Abstract
Understanding the cellular mechanisms of platelet activation and their pharmacologic modulation is of major interest for basic and clinical research. Here we introduce a comprehensive human platelet repository (PlateletWeb) for systems biologic analysis of platelets in the functional context of integrated networks. Functional, drug, and pathway associations provide a first systemic insight into various aspects of platelet functionality and pharmacologic regulation. Detailed manual curation of recent platelet proteome and transcriptome studies yielded more than 5000 platelet proteins. Integration of protein-protein interactions with kinase-substrate relationships unraveled the platelet signaling network involving more than 70% of all platelet proteins. Analysis of the platelet kinome in the context of the kinase phylogenetic background revealed an over-representation of tyrosine kinase substrates. The extraction and graphical visualization of specific subnetworks allow identification of all major signaling modules involved in activation and inhibition. An in-depth analysis of DOK1 signaling identifies putative signal modulators of the integrin network. Through integration of various information sources and high curation standards, the PlateletWeb knowledge base offers the systems biologic background for the investigation of signal transduction in human platelets (http://plateletweb.bioapps.biozentrum.uni-wuerzburg.de).
Introduction
Platelets play a key role in hemostasis and represent a central target for research in many pathophysiologic processes, including cardiovascular diseases, inflammation, host immune response, and metastasis.1,2 These anucleate blood cells originate from megakaryocytes and have a short life span of approximately 10 days. They are activated by injury of the vessel wall, which causes platelets to adhere to the injured surface, aggregate, and build a firm thrombus with the help of surface-adhesion molecules primarily from the family of integrins. By secretion of factors, such as TXA2 and ADP, more platelets are gathered at the damaged endothelium. In a tight balance, platelets control the initial steps of hemostasis and thrombus formation and play a key role in pathologic processes, such as atherosclerosis. The finely tuned balance between platelet activation and inhibition ensures the optimal functionality of hemostatic mechanisms. Disturbances of this system are involved in the most common cardiovascular diseases: thrombosis, stroke, and myocardial infarction.3 Platelets contain a pool of mRNA, which can be spliced and translated in a signal-dependent manner.4-7 Large-scale proteomic data are crucial for understanding the systems biologic background of cellular processes as well as for identifying potential new biomarkers in human diseases as recently shown for atherothrombosis.8,9 Proteomic analyses of platelet signaling have been spurred by the development of novel mass spectrometry-based technologies for proteomics and phosphoproteomics.10 Complementing studies of the entire platelet proteome, several investigations on platelet subcompartments have been performed, such as microparticles, α-granules, membranes, and the secretome.11 Additional studies were focused on the platelet phosphoproteome12 ; however, the signaling changes have not yet been analyzed in a network context. Considerable research efforts have been invested in unraveling platelet signaling, most of it focusing on specific molecules or subparts of signaling pathways.13 New studies are constantly performed, and there is a growing need for a platform combining multiple sources of platelet data. Bioinformatic integration is a prerequisite for the systems biologic analysis of human platelets, as done previously for a large variety of human cells and tissues.14-16 We present here a knowledge base on the integration of various large-scale datasets yielding an in-depth catalog of human platelet proteins, network modules, and regulators of platelet functionality. Platelet source data complemented with references enable the evaluation of data quality, whereas the various query modes allow data integration, thus providing insights on multiple options, such as pharmacologic modulation and network analysis. The systems view allows not only an integrative approach for platelet investigation but also a deeper understanding of the dynamic modifications of platelet signaling networks with their different systems states and pathophysiologic roles.
Methods
Platelet interactome, proteome, and phosphoproteome
Information on human protein-protein interactions (PPIs) was obtained from the Human Protein Reference Database (HPRD; Version 9.0, April 2010)14 and the Entrez Gene National Center for Biotechnology Information (NCBI) server17 (accessed December 2010). It was combined with data on protein phosphorylation from HPRD (Version 9.0) and PhosphoSite (accessed January 2011)15 as well as kinase predictions for platelet-specific phosphoproteome data12 using the NetworKIN algorithm.18,19 From this an interaction network was created using data for interacting proteins. Similarly, a phosphorylation network consisting of all kinases and their substrates (which may also be kinases) has been assembled. Here the degree of each kinase represents the number of substrates it phosphorylates. In total, the complete human PPI network contains 54 218 simple interactions, 4406 phosphorylation events, and 135 dephosphorylation events between 10 916 human proteins.
Based on a first catalog of the platelet proteome,20 data from various mass spectrometry studies published over the last 10 years have been assembled. These contain studies of unfractionated platelets as well as studies of specific platelet subcompartments, including plasma membrane, secretome, and microparticles. Furthermore, literature-curated information was extracted from the NCBI GeneRifs17 and filtered for new platelet proteins. Platelet transcriptome data included a previously performed Serial Analysis of Gene Expression (SAGE) analysis of human platelets.5 A detailed listing of all platelet data sources used is available in supplemental Table 1 (see the Supplemental Materials link at the top of the article). Platelet-specific, experimentally validated phosphorylation sites were assembled from a recent mass spectrometry analysis of resting human platelets (533 phosphosites)12 and from experiments described in the literature (73 734 phosphosites). Phosphorylation sites from platelet-specific experiments are referred to as “platelet phosphorylations,” whereas all sites from databases and literature sources are referred to as “human phosphorylations.”
Platelet protein information
Protein name and summary information was extracted from the NCBI. Physical properties of the protein (protein length, molecular weight, isoform, protein sequence, and mRNA sequence) were assembled from HPRD. To attain cross-references between multiple databases and their datasets, the International Protein Index was used internally. Approved gene symbols for the corresponding proteins have been obtained from HUGO Gene Nomenclature Committee. Information on proteins domains and motifs has been collected from HPRD. Transmembrane domains have been predicted using the TMHMM Server, Version 2.0,21 yielding a total of 5107 transmembrane proteins, of which 1158 are platelet proteins. Tissue expression data from HPRD were used to determine further platelet proteins in the database.
Kinase and phosphatase information
A comprehensive list of human kinases was extracted from Manning et al22 and used for reference and validation of the HPRD phosphorylation data. All kinases were mapped to the human kinome tree created by Miller et al19, and visualized using the interactive online tool Tree Of Life.23
The catalog of human phosphatases was acquired from the Human Protein Phosphatases PCR Array (QIAGEN; 82 phosphatases) and the assembly of protein tyrosine phosphatases in the human genome24 (103 phosphatases). The rest of the phosphatases were added by manual search in the PlateletWeb for proteins with the term “protein phosphatases” in their description. The total number of human protein phosphatases adds up to 191 (phosphatases associated with a substrate, 39; platelet phosphatases, 73; platelet phosphatases with a substrate, 24).
