiesis, and that the defect in erythropoiesis in MT1-MMP−/− mice might be due, in part, to impaired Epo production.
Granulocytic cells of adult mouse BM express CD11b and high amounts of Gr-1. The absolute number of CD11b+/Gr-1+ granulocytes was lower in MT1-MMP−/− than in MT1-MMP+/+ BM cells (Figure 3D). G-CSF is a growth, differentiation, and activating factor for neutrophils and their precursors. Regardless of the observed neutropenia, no difference in G-CSF (Figure 3E) or thrombopoietin (Tpo; data not shown) expression was observed in the BM cells of these mice, indicating that other factors might be responsible for the impaired myelopoiesis.
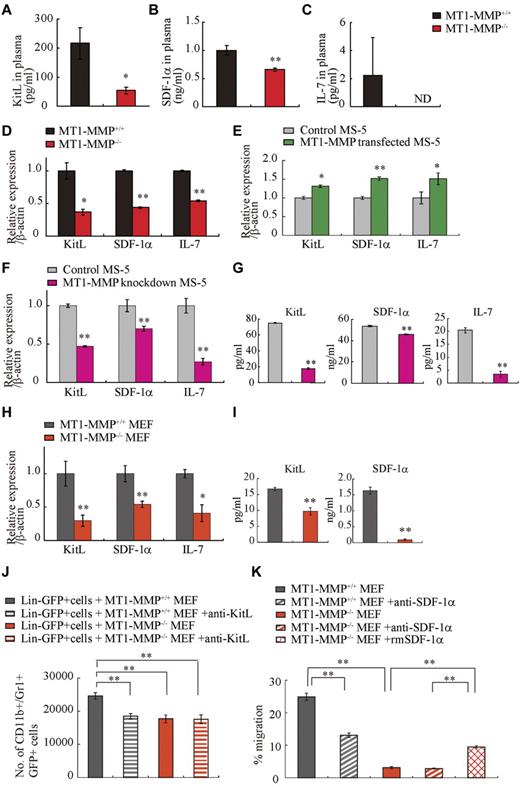
MT1-MMP deficiency reduces stromal cell–derived cytokine production
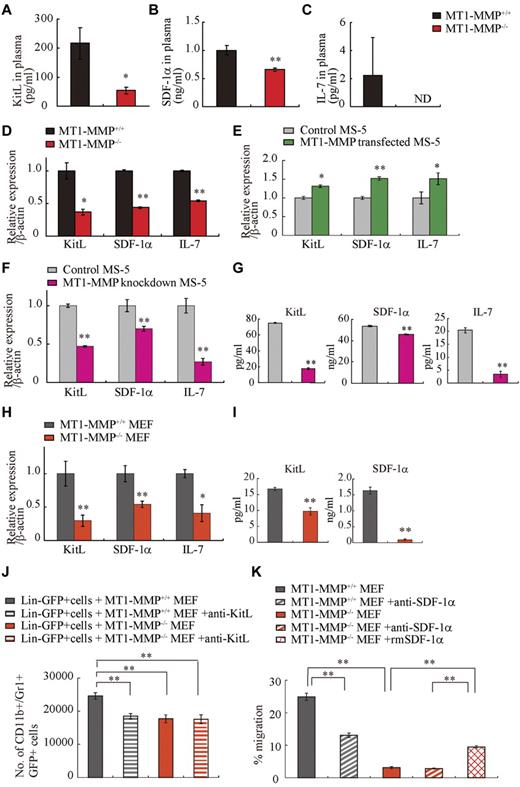
Chimera experiments indicated that the observed hematopoietic defect in MT1-MMP–deficient mice was in part because of a niche defect, suggesting that dysregulated production of stromal cell–derived cytokine(s) may be responsible for the hematopoietic differentiation block observed in MT1-MMP−/− mice. BM and thymic stromal cells produce IL-7.36 Plasma levels of the stromal cell–derived factors KitL, SDF-1α, and IL-7 were lower in MT1-MMP−/− mice than in MT1-MMP+/+ mice (Figure 4A-C). We observed a reduced number of nestin+ niche cells in MT1-MMP−/− mice and, interestingly, nestin+ niche cells highly express KitL and SDF-1.31 In accordance with these data, MT1-MMP−/− BM cells and MT1-MMP−/− primary stromal cells showed lower mRNA expression of IL-7, KitL, and SDF-1α/CXCL12 compared with MT1-MMP+/+ cells (Figure 4D, supplemental Figure 1). In addition, very few primary stromal cells grew in cultures established using MT1-MMP−/− BM cells (supplemental Figure 1).
