Abstract
IGHV1-69/51p1 is expressed by ∼ 30% of unmutated chronic lymphocytic leukemia (U-CLL) and combines with selected IGHD and IGHJ genes generating stereotypes if HCDR3 amino acid homology is > 60%. We had previously revealed stereotypic IGHV1-69/IGHJ6 rearrangements in normal naive B cells, thereby identifying potential counterparts of U-CLL. A different stereotypic IGHV1-69/IGHD3-16(RF2)/IGHJ3 rearrangement carrying the CAR(GGx)YD motif in the N1-region, recurrent in 6% IGHV1-69+ve CLL, is exceptionally sequence restricted, strongly suggestive of shared antigen recognition. We have now analyzed IGHV1-69/IGHJ3 rearrangements in circulating B cells of healthy individuals using several PCR-based approaches with IGHV1-69/IGHJ3 CLL sequences for reference. Stereotypes were found, but all were distinct from CLL. Remarkably, even a highly sensitive semi-nested PCR, specific for the CLL-expressed IGHV1-69/IGHD3-16(RF2)/IGHJ3 stereotype, failed to identify the CAR(GGx)YD sequence, although similar motifs were found. These highly specific B cells are not apparent in the accessible normal repertoire and may expand in response to rarely expressed antigens important in the pathogenesis of CLL.
Introduction
The IGHV1-69/51p1 allele is expressed by ∼ 30% of unmutated chronic lymphocytic leukemia (U-CLL).1–3 In these cases, IGHV1-69 rearranges to a restricted number of IGHD and IGHJ, generating conserved sequences in the third complementarity-determining regions (HCDR3). Using a 60% amino-acid (AA) homology cut-off, ∼ 50% of cases can be assigned to stereotypic subsets suggestive of shared antigen recognition.2
The B cell of origin of CLL has long been sought and these conserved sequences provide tools for probing the normal adult expressed repertoire. We previously revealed a significant proportion (∼ 33%) of stereotypes involving IGHV1-69/IGHJ6 rearrangements among normal circulating naive B cells, several (∼ 20%) being similar to those of U-CLL.4 However, these incorporate the long IGHJ6, which accounts for a large proportion of the similarity.2,5,6 A more restricted HCDR3 is found in the CLL subset 6,2 where IGHV1-69, combined with IGHD3-16 in reading-frame 2 (RF2) with the shorter IGHJ3, generates almost identical sequences between cases. This is highly suggestive of shared antigen recognition,2,7 especially as there is unusual conservation of the nontemplated N-regions, generating the characteristic CAR(GGx)YD N1 and SYR(xND)AFDI N2 junctional motifs.7–9 Frequent combination with the same IGKV3-20/A27 adds strength to this conclusion.7 Here we have probed IGHV1-69/IGHJ3 rearrangements in the circulating B cells of healthy donors, specifically seeking counterparts of subset 6.
Methods
Amplification of IGHV1-69/51p1–related rearrangements from healthy donors
IGHV1-69/51p1–related rearrangements were amplified, cloned and sequenced from peripheral blood lymphocyte cDNA of 3 healthy individuals,4,10 in 2 centers. First, the forward IGHV1-69/HCDR2 primer4 was used with C μ100 or IGHJ3 primers (prJ3, prDJ3, prNDJ3) with different levels of extension into the CLL-specific N2-region (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The IGHJ3 primers were designed to provide a hierarchy from no (prJ3) to high specificity (prNDJ3) for subset 6–related N2-region containing the asparagine-aspartic acid (ND) motif.7,8 Second, a semi-nested PCR/cloning approach was used to detect subset 6 sequences in donors. This included a first PCR with forward IGHV1-69/HCDR2 primer and IGHJ3 reverse primers prDJ3 or prNDJ3 followed by a second round with the reverse prs6D3-16 primer specific for subset 6 (supplemental Figure 1). This approach increased the sensitivity of detection of subset 6–related rearrangements to < 1 in 107 B cells (supplemental Figure 2).
Analysis of IGHV1-69/IGHJ3 combinations, HCDR3 clustering and search for subset 6 IGHV1-69/IGHD3-16(RF2)/IGHJ3 rearrangements in healthy donors
A CLL database (n = 653) of 256 previously published,4 and 397 new IGHV1-69-D-J rearrangements centralized at 4 different institutions was used for comparative sequence analysis. First, we compared the 77 CLL derived rearrangements using IGHJ3 (supplemental Table 2), with those from the healthy donors. All sequences were aligned to the ImMunoGeneTics (IMGT) directory and analyzed for IGHV1-69 mutational status, IGHD, IGHJ, HCDR3 junctional characteristics, and stereotypes.2,3,6,11 Identical IGHD-J usage and IGHD reading-frame (RF), ClustalW2 score > 60, AA identity > 60% and < 3 AA difference in length were required, exactly as published.2–4,12 Numbering of subsets was updated according to previous criteria.2,4 Second, 65 CLL subset 6 sequences (see supplemental Table 2 and publications)2,3,5,7,8,12–20 were compared with those obtained from the sensitive semi-nested PCR/cloning procedures used to extract subset 6 sequences from healthy individuals.