Drugs and diseases
Drug data were downloaded from DrugBank Version 3.0,25 which includes detailed information on drugs as well as on drug targets. The drug-target relationships, provided in this catalog, are composed of physical drug-target interactions as well as indirect functional effects. The drugs are divided into experimental and US Food and Drug Administration–approved. The database contains 4311 human drugs, which have a human drug target in the PlateletWeb knowledge base (approved, 1195; experimental, 3015) and act on 2106 distinct human proteins. There are 950 platelet proteins among these drug targets. Genetic disease information was extracted from HPRD and is available for 701 platelet proteins.
GO functional annotation
Gene Ontology (GO) information was extracted from the GO database26 (Web site accessed December 2010) and used for functional enrichment analysis. There are 4728 platelet proteins annotated with a GO function, which accounts for a coverage of 94%.
GO enrichment analysis was performed by the BINGO plug-in Version 2.4427 of the network analysis software Cytoscape.28 For the full GO annotation comparison, all platelet proteins with a GO functional annotation in the network were considered: Biological Process, 3263; Molecular Function, 3412; Cellular Component, 3394 of total 5025 platelet proteins). Statistically significant categories (P < .0001) were selected according to their corrected P values, using a hypergeometric test. Visualization of the Biologic Process, Molecular Function, and Cellular Component results was performed for selected GO terms with less than 600 proteins. The top 25 GO terms were then used for visualization and colored according to the common parent with high information content in the hierarchical GO tree. Common parents were extracted using Visualize Go from the GO ontology page.
KEGG pathway data and analysis
KEGG pathways were downloaded from the KEGG database (Release 57.0, January 1, 2011).16 The Advanced Pathway Painter Version 2.26 was used for the visualization of KEGG pathways in the PlateletWeb knowledge base. Enrichment analysis of pathways was performed using Fisher exact test comparing the number of platelet proteins in the pathway against the number of all platelet proteins annotated in KEGG pathways.
Statistical analyses
Fisher exact test (2-sided for kinase enrichment) was used for all enrichment analyses. All P values were adjusted for multiple testing families using the Benjamini and Hochberg approach. Adjusted P values < .05 were considered significant. All statistical analyses were performed with R Version 2.13.0 statistical analysis software.29
Results
Assembly of the PlateletWeb resource
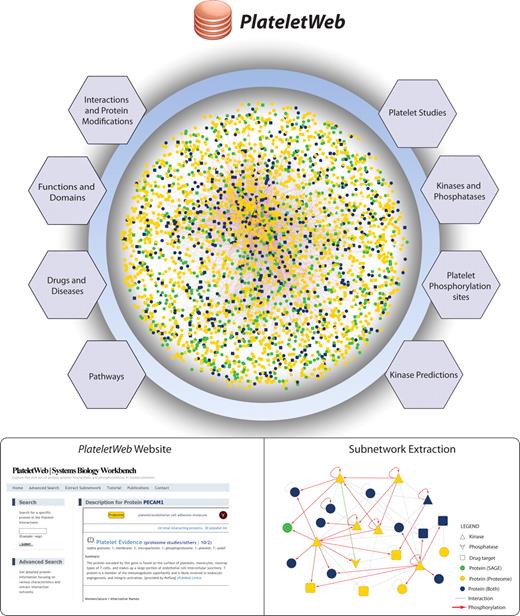
In a previous study,20 we collected platelet proteome and transcriptome data along with human PPIs to derive a first draft of the platelet interactome. Here we present a comprehensive functional knowledge base (PlateletWeb, Systems Biology Workbench) for the analysis of signaling in human platelets (Figure 1). Besides extended interactome and proteome data, it additionally provides a systems biologic perspective using site-specific phosphorylation reactions, detailed information on each protein, assessment of data quality, as well as ample information on pathways, disease associations, drugs, and functional annotations. All data are available online as an Internet platform with a broad and intuitive interface for functional network analysis, including advanced data-mining capabilities and the visualization of subnetworks with integrated information on phosphorylations and interactions.
Assembly of an integrated platelet network. Data from various sources were combined, creating a comprehensive network, which serves as a foundation for the integrated network analysis of signaling pathways, kinase distribution, and functional enrichment in platelets. A comprehensive resource of human platelets was created using data from protein-protein interactions and modifications complemented with drug and disease association data, pathway information, kinase-substrate data, and predictions for kinases and transmembrane domains. Based on this, a systems biology workbench was created for easy access to platelet proteins and their related information. Subnetwork extraction allows in-depth analysis of multiple proteins with their modification type, kinase-substrate relationships, and drug association.
Assembly of an integrated platelet network. Data from various sources were combined, creating a comprehensive network, which serves as a foundation for the integrated network analysis of signaling pathways, kinase distribution, and functional enrichment in platelets. A comprehensive resource of human platelets was created using data from protein-protein interactions and modifications complemented with drug and disease association data, pathway information, kinase-substrate data, and predictions for kinases and transmembrane domains. Based on this, a systems biology workbench was created for easy access to platelet proteins and their related information. Subnetwork extraction allows in-depth analysis of multiple proteins with their modification type, kinase-substrate relationships, and drug association.
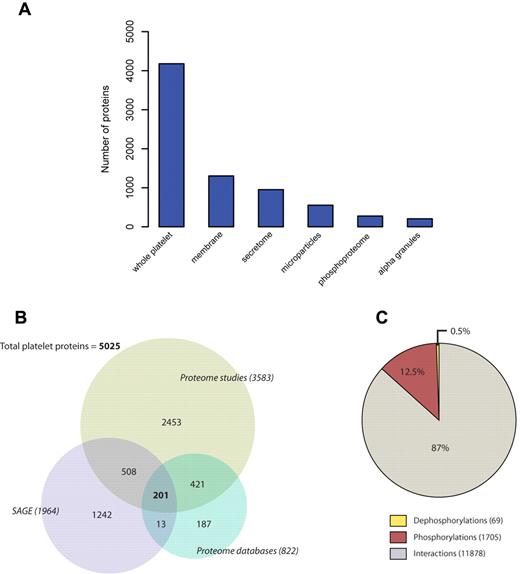
Platelet proteome data have been collected manually from all available literature and proteomic databases (supplemental Table 1) and combined with transcriptome data (SAGE).5 Detailed annotation resulted in a set of 5025 platelet proteins, including 22 major platelet datasets in the database (supplemental Figure 1; supplemental Table 1), creating a comprehensive and reliable backbone for platelet-specific information. Proteins were also separated according to the cellular subcompartment from which they were isolated, with membranes and secretome being the most abundant fractions after whole platelet analyses (Figure 2A). Proteins were analyzed individually depending on the source of platelet information (proteome and transcriptome). The proteome group was in turn subdivided into proteome studies (large-scale proteomic analysis) and proteome databases (bioinformatic databases). The majority of proteins (3783 proteins, 75%) have been described on the proteome level, 14% (722 proteins) have additional evidence on the transcriptome level, and 25% (1242) of all platelet proteins have been detected exclusively on the mRNA level (Figure 2B).