MT1-MMP deficiency prevents transcription of niche chemokines/cytokines. (A-C) KitL, SDF-1α, and IL-7 plasma levels in MT1-MMP+/+ and MT1-MMP−/− plasma were measured by ELISA (n > 6). KitL, SDF-1α, and IL-7 gene expression in (D) total BM cells, (E) MS-5 control and MT1-MMP–overexpressing cells, and (F) MT1-MMP knockdown (KD) and control MS-5 cells were analyzed using real-time PCR. The results are expressed relative to expression of a β-actin. (G) KitL, SDF-1α, and IL-7 protein levels in MT1-MMP knockdown (KD) and control MS-5 cell-culture supernatants were determined by ELISA. (H) KitL, SDF-1α, and IL-7 gene expression in MT1-MMP+/+ and MT1-MMP−/− MEF cells was determined using real-time PCR. The results are expressed relative to expression of β-actin. (I) KitL and SDF-1α protein levels in the indicated MEF cell-culture supernatants were evaluated by ELISA. (J) Lin−GFP+ cells were cultured on MT1-MMP+/+ and MT1-MMP−/− MEF cells in the presence or absence of neutralizing Abs against KitL. The number of CD11b+Gr1+ cells was assessed after 7 days by FACS (n = 5). (K) Lin− cells were plated in transwells. MT1-MMP+/+ and MT1-MMP−/− MEF cell-culture supernatants supplemented with recombinant SDF-1α were added to the lower chamber. Neutralizing Abs against SDF-1α were added to both chambers. The percentage of migrated cells was determined (n = 9 from 2 independent experiments). Errors in bar graphs are SEM; *P < .05, **P < .01. Data shown are representative of 3 to 4 independent experiments.
MT1-MMP deficiency prevents transcription of niche chemokines/cytokines. (A-C) KitL, SDF-1α, and IL-7 plasma levels in MT1-MMP+/+ and MT1-MMP−/− plasma were measured by ELISA (n > 6). KitL, SDF-1α, and IL-7 gene expression in (D) total BM cells, (E) MS-5 control and MT1-MMP–overexpressing cells, and (F) MT1-MMP knockdown (KD) and control MS-5 cells were analyzed using real-time PCR. The results are expressed relative to expression of a β-actin. (G) KitL, SDF-1α, and IL-7 protein levels in MT1-MMP knockdown (KD) and control MS-5 cell-culture supernatants were determined by ELISA. (H) KitL, SDF-1α, and IL-7 gene expression in MT1-MMP+/+ and MT1-MMP−/− MEF cells was determined using real-time PCR. The results are expressed relative to expression of β-actin. (I) KitL and SDF-1α protein levels in the indicated MEF cell-culture supernatants were evaluated by ELISA. (J) Lin−GFP+ cells were cultured on MT1-MMP+/+ and MT1-MMP−/− MEF cells in the presence or absence of neutralizing Abs against KitL. The number of CD11b+Gr1+ cells was assessed after 7 days by FACS (n = 5). (K) Lin− cells were plated in transwells. MT1-MMP+/+ and MT1-MMP−/− MEF cell-culture supernatants supplemented with recombinant SDF-1α were added to the lower chamber. Neutralizing Abs against SDF-1α were added to both chambers. The percentage of migrated cells was determined (n = 9 from 2 independent experiments). Errors in bar graphs are SEM; *P < .05, **P < .01. Data shown are representative of 3 to 4 independent experiments.