The study was approved by IRBs at the Universities of Southampton (228/02/t) and Siena (AIRC IG-5298, ITT-2008). Patients and donors provided informed consent before inclusion in the study in accordance with the Declaration of Helsinki.
Results and discussion
IGHV1-69 recombinations in immunoglobin M (IgM)–positive normal blood B cells involve a range of IGHJ genes with IGHJ3 accounting for ∼ 20%,10 and this was confirmed as 16.8% (23/137) in the present study (supplemental Table 3). The frequency in the current CLL database was 11.8% (77/653; supplemental Table 4), as found previously.2,4,10 From normal B cells, we obtained 23 IGHV1-69/IGHJ3 sequences using the IGHV1-69 and Cμ100 primer pair. To increase the number of sequences expressing IGHJ3, we took advantage of our previous observation that B cells expressing IGHV1-69, detectable with the G6 monoclonal antibody, were within the naive IgM-positive circulating population.4 We therefore moved to a IGHJ3-specific primer, confident that amplimers were derived from these B cells. This allowed collection of 165 IGHV1-69/IGHJ3 rearrangements (supplemental Table 5) for comparison with the 77 from CLL.
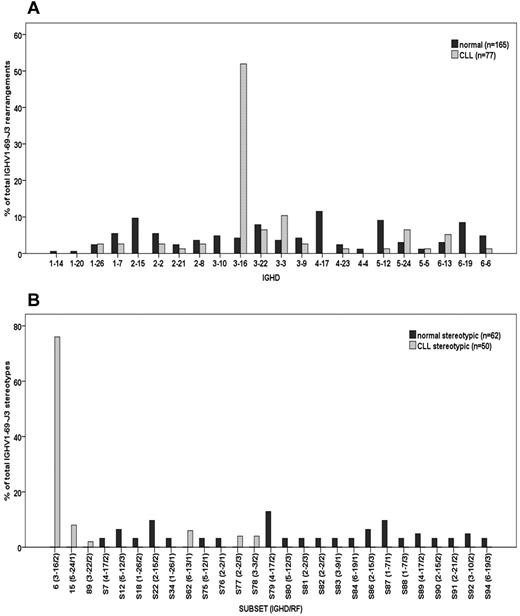
Almost all normal B cell sequences were unmutated (163/165, 98.8%) as were most from CLL (65/77, 84.4%). Cases assigned to subset 15 (CLL only) or to no subset (either normal or CLL) were the exceptions.2 However a striking difference in IGHD usage was evident, with normal B cells expressing a wide range, but CLL cells predominantly (40/77, 51.95%) expressing IGHD3-16 (Figure 1A). Stereotypic sequences defined by pre-established criteria, including 60% homology,2–4,12 were evident in normal B cells (62/165, 37.6%) and, as expected from IGHD-gene usage, were wide-ranging (Figure 1B). In contrast, CLL stereotypes reflected the preferential usage of IGHD3-16(RF2), with 38/77, 49.4% (76% of total stereotypes; Figure 1B) in subset 6, and 12/77 assigned to other subsets. Strikingly, none of the stereotypic sequences from normal B cells clustered with any CLL subsets (Figure 1B). Among the few normal cells using IGHD3-16 (7/165, 4.24%), RF2 was detected in 5 (71.4%), but none of the sequence patterns fulfilled the strict pre-established criteria for assignment to subset 6.2–4,12
IGHD and Stereotype distribution in normal and CLL B cells using IGHV1-69/IGHJ3 rearrangements. Bars identify percent sequences (y-axis) using IGHD (A) or assigned to each subset (B) of all normal (dark gray) or CLL (light gray) IGHV1-69/IGHJ3 rearrangements. (A) Percent IGHD distribution among normal and CLL B cells refers to all IGHV1-69/IGHJ3 rearrangements analyzed (stereotypic and nonstereotypic). (B) Percent stereotypic pattern distribution refers to those normal and CLL B cells using IGHV1-69/IGHJ3 rearrangements assigned to subsets only. For each subset, IGHD gene and reading frame are also indicated (in parentheses separated by a slash) in the x-axis.