Analysis of the platelet proteome. (A) Platelet proteins were detected in multiple fractions depending on the type of study. Most studies included whole platelet lysates or membrane proteome analysis. (B) Distribution of transcriptome (SAGE) and proteome proteins in the PlateletWeb database. The proteome sources were further divided into proteome studies and proteome databases with a higher fraction of proteins extracted from proteome studies. (C) Interaction, phosphorylation, and dephosphorylation events among platelet proteins are presented as fractions of the total interactome.
Analysis of the platelet proteome. (A) Platelet proteins were detected in multiple fractions depending on the type of study. Most studies included whole platelet lysates or membrane proteome analysis. (B) Distribution of transcriptome (SAGE) and proteome proteins in the PlateletWeb database. The proteome sources were further divided into proteome studies and proteome databases with a higher fraction of proteins extracted from proteome studies. (C) Interaction, phosphorylation, and dephosphorylation events among platelet proteins are presented as fractions of the total interactome.
The platelet signaling network exhibits high connectivity
Using data on PPIs and site-specific phosphorylations as well as dephosphorylations, we assembled a densely connected central platelet network consisting of 3628 platelet proteins and 13 652 interactions between them, including 1704 described phosphorylations. This represents an evident increase in the number of proteins and interactions compared with earlier studies.20 The assembled network has a high average degree and good connectivity, reflected by a huge “largest connected component” of the network (supplemental Table 2), suggesting that the large number of recently included platelet proteins are highly connected with each other and the proteome data are well covered by the network. Based on the curated interactome information, platelet interactions were annotated according to the method used for their detection (yeast 2-hybrid, in vivo, in vitro). The interaction data are based on 7127 distinct experimental studies, assembled by high-quality databases. Decomposing the network into interactions, phosphorylations and dephosphorylations revealed that 13% of the platelet interactome consists of phosphorylation events (Figure 2C).
Analysis of the platelet phosphoproteome
To incorporate the central phosphorylation signaling framework into our resource, we extracted site-specific phosphorylations from public databases, which are based on experimental data of human cells and tissue experiments published in the literature.14,15 The entire set of human phosphorylations consists of 73 734 distinct phosphorylation sites on 10 441 human proteins. Phosphorylation sites of platelet proteins account for 39% of all phosphorylations (28 800). These literature-derived data were complemented by a set of 533 phosphosites experimentally measured in human platelets,12 of which 16 sites have not yet been described in the human proteome.
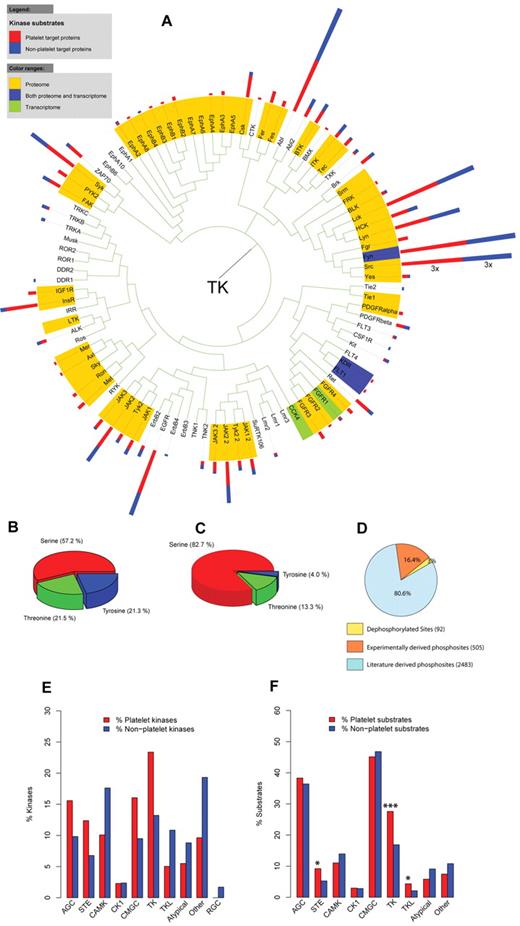
Analysis of residue-specific distribution of the literature phosphorylation sites on platelet proteins showed that the majority (57.2%) of these sites are on a serine residue followed by almost equal amounts of threonine (21.5%) and tyrosine phosphorylations (21.3%; Figure 3B), which is comparable to the distribution of all human phosphosites (data not shown). In contrast to this, the experimentally validated platelet phosphosites contain a substantially lower share of tyrosine phosphorylations (4.0%) combined with a higher percentage of serine phosphorylations (82.7%; Figure 3C).
Analysis of platelet kinases and their substrates. (A) Phylogenetic tree of the platelet and nonplatelet kinases belonging to the family of TK. Colors highlight the level of platelet expression: yellow represents proteome; green, transcriptome; and blue, both. The kinase substrates are represented as 2-colored bars next to the corresponding kinase label: red represents platelet substrates; and blue, nonplatelet substrates. (B) The distribution of pS-, pT-, and pY-phosphorylation sites in platelet proteins and (C) experimentally validated phosphorylation sites in platelets. Serine phosphorylations were most abundant, followed by threonine and tyrosine phosphorylations. (D) The distribution of phosphorylation and dephosphorylations sites associated with a kinase according to the source of protein modification. (E) Enrichment analysis of kinase families indicated no over-representation of any kinase family in platelets. (F) Enrichment analysis of platelet kinase substrates according to the kinase family revealed a significant enrichment of platelet substrates for kinases of the STE, TKL, and TK families. *P < .05.***P < .001. AGC indicates AGC kinase family; STE, STE kinase family; CAMK, calcium-calmodulin-dependent protein kinase family; CK1, casein kinase 1 family; CMGC, CMGC kinase family; TKL, tyrosine-kinase-like kinase family; TK, tyrosine kinase family; Atypical, kinase family of atypical protein kinases; and RGC, RGC kinase family.