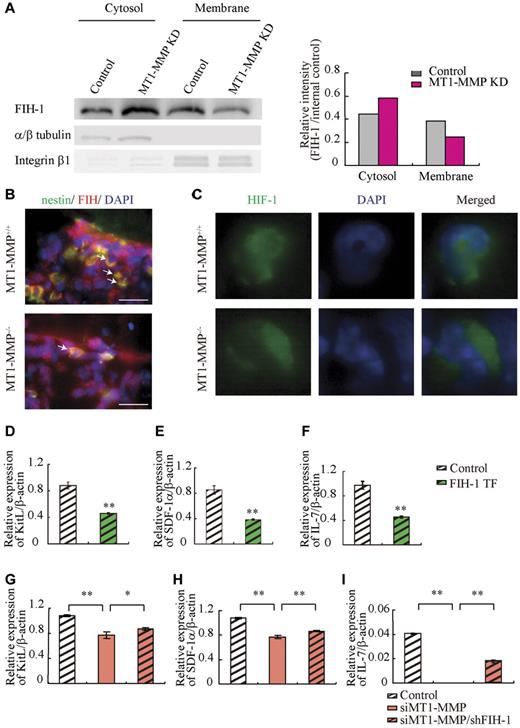
To exclude the possibility that the growth/chemokine factor decrease in MT1-MMP−/− mice was because of a lower number of the growth factor–producing stromal cells, we modulated MT1-MMP expression on a murine stromal cell line (MS-5). Overexpression of MT1-MMP in MS-5 stromal cells (MT1-MMP TF) increased KitL, SDF-1α, and IL-7 gene expression (Figure 4E). In contrast, MT1-MMP knock down in MS-5 murine stromal cells using shRNA (MT1-MMP KD) reduced the expression of KitL, SDF-1α, and IL-7 mRNA than control MS-5 cells (Figure 4F). Consistent with this result, less KitL, SDF-1α, and IL-7 protein was detected in supernatants of MT1-MMP KD cultures compared with control cultures as determined by ELISA (Figure 4G). The frequency of CXCR4 (the SDF-1 receptor) and c-Kit (the KitL receptor) expressing Sca-1+ MT1-MMP−/− BMMCs was not significantly changed (data not shown). These data indicate that loss of MT1-MMP activity in stromal cells reduced signaling by IL-7, KitL, and SDF-1α, factors known to regulate B and T lymphopoiesis and erythropoiesis.
We next examined the release and expression of these factors by primary MEFs. Confirming our BM cell and primary stromal cell cytokine/chemokine data, low expression of KitL, SDF-1α, and IL-7 was found in MT1-MMP−/− MEFs (Figure 4H-I).
To investigate whether the observed impaired production of cytokines/chemokines would also result in impaired function, we set up an MEF feeder–based culture supplemented with IL-3. Whereas Lin− cells differentiated in MT1-MMP+/+ MEF-supported cultures, the number of CD11b+/Gr-1+ cells in MT1-MMP−/− cultures did not change significantly (Figure 4J). Addition of neutralizing Abs against KitL confirmed that the observed myeloid cell differentiation was mainly because of the KitL that was released by the cultures.
We set up a migration assay to investigate SDF-1α function. Lin− cells derived from BM cells migrated toward an MT1-MMP+/+ MEF supernatant, a process that is mediated by SDF-1, as shown by using neutralizing Abs against SDF-1. In contrast, no migration of Lin− cells was observed toward an MT1-MMP−/− supernatant (Figure 4K).