IGHD and Stereotype distribution in normal and CLL B cells using IGHV1-69/IGHJ3 rearrangements. Bars identify percent sequences (y-axis) using IGHD (A) or assigned to each subset (B) of all normal (dark gray) or CLL (light gray) IGHV1-69/IGHJ3 rearrangements. (A) Percent IGHD distribution among normal and CLL B cells refers to all IGHV1-69/IGHJ3 rearrangements analyzed (stereotypic and nonstereotypic). (B) Percent stereotypic pattern distribution refers to those normal and CLL B cells using IGHV1-69/IGHJ3 rearrangements assigned to subsets only. For each subset, IGHD gene and reading frame are also indicated (in parentheses separated by a slash) in the x-axis.
To probe more specifically among normal B cells for the characteristic conserved HCDR3 sequence of subset 6, a primer was designed to move further into the IGHD3-16/N2/IGHJ3 junction (supplemental Table 1 and supplemental Figure 1). A single-step PCR (> 10−5 sensitivity) yielded no products, but semi-nested PCR with pr(N)DJ3 and prs6D3-16 primers (> 10−7 sensitivity) generated amplimers. We identified 37 sequences with unique HCDR3s together with multiple repetitive sequences, possibly reflecting limited available template (Figure 2 and supplemental Table 6). Among the wide range of HCDR3 sequences in normal B cells (Figure 2), the subset 6 defining motif CAR(GGx)YD (codons 107-111 counting from first G),7,8 characteristic of CLL sequences, where X varies (Figure 2), was not found. In contrast to the 3 AA (GGx) of the N1 junctional region, normal B cells displayed variable lengths ranging from 3-12 AA (Figure 2). Variability was because of single AA deletions/insertions and/or insertion of IGHD2-2 (sequences 3448-50) or another unknown segment (sequences 3140-1) upstream of IGHD3-16.
Amino acid sequence alignments of HCDR3 region from CLL-derived IGH rearrangements assigned to subset 6 and from healthy donor-derived IGH rearrangements identified with a semi-nested subset 6 specific primer approach. (A) Left column: the first number indicates sequence identification number (SIN.); DON1/2/3 indicate donor's source; x followed by a number (eg, x13) indicates the number of identical sequences (eg, 13 identical sequences) identified among different tumor rearrangements from CLL individuals or by cloning of PCR products from normal individuals. (.) indicates homology; and (−), lack of amino acid. Among CLL sequences represented with identical (GGx)YD motif in separate lines, different amino acids were observed at codon 111.2 and or N2 junctional region (codons 112.1-112). Among normal sequences, a different nucleotide sequence at codon 111.2, leading different amino acid translation outside primer sequence was observed between clones 3448 and 3450. Among the healthy donors, those amino acid sequences, identical from position 107 to 111.1 but different outside the represented amino acid sequence or in the degenerate prs6D3-16 primer sequence at position 111.2 in IGHD3-16 and/or 112-113 in the N2 junctional region, are indicated with different SIN. (B) Crude percent frequency by logo analysis representation. Top row: alignment for CLL subset 6 rearrangements. Bottom row: normal sequences identified by cloning after PCR amplification with HCDR2-J3 and subsequent semi-nested HCDR2-prs6D3-16 primers approach. Amino acid sequences are represented as sequence logos as described at http://weblogo.berkeley.edu/. The letters represent the amino acids used at each particular codon numbered according to IMGT criteria. When more than one amino acid was observed in a codon, the letters representing each change are displayed as a stack. The size of the amino acid symbol represents the crude percent frequency of any of the observed amino acid in that position. Sequences with no amino acid at a given position were excluded from the calculation of the frequency at that codon. Blank spaces represent absence of any amino acid in that particular position as a comparison between CLL and normal sequences. Codon numbering between IMGT positions 108 and 109 is omitted.
Amino acid sequence alignments of HCDR3 region from CLL-derived IGH rearrangements assigned to subset 6 and from healthy donor-derived IGH rearrangements identified with a semi-nested subset 6 specific primer approach. (A) Left column: the first number indicates sequence identification number (SIN.); DON1/2/3 indicate donor's source; x followed by a number (eg, x13) indicates the number of identical sequences (eg, 13 identical sequences) identified among different tumor rearrangements from CLL individuals or by cloning of PCR products from normal individuals. (.) indicates homology; and (−), lack of amino acid. Among CLL sequences represented with identical (GGx)YD motif in separate lines, different amino acids were observed at codon 111.2 and or N2 junctional region (codons 112.1-112). Among normal sequences, a different nucleotide sequence at codon 111.2, leading different amino acid translation outside primer sequence was observed between clones 3448 and 3450. Among the healthy donors, those amino acid sequences, identical from position 107 to 111.1 but different outside the represented amino acid sequence or in the degenerate prs6D3-16 primer sequence at position 111.2 in IGHD3-16 and/or 112-113 in the N2 junctional region, are indicated with different SIN. (B) Crude percent frequency by logo analysis representation. Top row: alignment for CLL subset 6 rearrangements. Bottom row: normal sequences identified by cloning after PCR amplification with HCDR2-J3 and subsequent semi-nested HCDR2-prs6D3-16 primers approach. Amino acid sequences are represented as sequence logos as described at http://weblogo.berkeley.edu/. The letters represent the amino acids used at each particular codon numbered according to IMGT criteria. When more than one amino acid was observed in a codon, the letters representing each change are displayed as a stack. The size of the amino acid symbol represents the crude percent frequency of any of the observed amino acid in that position. Sequences with no amino acid at a given position were excluded from the calculation of the frequency at that codon. Blank spaces represent absence of any amino acid in that particular position as a comparison between CLL and normal sequences. Codon numbering between IMGT positions 108 and 109 is omitted.