Analysis of platelet kinases and their substrates. (A) Phylogenetic tree of the platelet and nonplatelet kinases belonging to the family of TK. Colors highlight the level of platelet expression: yellow represents proteome; green, transcriptome; and blue, both. The kinase substrates are represented as 2-colored bars next to the corresponding kinase label: red represents platelet substrates; and blue, nonplatelet substrates. (B) The distribution of pS-, pT-, and pY-phosphorylation sites in platelet proteins and (C) experimentally validated phosphorylation sites in platelets. Serine phosphorylations were most abundant, followed by threonine and tyrosine phosphorylations. (D) The distribution of phosphorylation and dephosphorylations sites associated with a kinase according to the source of protein modification. (E) Enrichment analysis of kinase families indicated no over-representation of any kinase family in platelets. (F) Enrichment analysis of platelet kinase substrates according to the kinase family revealed a significant enrichment of platelet substrates for kinases of the STE, TKL, and TK families. *P < .05.***P < .001. AGC indicates AGC kinase family; STE, STE kinase family; CAMK, calcium-calmodulin-dependent protein kinase family; CK1, casein kinase 1 family; CMGC, CMGC kinase family; TKL, tyrosine-kinase-like kinase family; TK, tyrosine kinase family; Atypical, kinase family of atypical protein kinases; and RGC, RGC kinase family.
Distinct phosphosites guide platelet responses
Phosphorylation sites can be analyzed in a network only in the context of a given kinase-substrate relationship, supplying information about the responsible kinase. In this regard, we focused on phosphorylated platelet proteins associated with a kinase, which represent only 23% (n = 814) of all phosphorylated platelet proteins (n = 3532). Transferring this question onto distinct phosphorylation sites, kinase and phosphatase information was available for a total of 3080 sites. However, kinase data for experimentally validated phosphorylations were available for only 69 phosphosites. We used a novel network-based algorithm to predict potential kinases for these sites,18,19 which resulted in kinase predictions for a further 436 sites, yielding a total kinase annotation for 505 (94.5%) phosphosites. When introduced into the entire platelet phosphoproteome, these predictions contribute to 16% of all modification events with available kinase or phosphatase information (Figure 3D).
The entire platelet proteome dataset contains 229 kinases (43.5% of 526 total human kinases), 162 (70.7%) of which have well-described substrates in the platelet proteome. Nearly all (216, 94%) have documented phosphorylation sites. The kinase families tyrosine kinase (TK), CMGC, and AGC contain the highest percentage of platelet kinases (supplemental Table 3). Dephosphorylation is achieved by 73 platelet phosphatases (38.2% of 191 total human phosphatases; supplemental Table 4), 24 of which have characterized substrates. A graphical representation of the platelet kinases and phosphorylated proteins within the network is available in supplemental Figure 2.
A central role of tyrosine phosphorylations in platelet signaling
To analyze platelet kinases and their substrates in a phylogenetic context, all assembled platelet kinases were mapped onto the human kinome tree.19 The kinase data were then visualized in combination with the level of platelet expression (proteome, transcriptome, or both) and the number of platelet and nonplatelet substrates (supplemental Figure 3). The majority of platelet tyrosine kinases were found on the proteome level. The tyrosine kinase subtree was extracted for detailed analysis from the main phylogenetic tree (Figure 3A). This subtree reveals a strong representation of Src-family kinases, which have many characterized substrates in platelets and mediate platelet activation through numerous phosphorylation events. In contrast, a clear absence of kinase groups, such as the neurotrophic tyrosine kinases (TRKA, TRKB, TRKC) and neural growth kinases (ROR1), can be observed, as these are kinases with a high specificity for neuronal tissues.
The resource allows detailed investigation of kinase families in the platelet kinome based on the kinase family classification presented by Manning et al22 (supplemental Table 5). Although kinases of the tyrosine and AGC family are relatively abundant in human platelets, no significant over-representation of a particular kinase family could be detected after multiple testing correction (Figure 3E; supplemental Figure 4). Analogously, we investigated the enrichment of platelet kinase substrates according to kinase family and residue specificity (Figure 3F; supplemental Figure 4). The analysis yielded a significant enrichment of platelet substrates of STE (P = .01), TKL (P = .03), and predominantly substrates of the TK kinase family (P = 1.37 × 10−4), also reflected on the residue level by a significant enrichment of tyrosine-specific kinase substrates. The STE family contains a broad repertoire of kinases important for the upstream signaling in the MAPK/ERK pathway (MAPKKK kinases). The TK group contains the main activatory kinases of the Src-family for which a large number of platelet substrates have been described (compare Figure 3A), and they participate in processes, such as collagen and VWF-induced platelet activation.30 Further analysis indicated that gated interaction-mediating domains SH2 (supplemental Figure 5) are over-represented in platelets, consistent with their role as counterparts of tyrosine kinases through recognition of tyrosine-phosphorylated residues.31