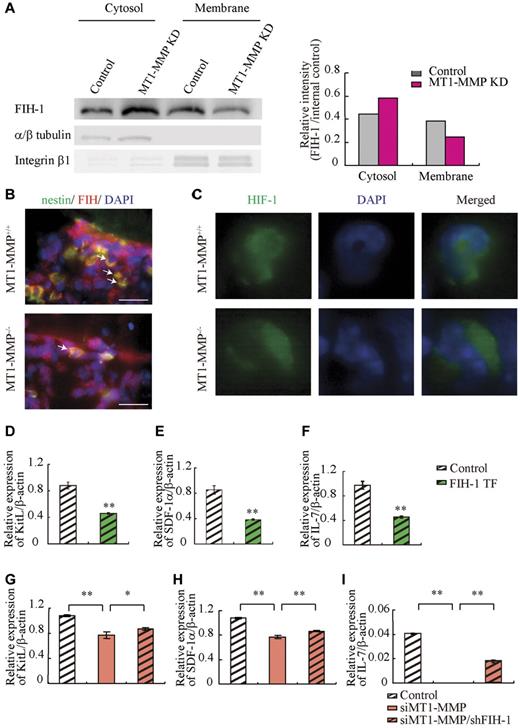
MT1-MMP increases cytokine/chemokine production in niche cells by suppressing HIF-1α
The HIF gene is known to regulate VEGF, placental growth factor, angiopoietin 2, platelet-derived growth factor-B, SDF-1α, Epo, and KitL/SCF expression.7 HIF-1α activity under normoxia depends on the FIH-1. FIH1-mediated hydroxylation disrupts a critical interaction between HIFα and the coactivators p300/CBP, impairing HIF transcriptional activity.37,38 To determine whether HIF signaling is involved in MT1-MMP modulation of hematopoietic differentiation, we assessed HIF-1α, HIF-2α, and FIH-1 protein levels in MT1-MMP knockdown and control MS-5 cells. MT1-MMP knockdown did not affect HIF-1α or HIF-2α protein expression (supplemental Figure 2), but did up-regulate FIH-1 protein expression within the cytosol of MS-5 cells (Figure 5A), where FIH-1 can prevent HIF-1α binding to the transcriptional coactivator p300/CBP thereby blocking HIF-1-induced transcription of genes such as SDF-1 and KitL.39 Immunohistochemical analysis showed that less nestin+ stromal cells coexpressing FIH-1 were found in MT1-MMP−/− BM cells than in MT1-MMP+/+ BM cells (Figure 5B). These findings are consistent with previous data that, in monocytes, the cytoplasmic tail of MT1-MMP binds to FIH-1, leading to inhibition of FIH-1 activity by its inhibitor, Mint3/APBA3.23
MT1-MMP deficiency prevents HIF-mediated transcription of nichefactors. (A) FIH-1 expression in subcellular fractions was analyzed by Western blotting. α/β tubulin and integrin-β1 are representative cytosolic and membrane proteins, respectively. (B) Representative images of immunofluorescent staining of nestin (green fluorescence) and FIH-1 (red fluorescence) in BM sections derived from MT1-MMP+/+ and MT1-MMP−/− mice. The arrows indicate nestin+/FIH-1 cells. Nuclei were counterstained with DAPI (blue; bars, 100 μm). (C) Representative images of immunofluorescent staining of HIF-1α (green fluorescence) in BM cells derived from MT1-MMP+/+ and MT1-MMP−/− mice. Nuclei were counterstained with DAPI (blue). (D-I) KitL, SDF-1α and IL-7 gene expression in (D-F) MS-5 cells overexpressing FIH-1, and (G-I) in MT1-MMP–deficient MS-5 cells with or without FIH-1 knockdown as analyzed using real-time PCR. The results are expressed relative to expression of a β-actin, which was set at 1.0. Data shown are representative of 3 to 4 independent experiments.
