Potentially effective and life-saving treatment for patients with systemic amyloidosis relies on the astute clinician recognizing the signs and symptoms, histochemical identification of fibrils, and accurate diagnosis of amyloid type. In this issue of Blood, Brambilla et al report a new methodology for accomplishing the key third step in this process.1
Accurate diagnosis of systemic amyloidosis is an essential step in therapeutic decision-making, in particular, avoiding the inappropriate use of chemotherapy in patients who do not have immunoglobulin light chain (AL) amyloidosis. The clinical presentation and the protein deposition process in AL can phenotypically resemble those of AF (familial) and AA (secondary) amyloidosis. However, AF is caused by inheritance of a mutant germ line gene encoding an abundant serum protein such as transthyretin (TTR), and AA is due to up-regulation of production of serum amyloid A (SAA) protein in the liver, associated with chronic infection or inflammation.
By the seventh edition of the textbook Clinical Hematology published in 1974 by Max Wintrobe and colleagues, “primary” (AL) amyloidosis was described with accuracy.2 However, on page 1635, Wintrobe refers to “AUO,” or “amyloid of unknown origin”: cases of nonimmunoglobulin amyloid occurring secondary to other diseases or associated with tumors such as insulinoma or medullary carcinoma of the thyroid. Distinguishing these was not particularly essential, as Wintrobe was nihilistic about treatment, noting that, “In rare instances of ‘secondary amyloidosis’, regression of amyloid deposits has been noted after successful therapy of the underlying disease…. With the exception of the favorable effects of therapy in these few patients, no specific and efficacious form of therapy is currently available. In some patients with amyloidosis associated with plasma cell dyscrasias, attempts have been made to treat both conditions with cyclophosphamide or melphalan, but while it has been possible to lower the concentration of M-components in some patients, the clinical results have been disappointing.”2 page 1635
We have come far in 37 years. Biochemical, biophysical, and genetic studies have shed light on the pathogenesis of the AUO diseases, through multidisciplinary science that here we term “amyloidomics” (see figure). We have identified many of the mutations that can cause AF, and many interesting inherited and acquired syndromes associated with cytokine overproduction leading to AA.3 In addition, we have made great strides in therapeutics for the systemic amyloidoses. In AL, oral or high-dose IV melphalan, proteasome inhibitors, and immunomodulators can produce effective hematologic responses, extinguishing light chain production and preventing further fibril formation. For AA, a small molecule therapeutic, eprodisate, has been found to delay the progression of renal amyloidosis4 ; it is hoped that another phase 3 study will lead to FDA approval. For ATTR, 2 structurally related small molecules have been studied: a parent compound, the nonsteroidal diflunisal; and a congener, tafamidis. A reduction in progression of familial amyloidotic polyneuropathy has been reported with tafamidis at the last international amyloidosis symposium (Rome, 2010), and regulatory approval is being sought. The diflunisal trial is ongoing.
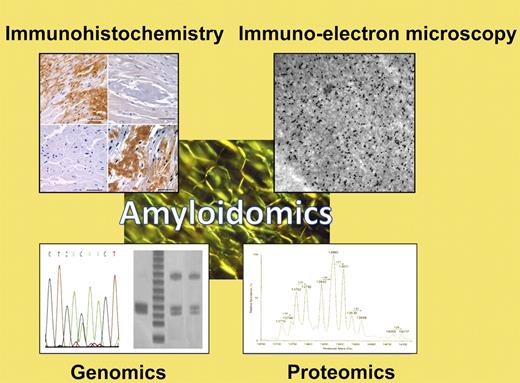
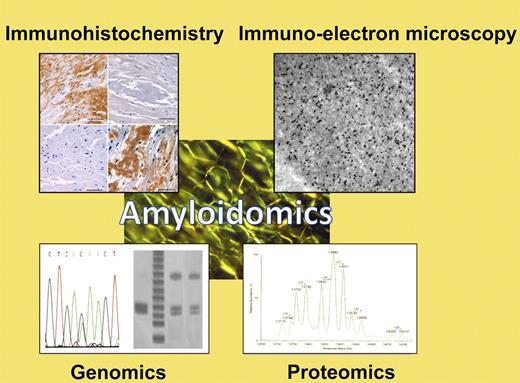
Apple-green birefringence of fibrillar protein deposits is the sine qua non of amyloidosis. To type amyloid fibrils and guide appropriate therapeutic decision-making, a variety of specialized techniques may be employed, collectively here termed “amyloidomics.” These include immunologic techniques such as immunohistochemistry or immuno-electron microscopy; genomics techniques including analysis of restriction fragment linked polymorphisms (RFLPs), single nucleotide polymorphisms (SNPs), or gene sequencing; or mass spectrometry-based proteomics techniques such as the one described by Brambilla et al in this issue.1 (Examples of diagnostic techniques courtesy of Drs Skinner, Connors, O'Hara, Costello, and colleagues in the Gerry Amyloid Reference Laboratory at Boston Medical Center.)