Platelet functionality and pathway analysis
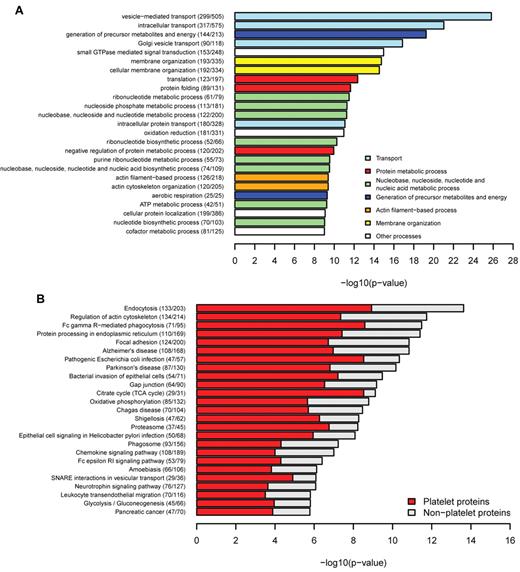
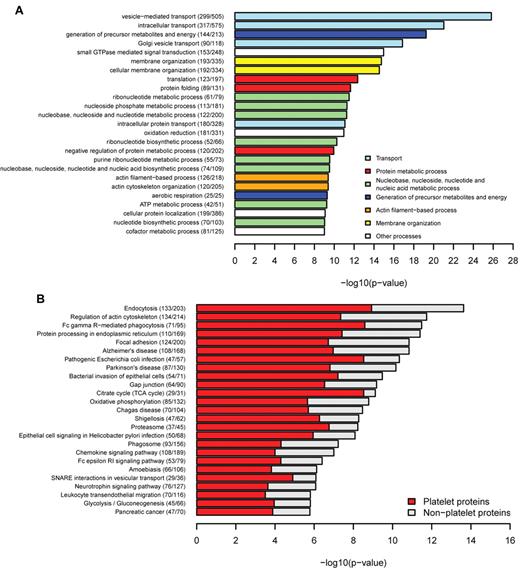
Functional characteristics of platelet genes and gene products are stored in the knowledge base, including the GO categories.26 This allows a functional characterization of the platelet network. GO enrichment analysis of the platelet interactome compared with the entire human interaction network27 revealed a significant enrichment of platelet relevant biologic processes, such as “vesicle-mediated transport” (P = 1.54 × 10−26), “small GTPase-mediated signal transduction” (P = 1.08 × 10−15), and “actin cytoskeleton organization” (P = 4.09 × 10−10), processes known to play a central role in platelet function (Figure 4A). Similarly, enriched molecular functions mediate key platelet processes, such as “actin binding” (P = 8.51 × 10−12) and “cytoskeletal protein binding” (P = 1.09 × 10−11; supplemental Figure 6A), whereas cellular components were enriched for mitochondrial compartments and membrane components (supplemental Figure 6B). Membrane proteins perform key roles in cell-cell signaling and facilitate the initial steps in cell signaling activation; therefore, predictions for transmembrane domains were performed for all human proteins in the database. The number of transmembrane domains containing proteins in platelets (1158) represents 23% of the platelet proteome, which is comparable with the number of all human transmembrane proteins constituting 26% of the human interactome. These results correlate with the 27% transmembrane proteins estimated in the human genome by Almen et al.32 In accordance with the GO enrichment results, the KEGG pathway analysis (Figure 4B) yielded “endocytosis” as the top enriched pathway (P = 2.35 × 10−14), followed by “regulation of cytoskeleton” (P = 1.82 × 10−12) and “Fc γ receptor mediated phagocytosis” (P = 3.28 × 10−12). Integrin signaling was present in the focal adhesion pathway, emphasizing its importance in platelet activation. Pathways associated with inflammation (“Leukocyte transendothelial migration” and “Chemokine signaling pathway”) were also enriched for platelet proteins, consistent with the platelet role in inflammatory responses.2 Pathways highly specific for other tissues, such as “Neuroactive ligand-receptor interaction” (P = 2.06 × 10−15) and “Olfactory transduction” (P = 4.36 × 10−50), were strongly under-represented in the platelet proteome (supplemental Table 6).
Functional enrichment of platelets. (A) Biological processes were tested for enrichment in platelets. The top most significantly enriched specific terms were plotted against the negative log10 of the P value and colored according to their general functional category. Number of platelet proteins and total proteins for each term are given in parentheses. There is enrichment of terms related to transport, membrane organization, actin-filament based processes, and GTPase-mediated signal transduction. (B) KEGG pathway enrichment. Endocytosis, regulation of actin cytoskeleton, and focal adhesion were highly enriched for platelet proteins.
Functional enrichment of platelets. (A) Biological processes were tested for enrichment in platelets. The top most significantly enriched specific terms were plotted against the negative log10 of the P value and colored according to their general functional category. Number of platelet proteins and total proteins for each term are given in parentheses. There is enrichment of terms related to transport, membrane organization, actin-filament based processes, and GTPase-mediated signal transduction. (B) KEGG pathway enrichment. Endocytosis, regulation of actin cytoskeleton, and focal adhesion were highly enriched for platelet proteins.
Topologic investigation of drug targets and disease proteins
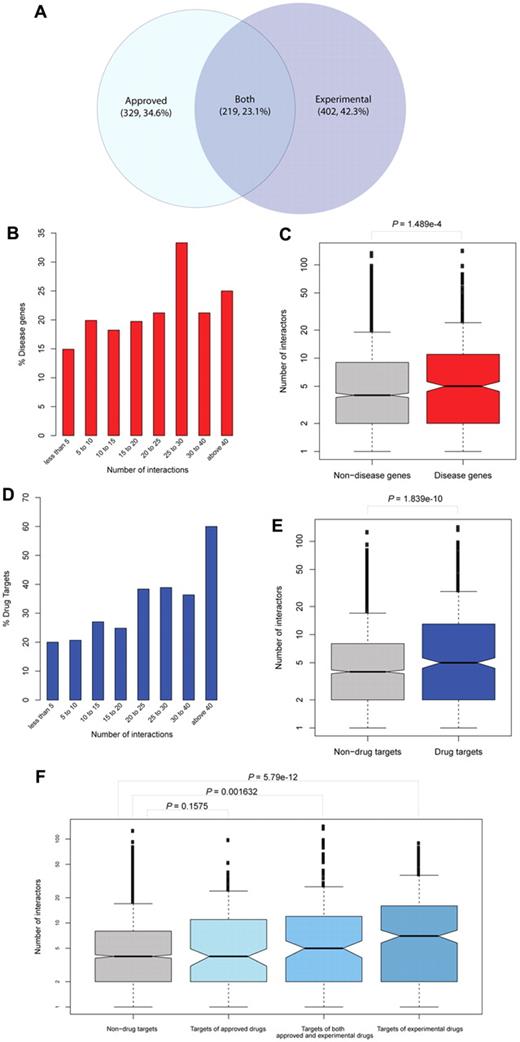
A systems biologic viewpoint can exploit key features of the platelet signaling network for drug-protein interactions by combining drug target information with phosphorylation and interaction data. For extensive analysis of the platelet drug target network, data on human drugs and drug targets of both approved (1195) and experimental (3015) types were extracted from the DrugBank database.25 This yielded a dataset of 4311 human drugs, half of which (2706) act on platelet proteins. Of 2106 human proteins associated with drugs, 950 are platelet drug targets (19% of all platelet proteins). For topologic exploration of these drug targets, we examined their connectivity within the network. Proteins were grouped by degree of interaction into nonoverlapping intervals, and the fraction of drug targets (of approved and experimental drugs) in each group was calculated (Figure 5A). The resulting distribution of drug targets indicated an overall increase in drug targets among highly connected proteins in the network (Figure 5D), and the number of interactors of drug-targeted proteins (mean = 11.6) were significantly higher than those not associated with drugs (mean = 6.8; P = 1.84 × 10−10, Figure 5E).