MT1-MMP deficiency prevents HIF-mediated transcription of nichefactors. (A) FIH-1 expression in subcellular fractions was analyzed by Western blotting. α/β tubulin and integrin-β1 are representative cytosolic and membrane proteins, respectively. (B) Representative images of immunofluorescent staining of nestin (green fluorescence) and FIH-1 (red fluorescence) in BM sections derived from MT1-MMP+/+ and MT1-MMP−/− mice. The arrows indicate nestin+/FIH-1 cells. Nuclei were counterstained with DAPI (blue; bars, 100 μm). (C) Representative images of immunofluorescent staining of HIF-1α (green fluorescence) in BM cells derived from MT1-MMP+/+ and MT1-MMP−/− mice. Nuclei were counterstained with DAPI (blue). (D-I) KitL, SDF-1α and IL-7 gene expression in (D-F) MS-5 cells overexpressing FIH-1, and (G-I) in MT1-MMP–deficient MS-5 cells with or without FIH-1 knockdown as analyzed using real-time PCR. The results are expressed relative to expression of a β-actin, which was set at 1.0. Data shown are representative of 3 to 4 independent experiments.
HIF-1 immunostaining showed that HIF-1, in contrast to its expression in MT1-MMP+/+ BM cells, was preferentially expressed in the cytoplasm of MT1-MMP−/− BM cells (Figure 5C).
Our data suggested that MT1-MMP can activate the HIF1 pathway in stromal cells as it does in monocytes. We therefore hypothesized that FIH-1 overexpression in stromal cells would cause a reduction in cytokine/chemokine production, which was indeed the case (Figure 5D-F). These data suggested that FIH-1 overexpression reduced SDF-1, KitL, and IL-7 gene transcription in stromal cells. Furthermore, although the knockdown efficiency of FIH-1 by shRNA was only 30%, FIH-1 knockdown rescued KitL, SDF-1α, and IL-7 gene expression in MT1-MMP knockdown (70% reduction by siRNA) MS-5 cells (Figure 5G-I) indicating that the decreased HIF-mediated cytokine gene transcription in MT1-MMP knockdown stromal cells can be partially rescued by blocking FIH-1 activity. These data highlight the importance of MT1-MMP–mediated HIF-1 activation for the transcriptional regulation of critical hematopoietic niche factors.
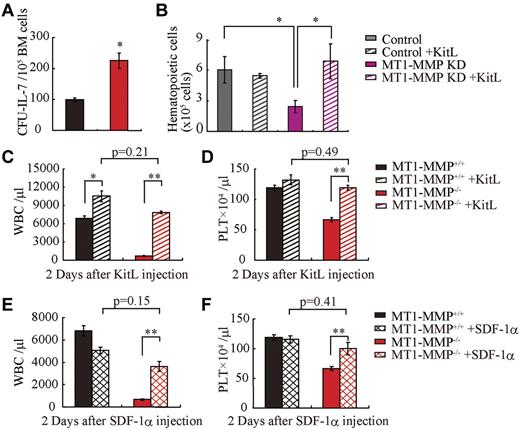
Exogenous SDF-1α and KitL addition restores leukopenia and thrombopenia in MT1-MMP−/− mice
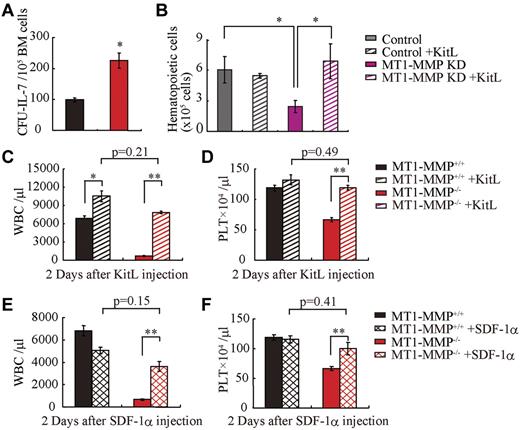
Finally, we asked whether growth factor addition would rescue the observed block in leukopenia and thrombopenia because of MT1-MMP deficiency. Indeed, addition of recombinant IL-7 to MT1-MMP−/− or wild-type BMMC cultures induced a similar number of B-cell colonies (CFU–IL-7) in the MT1-MMP−/− as in the wild-type cells (Figure 6A).