Apple-green birefringence of fibrillar protein deposits is the sine qua non of amyloidosis. To type amyloid fibrils and guide appropriate therapeutic decision-making, a variety of specialized techniques may be employed, collectively here termed “amyloidomics.” These include immunologic techniques such as immunohistochemistry or immuno-electron microscopy; genomics techniques including analysis of restriction fragment linked polymorphisms (RFLPs), single nucleotide polymorphisms (SNPs), or gene sequencing; or mass spectrometry-based proteomics techniques such as the one described by Brambilla et al in this issue.1 (Examples of diagnostic techniques courtesy of Drs Skinner, Connors, O'Hara, Costello, and colleagues in the Gerry Amyloid Reference Laboratory at Boston Medical Center.)
Thus, with potential therapeutic interventions available, the obligation to diagnose systemic amyloidosis in a timely and accurate fashion has increased significantly. The first step in this process is recognition of the clinical manifestations of disease; raising suspicion that amyloidosis is present. The second step is demonstration of fibrils in an abdominal fat aspirate or in a biopsy from an involved organ. The third critical step is accurate typing. Historically, techniques to accomplish this have included immunologic identification of fibril constituents, screening for clonal plasma cell process, and sequencing to identify a germ line mutation in an amyloidogenic gene. In many circumstances, these techniques are adequate. However, in some cases, we can be misled by overlapping clinical presentation or by co-existing conditions; in up to 10% of cases, misdiagnosis can occur.5 In the elderly, monoclonal gammopathies are common, and can be present in a patient who actually has AF.6 To make matters more baffling, in senile systemic amyloidosis patients, the deposited TTR in fibrils is unmutated and cannot be detected by gene sequencing. Treating such patients with chemotherapy would be ineffective and potentially harmful. Conversely, there are populations in which TTR mutations are prevalent in which AL can occur. For example, the V122I mutation in TTR is carried by ∼ 4% of blacks, but 11% of black patients with V122I TTR and cardiomyopathy actually turn out to have AL.7
Thus, for many patients direct identification of the amyloid fibril constituents is essential. Again, not an easy undertaking! Amyloid fibrils are notoriously sticky, and the deposits are composed of the fibrillogenic protein, associated serum amyloid P component, glycosaminoglycans, apoplipoproteins, and many other constituents; antibody reagents can bind nonspecifically. So for protein-based diseases, protein-based diagnostics make sense. In 2008, in experiments carried out by investigators in Boston and the Pavia Amyloidosis Research and Treatment Center, amyloid proteins were extracted from fat specimens, separated using two-dimensional polyacrylamide gels, and identified by matrix-assisted laser desorption/ionization mass spectrometry and peptide mass fingerprinting.8 In the current report, this technique has been refined to work on easily obtained fat aspirate specimens without the need for electrophoretic separation, and used to accurately identify the amyloid protein in 26 patients.1 In 2009, Vrana et al reported on the use of laser capture microdissection to isolate deposits from which amyloid fibrils were extracted and subjected to tandem mass spectrometry to identify the amyloid proteins.9 In 40 of 41 samples, the likely amyloid protein was successfully identified. This technique is available commercially through the Mayo Clinic laboratories. Bioinformatics is a key component of these analyses, as the amyloid protein is not typically the most abundant protein obtained in the biopsy specimen. One of the advances in the current report is the development of an algorithm to confidently assign the amyloid protein type, based on “MudPIT,” or multidimensional protein identification technology, a technique first developed for analyzing the yeast proteome.10 This approach adds to our armamentarium for accurate clinical diagnostics. As an additional scientific benefit, proteomic approaches such as this provide information about pathophysiology and biomarkers of disease.
Conflict-of-interest disclosure: The authors declare no relevant financial interest. ■