Topologic network analysis of drug targets and disease genes. (A) Distribution of platelet drug targets according to the type (group) of drugs with which they are associated (approved or experimental). (B) Proteins were grouped by their number of interactions. Network analysis indicates a higher number of disease-associated genes among well-connected proteins. (C) The datasets of disease proteins and nondisease proteins are presented in box-plots. The number of interactors is shown in a logarithmic scale on the y-axis. Their median is higher in the group of disease proteins compared with nondisease proteins. (D) Topologic network analysis of drug targets reveals an increase in the number of drug targets among highly connected platelet proteins. (E) Drug targets and nondrug targets are presented in box-plots according to their number of interactors. The median is higher in the group of drug targets, indicating enrichment of highly connected proteins in drug targets. (F) Detailed analysis of drug targets distinguished by the type of targeting drug. Proteins associated exclusively with experimental drugs are more likely to have a higher number of interactions compared with targets of approved drugs or both approved and experimental drugs.
Topologic network analysis of drug targets and disease genes. (A) Distribution of platelet drug targets according to the type (group) of drugs with which they are associated (approved or experimental). (B) Proteins were grouped by their number of interactions. Network analysis indicates a higher number of disease-associated genes among well-connected proteins. (C) The datasets of disease proteins and nondisease proteins are presented in box-plots. The number of interactors is shown in a logarithmic scale on the y-axis. Their median is higher in the group of disease proteins compared with nondisease proteins. (D) Topologic network analysis of drug targets reveals an increase in the number of drug targets among highly connected platelet proteins. (E) Drug targets and nondrug targets are presented in box-plots according to their number of interactors. The median is higher in the group of drug targets, indicating enrichment of highly connected proteins in drug targets. (F) Detailed analysis of drug targets distinguished by the type of targeting drug. Proteins associated exclusively with experimental drugs are more likely to have a higher number of interactions compared with targets of approved drugs or both approved and experimental drugs.
For further analysis, all drug targeted proteins were separated into 3 groups: proteins targeted by experimental drugs, by approved drugs, or by both drug types. All 3 groups were compared with the proteins not associated with drugs. Experimental drug targets were found significantly more often among well-connected proteins (mean = 13.1; P = 5.79 × 10−12, Figure 5F). However, this effect notably decreased compared with drug targets affected by both approved and experimental drugs (mean = 13.3; P = .0016). No significant difference could be detected for approved drug targets only (mean = 8.0; P = .16).
These results suggest that drugs under development (experimental) are more often associated with highly connected proteins (hubs) in the platelet network. To further elucidate this effect, we performed a functional enrichment analysis on experimental drug targets, which resulted in an over-representation of the process “phosphorylation” indicating kinase activity (supplemental Figure 7). Kinase involvement could be further validated with topologic analysis performed analogously on the kinase drug targets in the platelet phosphorylation network (supplemental Figure 8).
Another essential aspect with high clinical relevance is the investigation of platelet proteins associated with known genetic diseases. Genetic disease associations were extracted from HPRD revealing 701 platelet proteins from a total of 1933 human genes connected to a genetic disease for the platelet dataset. Topologic exploration of disease associated genes suggested that the products of these genes interact more often with other proteins (Figure 5B-C; P = 1.49 × 10−4).
Case study on integrated network analysis: DOK1 phosphorylation as a switch for integrin activation
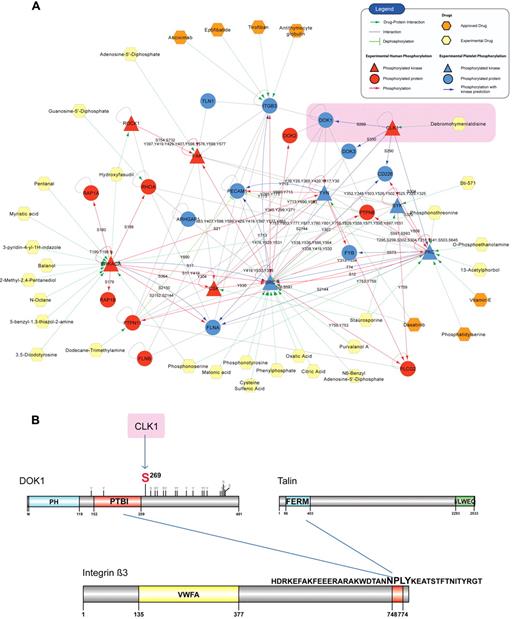
Integrins are bidirectional molecules that require a conformational change for activation. This process is called inside-out signaling and is mediated by the protein talin, which binds to the β3 integrin (ITGB3) tail via its FERM domain and facilitates integrin activation.33 Integrin signaling is one of the major pathways in platelet activation and leads to downstream responses, such as firm adhesion and platelet aggregation during hemostasis.1 The over-representation of the focal adhesion pathway in the pathway enrichment analysis indicated the predominant role of integrin signaling in platelets (Figure 4B). The PlateletWeb knowledgebase offers the possibility for an integrated analysis of the proteins from this pathway using combined information from the platelet interactome, phosphorylation signaling network, and functional data. From the entire platelet network, we extracted a subnetwork of integrin signaling near the α2bβ3 receptor (Figure 6A). We generated an overview of phospho-signaling events in the integrin pathway, including literature phosphorylations and kinase predictions,18,19 for experimental phosphorylation sites measured in platelets12 (Figure 6A; supplemental Tables 7 and 8).