MT1-MMP–deficient mice have a defective BM stromal niche with impaired terminal differentiation because of impaired release of HIF-1α–associated factors. (A) Number of colonies in IL-7–containing cultures of MT1-MMP+/+ and MT1-MMP−/− BM cells (n = 3). (B) Coculture of wild-type Lin− BM GFP+ cells on confluent MT1-MMP knockdown or control MS-5 cells with/without KitL. (C-D) PB WBCs (C) and PLT (D) after KitL injections into MT1-MMP+/+ (n ≥ 10) and MT1-MMP−/− mice (n = 3). (E-F) PB WBCs (E) and PLT (F) counts 2 days after initiation of SDF-1 treatment of MT1-MMP+/+ (n ≥ 10) and MT1-MMP−/− mice (n = 2). Errors in bar graphs are SEM; *P < .05, **P < .01.
MT1-MMP–deficient mice have a defective BM stromal niche with impaired terminal differentiation because of impaired release of HIF-1α–associated factors. (A) Number of colonies in IL-7–containing cultures of MT1-MMP+/+ and MT1-MMP−/− BM cells (n = 3). (B) Coculture of wild-type Lin− BM GFP+ cells on confluent MT1-MMP knockdown or control MS-5 cells with/without KitL. (C-D) PB WBCs (C) and PLT (D) after KitL injections into MT1-MMP+/+ (n ≥ 10) and MT1-MMP−/− mice (n = 3). (E-F) PB WBCs (E) and PLT (F) counts 2 days after initiation of SDF-1 treatment of MT1-MMP+/+ (n ≥ 10) and MT1-MMP−/− mice (n = 2). Errors in bar graphs are SEM; *P < .05, **P < .01.
MS-5 cells support myeloid cell differentiation of Lin− wild-type cells in vitro. However, fewer GFP+ hematopoietic cells were generated from Lin− wild-type BM cells on MT1-MMP KD MS-5 stromal cells than on control MS-5 cells. Addition of KitL restored hematopoietic cell growth on the MT1-MMP KD MS-5 feeder cultures (Figure 6B).
Similarly, KitL treatment restored WBC and PLT numbers in the PB in MT1-MMP−/− mice to the values of wild-type controls (Figure 6C-D) and a single injection of SDF-1α increased both WBC and PLT counts in the PB of MT1-MMP−/− mice 2 days after injection (Figure 6E-F). Although hematopoietic cell growth was restored in MT1-MMP−/− mice by injection of KitL or SDF-1, survival was not improved (data not shown).
Collectively, these results suggest that MT1-MMP alters the HSC niche by modulating HIF signaling, which promotes cytokine production and enhances cell differentiation and migration.