Further probing revealed several sequences involving IGHD3-16/IGHJ3 which, however, did not meet the strict subset 6-specific criteria of identity to the CAR(GGx)YD motif. The closest sequences were CAS(QG_)YD (Clone 3506), CAS(QGx)YD (3481) CAS(QGx)YD (3479) and CAS(VGx)YD (3431-34) that all have 2-3 AA in N1, including at least 1 G residue and variable X, as for CLL subset 6 (Figure 2).
This analysis of IGHV1-69/IGHJ3 recombinants in the normal naive B-cell repertoire reveals heterogeneity of HCDR3 sequences, as expected in innate B cells programmed to recognize a wide range of common pathogens. Similar heterogeneity was observed in the IGHV1-69/IGHJ6 recombinants.4 However, the range of these unmutated sequences appears relatively limited, possibly reflecting the low affinity and consequent cross-reactivity of antigen binding.21 This limitation leads to stereotypes, subsequently reflected in transformed B cells of CLL. The unusually restricted HCDR3 of IGHV1-69/IGHD3-16(RF2)/IGHJ3 (subset 6) represents one end of the spectrum. The fact that we found similar, albeit not identical, sequences again indicates the limited range of the repertoire. The reason for some expansion of this subset in CLL is likely to involve antigen drive, but one possibility is that the sequences have to interact both with the initiating pathogen and with an autoantigen. The nonmuscle myosin heavy chain IIA antigen exposed on apoptotic bodies is recognized by CLL subset 6 immunoglobulins,8 and may promote expansion and survival of CLL cells. Similar cross-reactivity has been proposed for other CLL derived immunoglobulins,22 and has potential implications for driving proliferation in vivo and therefore clinical outcome.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Victoria Bryant for input to sequence analysis. They especially thank Dr Kostas Stamatopoulos (Democritus University of Thrace, Alexandroupolis, Greece) and Dr Nikos Darzentas (Institute of Agrobiotechnology, Thessaloniki, Greece) for the help with stereotypic analysis.
This study was supported by Cancer Research UK (CR-UK) and the Southampton CR-UK Center; Experimental Cancer Medicine Center, Tenovus Solentside, Associazione Italiana per la Ricerca sul Cancro (AIRC IG-5298 and Special Program Molecular Clinical Oncology, 5 × 1000, No. 10007), Milan, Italy; Istituto Toscano Tumori, Italy; Siena-AIL (Associazione Italiana contro le Leucemie, Linfomi e Mielomi, Siena Section); Hairy Cell Leukemia Research Foundation (Illinois); Fondazione MPS 2009 (Siena, Italy); Ministry of University and Research-Project of national interest 2009 (PRIN-MUR 2009).
Authorship
Contribution: F.F. designed the study, interpreted data, performed analysis, and wrote the manuscript; K.N.P. designed the study, interpreted data, performed analysis, and contributed to writing the manuscript; E.S., E.C., and I.H. performed research and IGHV analysis and interpreted data; R.B. and D.R. provided well-characterized biologic samples and molecular and clinical data of the reference cohort; V.G., G.G., and G.P. contributed to interpretation of the data; and F.K.S. designed the study, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Francesco Forconi, Cancer Research UK Centre, Cancer Sciences Unit, University of Southampton Faculty of Medicine, Southhampton University Hospital Trust, Tremona Road, S016 6YD, Southampton, United Kingdom; e-mail: f.forconi@soton.ac.uk; or Hematology, Department of Clinical Medicine and Immunological Sciences, University of Siena & Department of Oncology, AOUS, Viale Bracci, 16, 53100, Siena, Italy; e-mail: forconif@unisi.it; or Prof Freda Stevenson, Genetic Vaccine Group, Cancer Sciences Division, Southampton University Hospitals Trust, Southampton General Hospital, Tremona Road, Southampton, SO16 6YD, United Kingdom; e-mail: fs@soton.ac.uk.