Case study on integrated network analysis. (A) A graphical representation of the core integrin signaling created by integrating information on phosphorylations, dephosphorylations (green arrows), and interactions (gray lines). Phosphorylations are shown according to the source of detection: red arrows indicate phosphorylations reported from human cells (HPRD), and blue arrows connect platelet proteins with experimentally verified phosphosites and their predicted kinase. The protein nodes are colored according to the source of phosphorylation (red represents phosphorylated in human cells; blue, phosphorylated in platelets; and yellow, a platelet nonphosphorylated protein), and the phosphorylation site is marked on each directed edge. Drugs are visualized with different colors according to the type of drug: investigational (experimental) or approved. DOK1, a docking protein associated with ITGB3 binding, is phosphorylated on Ser269 (highlighted). By further integrating kinase prediction and drug target information, the platelet kinase CLK1 has been proposed, and a putative therapeutic approach using the inhibitor debromohymenialdisine can be suggested. (B) Schematic representation of the DOK1 Ser269 phosphorylation site and the competitive binding between DOK1 and talin for the NPLY motif of the integrin β3-tail. DOK1 binds to the integrin and prevents talin from binding and activating the receptor.
Case study on integrated network analysis. (A) A graphical representation of the core integrin signaling created by integrating information on phosphorylations, dephosphorylations (green arrows), and interactions (gray lines). Phosphorylations are shown according to the source of detection: red arrows indicate phosphorylations reported from human cells (HPRD), and blue arrows connect platelet proteins with experimentally verified phosphosites and their predicted kinase. The protein nodes are colored according to the source of phosphorylation (red represents phosphorylated in human cells; blue, phosphorylated in platelets; and yellow, a platelet nonphosphorylated protein), and the phosphorylation site is marked on each directed edge. Drugs are visualized with different colors according to the type of drug: investigational (experimental) or approved. DOK1, a docking protein associated with ITGB3 binding, is phosphorylated on Ser269 (highlighted). By further integrating kinase prediction and drug target information, the platelet kinase CLK1 has been proposed, and a putative therapeutic approach using the inhibitor debromohymenialdisine can be suggested. (B) Schematic representation of the DOK1 Ser269 phosphorylation site and the competitive binding between DOK1 and talin for the NPLY motif of the integrin β3-tail. DOK1 binds to the integrin and prevents talin from binding and activating the receptor.
Focusing on the direct interactors of the ITGB3 tail, a yet functionally uncharacterized phosphorylation site was detected in unstimulated human platelets at Ser269 of DOK1 (Figure 6B), a protein containing a pleckstrin domain (PH) and a phosphotyrosine binding (PTB) domain.34 It has been detected in various tissues and cell types and is connected to integrin signaling, as there has been both in vivo and in vitro evidence for DOK1 binding to the cytoplasmatic tail of ITGB3.35 The Ser269 phosphorylation site is in close vicinity to the IRS-type PTB domain, which facilitates binding of DOK1 to ITGB3 (NPLY motif, Figure 6B) and inhibits its activation.36 The integrin activating molecule talin competitively binds to the same motif, which suggests that the phosphorylation site might influence the balance between DOK1 and talin binding.
To further explore the functional role of this modification, we performed a bioinformatic kinase prediction analysis. The applied prediction method18,19 pointed out CLK1 as the kinase responsible for phosphorylating Ser269 on DOK1. The predicted kinase-substrate relationships in the network were supported by experiments in other human cells, such as PKA phosphorylating SRC at Ser1737 and SRC phosphorylating ITGB3 at Tyr77338 (same as Tyr747, new nomenclature from HPRD). CLK1 is a kinase found in megakaryocytes, proplatelets, and platelets.6 It is inhibited by the chemical compound debromohymenialdisine39 (a marine sponge alkaloid), which is in the stage of development and used in phase 1 clinical trials as an anti–Alzheimer and anti–osteoarthritis agent. Thus, integrated network analysis coupled with kinase predictions can be a useful approach for functional exploration of candidate proteins and phosphorylations in cases, such as DOK1, which are not easily accessible experimentally.
PlateletWeb as a combined knowledge base
The PlateletWeb knowledge base presents a variety of advanced options for systems biologic analysis of platelet signaling. Each protein has been provided with characteristic features from the functional and network context along with the technique for identification in platelets (level of detection). The phosphorylation status of all proteins can be analyzed, distinguishing individually between phosphorylations derived from the published literature and those directly measured in human platelets. Furthermore, information on physical properties, such as isoform-specific sequence information, presence of transmembrane domains, isoelectric point, and molecular weight data, are provided and searchable, which could be a helpful tool for the analysis of Western blot and 2-dimensional gel experiments. Associations with diseases can be found using a key word functionality search in the description of proteins. The resource even allows users to combine various search options into a more complex advanced search concentrating on specific platelet proteins (Figure 7).
PlateletWeb knowledge base as system biologic workbench. The PlateletWeb knowledge base provides multiple options for functional platelet analysis. Various aspects of platelet function can be investigated using optimized search criteria based on physical and functional properties, pathway information, drug association, protein modifications, or key words related to the protein.
PlateletWeb knowledge base as system biologic workbench. The PlateletWeb knowledge base provides multiple options for functional platelet analysis. Various aspects of platelet function can be investigated using optimized search criteria based on physical and functional properties, pathway information, drug association, protein modifications, or key words related to the protein.
To better understand the platelet commitment in processes, such as immune defense, a functionality search (using GO terms) results in proteins involved in “immune response” and associated functions. In addition, it is possible to combine GO terminology with the phosphorylation state, level of detection, and presence of particular protein domains to retrieve groups of proteins fulfilling the search-defined criteria. Drug information provides additional insight about platelet proteins associated with specific drugs. As an example, results on pharmacologic modification by inhibiting prostacyclin receptors retrieve analogues of prostacyclin (epoprostenol, iloprost, treprostinil). The knowledge base thus allows a comprehensive and detailed analysis of the platelet not only on a single protein level but also on the scale of network regulation and functional association of signaling components. A detailed tutorial on the usage of the platform is presented in the supplemental Appendix (PlateletWeb user guide).
Discussion
The PlateletWeb knowledge base helps investigate the signaling mechanisms and modular functions in a comprehensive way, not limited to hemostasis alone, but also regarding other physiologic and pathophysiologic processes, such as immune defense against bacteria, sepsis, rheumatoid arthritis, and cancer.2 Multitasking and functional modularity of the platelet in various aspects are achieved by visualizing the interaction partners. Newly identified anti–thrombotic targets, such as LXR40 or the signaling modulator PECAM1,41 can be easily investigated in the context of interacting partners and phosphorylation events (supplemental Figures 9 and 10). The logical sequence of interactions and switches on activatory and inhibitory cascades rewires the network dynamically, as can be studied in domain resolution (eg, for vasodilator-stimulated phosphoprotein and its interactors42 ) or for the relationship between BCL2/BCL-X(L)-inhibition, apoptosis, and platelet activation.43 Furthermore, we can see in detail that this rewiring involves phosphorylations and change of substrates; otherwise, the multitasking of the platelet would not be possible: switching functional integrin modules (eg, around DOK1, above) on and off. In this regard, PlateletWeb is a valuable resource as it provides a systems biologic overview of the platelet interactome and phosphoproteome, including validated interactions and phosphorylations based on published studies and experimentally validated phosphorylation sites in platelets. This wealth of information has been complemented with drug target data, allowing a systemic analysis of pharmacologic regulation in platelets. Advanced search options are available for drugs, diseases, and functional annotations, which effectively enable the implementation of “data mining” strategies for the detection of novel platelet-specific targets. In addition, all proteins are cross-linked with their drug associations, allowing bidirectional navigation through the network of drug-target relationships.