Discussion
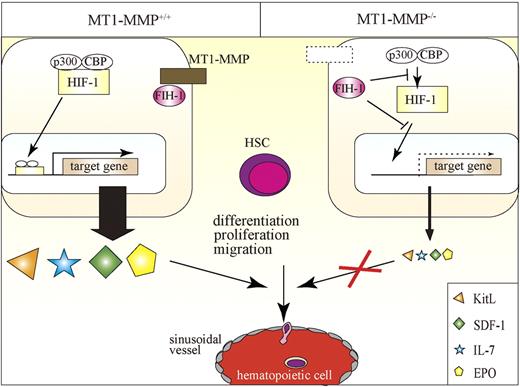
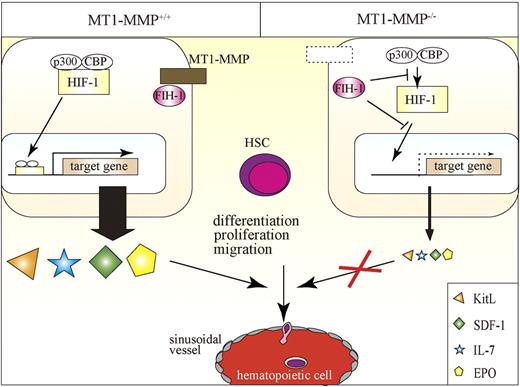
In this report, we identified MT1-MMP as a key player of postnatal hematopoiesis. We demonstrate that MT1-MMP–expressing cells serve as key links between the HIF-1 regulatory system and transcriptional regulation of vital niche chemokines/cytokines necessary for HSC maintenance and cell differentiation (Figure 7). Specifically, we show that MT1-MMP deficiency leads to impairment of steady-state hematopoiesis because of a reduced HSC pool and a trilineage differentiation block. Mechanistically, we provide evidence that MT1-MMP deficiency in niche/stromal cells increases cytosolic FIH1 on expense of the membrane-associated FIH1 expression, thereby preventing transcription of the HIF-responsive genes EPO, KitL, IL-7, and SDF-1α. Thus, this study identifies MT1-MMP as a key molecular link between hypoxia and the regulation of vital HSC niche factors.
Proposed model of the role of MT1-MMP–expressing niche cells, which serve as key links between the HIF-1 regulatory system and transcriptional regulation of vital niche chemokines/cytokines necessary for HSC maintenance and cell differentiation. MT1-MMP deficiency in niche cells up-regulates FIH1 expression, thereby preventing transcription of the HIF-responsive genes EPO, KitL, IL-7, and SDF-1α. MT1-MMP deficiency leads to impaired steady-state hematopoiesis because of a trilineage differentiation block.
Proposed model of the role of MT1-MMP–expressing niche cells, which serve as key links between the HIF-1 regulatory system and transcriptional regulation of vital niche chemokines/cytokines necessary for HSC maintenance and cell differentiation. MT1-MMP deficiency in niche cells up-regulates FIH1 expression, thereby preventing transcription of the HIF-responsive genes EPO, KitL, IL-7, and SDF-1α. MT1-MMP deficiency leads to impaired steady-state hematopoiesis because of a trilineage differentiation block.
We reported that MMP-9 and plasminogen activation is important for the ectodomain shedding of the hematopoietic growth factor like KitL.10,19 MT1-MMP can activate various proteases, including plasminogen or MMP-2, which in turn can activate MMP-9. It has been reported that MMP-2 activation can process SDF-1/CXCL12.40 We are currently examining whether ectodomain shedding by MT1-MMP–activated MMPs could be another reason for the low cytokine levels observed in MT1-MMP−/− mice.
Other known modulators of the BM and of niches such as the cancer stem cell niche, including vascular endothelial growth factor-A, angiopoietin 2, placental growth factor, and platelet-derived growth factor B, are also HIF-regulated target genes and therefore might also depend on MT1-MMP function.7,8,41-43 But further studies will be needed to proof this concept.
Using BM chimeras generated using MT1-MMP−/− and MT1-MMP+/+ donor cells, we showed that the developmental T-cell differentiation arrest observed in MT1-MMP−/− mice was mainly because of a niche defect, and not because of the impaired stem cell pool observed in these mice. In MT1-MMP−/− mice, we found preferential differentiation into CD4 SP thymocytes, which is most likely because of impaired Notch signaling.44 Indeed, a recent study demonstrated that MT1-MMP directly cleaves Dll-1, a Notch ligand on BM stromal cells.45 The continued presence of Dll-1 is required for T-cell commitment and maintenance at the DN1 and DN2 stages of thymocyte development. In the absence of Notch signaling, the developing DN1 and DN2 thymocytes adopt a NK-cell fate by default, a phenomena that we indeed observed in the MT1-MMP−/− mice.46
IL-7 signaling and Notch signaling are implicated in B-cell lymphopoiesis.47 We found that HSC differentiation toward B lymphocytes was compromised in MT1-MMP–deficient BM cells. The long-term proliferation capacity of BM pre-B1 cells is known to be critically dependent on KitL and IL-7 expression and signaling, factors that we have shown require the presence of MT1-MMP for their expression.48,49 Furthermore, we provide evidence that MT1-MMP deficiency in stromal cells also impairs the expression of SDF-1α, which is a key regulator of B-cell lymphopoiesis and BM myelopoiesis.50 Our data are consistent with those of a previous report, which observed that defects in B lymphopoiesis are modulated by MT1-MMP–mediated cleavage of Dll-1 on BM stromal cells.45 Impaired BM myelopoiesis was found in MT1-MMP–deficient mice, regardless of the fact that G-CSF expression was normal. Factors such as KitL and SDF-1α have also been implicated in myelopoiesis, and therefore might be at least partially responsible for the observed phenotype.
In addition, in vivo administration of KitL and SDF-1α rescued the pancytopenia in MT1-MMP–deficient mice. As both growth factors can increase the egress and mobilization of mature hematopoietic cells, but also can promote hematopoietic cell differentiation, the restoration of PB counts in MT1-MMP–deficient mice could be because of improved hematopoietic cell migration and/or differentiation. We confirmed data by Vagima et al demonstrating that MT1-MMP is expressed on hematopoietic progenitor cells.21 This group showed that MT1-MMP is required for G-CSF–mediated hematopoietic progenitor cell mobilization.
SDF-1α is required for the maintenance of BM HSCs and is expressed by both perivascular and endosteal cells.11,13 Deficits in the maintenance of the HSC pool in the absence of CXCR4 are HSC autonomous. Indeed, we could show that MT1-MMP deficiency altered SDF-1α/CXCL12-CXCR4 signaling and impaired the stem cell pool.
Our studies on the role of MT1-MMP in hematopoietic niche/stromal cells provide the rationale for further exploration of MT1-MMP in other hypoxic niches, for example, the cancer niche or the “ischemia-associated niche,” as MT1-MMP seems to control the hematopoietic cell response in those diseases by controlling cytokine production.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the FACS core facility in the Institute of Medical Science (University of Tokyo) for their help. They also thank Dr Hideo Ema for critical advice. Stephanie C. Napier kindly provided editorial assistance to the authors during the preparation of this manuscript. Pauline O'Grady edited the manuscript.
This work was supported by grants from the Japan Society for the Promotion of Science and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT; K.H. and B.H.); a Grant-in-Aid for Scientific Research on Priority Areas from the MEXT (K.H.); the Mitsubishi Pharma Research Foundation (K.H.); a Grant-in-Aid for Scientific Research on Innovative Areas from the MEXT (B.H.); and the Program for Improvement of the Research Environment for Young Researchers (B.H.) funded by the Special Coordination Funds for Promoting Science and Technology of the MEXT, Japan. In addition, this work was supported by grants from ENSHIN Medical Research Foundation, from Kyowa Hakko Kirin Co Ltd, and from Daiichi Sankyo Company Ltd (K.H.).
Authorship
Contribution: C.N., B.H., and K.H. designed and performed experiments, analyzed and interpreted data, and wrote the manuscript; K.K., Y.T., I.G., A.S., M.O.-K., Y.M., M.N., T.S., N.K., T.K., and S.K.-K. participated in performing experiments, data analysis, and discussion; M.S. and H.N. interpreted data; and H.N., B.H., and K.H. assisted with experimental design and manuscript writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Koichi Hattori, MD, PhD, Center for Stem Cell Biology and Regenerative Medicine, Institute of Medical Science at the University of Tokyo, 4-6-1, Shirokanedai, Minato-ku, Tokyo 108-8639, Japan; e-mail: khattori@ims.u-tokyo.ac.jp.
References
Author notes
B.H. and K.H. share senior authorship.