Protein kinases are key regulators of platelet signaling. Kinase activity is crucial in a variety of human diseases, such as cancer, rheumatoid arthritis, and cardiovascular and neurologic pathologies, and is indeed associated with more than 400 human diseases.44 Therefore, knowledge about human kinases and their substrates in a network context, coupled with information on associated drugs, can indicate putative new pharmacologic targets. Our analyses underline the tight functional relationship of SH2 domain proteins and tyrosine kinases31 in platelets, which is well reflected by the over-representation of tyrosine kinase substrates and SH2 domains in the enrichment analyses. Modern phosphoproteomic studies deliver large amounts of valuable data on site-specific phosphorylation, including quantitative measurements of dynamic signaling events.10 Usually, however, they offer no information about the corresponding kinase, which thus remains the missing link in the reconstruction of the cellular signaling cascades from phosphoproteome data. Here, the bioinformatic predictions and literature-curated networks may provide the necessary kinase associations and deliver a cellular context for the analysis of the raw experimental data.
However, this integrated analysis is not restricted to a single protein level. Proteins of interest taken from a sample can be investigated as required by extracting a subnetwork from the human phosphoproteome, kinome, and interactome. PlateletWeb allows the visual representation of resulting subnetworks in vector graphics or export for further analysis in Cytoscape. Various pathways considered to play a key role in hemostasis (GPIb/VWF signaling, ADP signaling, supplemental Figure 11) can be extracted, visualized, and analyzed for signal regulation, as demonstrated in the integrin inside-out signaling pathway (Figure 6A). Alternatively, platelet proteins belonging to characteristic pathways (defined by KEGG) are highlighted and available for pathway network analysis.
Visualizing subnetworks using the PlateletWeb knowledge base can help to generate novel hypotheses as exemplified here for the integrin subnetwork. Integrin signaling has substantial clinical relevance, and patients with functional defects in the integrin α2bβ3 receptor (Glanzmann thrombasthenia) may have severe bleeding disorders. On the other hand, the integrin α2bβ3 molecule is a prominent drug target, and inhibitors such as abciximab have been developed and are routinely used in emergency coronary artery bypass grafting to reduce the risk of thrombosis.45 Based on our network analysis, we hypothesize here on possible new mechanisms for the regulation of the β3-cytoplasmic tail. The integrin α2bβ3 receptor as a regulatory scaffold has already been discussed in previous studies.36,46 In our analysis, we generate new hypotheses for the inside-out signaling of this integrin based on a detected phosphorylation site of the DOK1 molecule. DOK1 could act as a negative regulator of platelet function, as was already shown in Jurkat T cells, where it inhibits PLCγ1 phosphorylation, Erk1/2 activation, and Ca2+ mobilization.47 It is closely related to the DOK3 protein, which is also a part of the integrin signaling pathway, and it has been proposed that DOK1 and DOK3 negatively regulate α2bβ3 outside-in signaling in the complex with SHIP-1 and Grb2.48
Certainly, more investigation is needed to understand the functional role of DOK1 phosphorylation at Ser269 and its possible association with CLK1. This kinase has been reported to phosphorylate and activate the phosphatase PTPN1, which in turn facilitates platelet activity49 ; therefore, a possible activatory role of CLK1 in platelet signaling might be assumed. Whether the DOK1 module and CLK1 inhibition might be a potential target for future anti–platelet therapy cannot be decided based on current data alone. However, the functional implications of Ser269 phosphorylation on DOK1 and its role in integrin signaling can be analyzed experimentally as an inhibitor (debromohymenialdisine) of the predicted kinase is available. This compound may facilitate an in-depth experimental investigation of DOK1 signaling in platelets, and our resource provides a rich data basis for planning experiments as well as for subsequent evaluation of results. Nevertheless, bioinformatic kinase predictions can also be useful in cases where experimental testing is sparse or unavailable because of a lack of antibodies or a well-studied mouse knockout model with platelet phenotype.50
In conclusion, the PlateletWeb knowledge base is a comprehensive Internet-based resource for platelet researchers, which presents intriguing and novel options for a systems biologic investigation of platelet signaling. It also provides many opportunities for the investigation of dynamic network restructuring in regard to the recently recognized condition-dependent further tasks of the platelet in infection, inflammation, cancer, and sepsis.2 Future efforts in the development of PlateletWeb will focus on the integration of novel platelet studies (eg, new transcriptome information from recent RNA sequencing data7 ) and updated datasets from the source databases while maintaining the high standard of data curation. Furthermore, as mice have become a major animal model in platelet research and more data for this organism become available, the establishment of cellular signaling network of murine platelets becomes an interesting and realistic perspective in the near future.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)/Systems Biology of ADP Receptor Activition (D.B. and S.N.), Land Nordrhein-Westfalen (I.B.), Land Bavaria (T.D. and M.D.), and Sonderforschungsbereich 688/A2 (T.D.).
Authorship
Contribution: D.B. and S.N. assembled interactome data, interactions, and modulation options and were involved in all required programming tasks; M.D. and D.B. performed the statistical analysis; I.B. provided expert advice on clinical aspects and platelet pathophysiology; M.D. and T.D. led and guided the study; and all authors analyzed the various datasets, were involved in database curation, and wrote and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marcus Dittrich or Thomas Dandekar, Department of Bioinformatics, Biocenter, University of Wuerzburg, Am Hubland, D-97074 Wuerzburg, Germany; e-mail: marcus.dittrich@biozentrum.uni-wuerzburg.de or dandekar@biozentrum.uni-wuerzburg.de.
References
Author notes
D.B. and S.N. contributed equally to this study.