Abstract
Runx1P1N/P1N mice are deficient in the transcription factor distal promoter-derived Runt-related transcription factor 1 (P1-Runx1) and have a > 90% reduction in the numbers of basophils in the BM, spleen, and blood. In contrast, Runx1P1N/P1N mice have normal numbers of the other granulocytes (neutrophils and eosinophils). Although basophils and mast cells share some common features, Runx1P1N/P1N mice have normal numbers of mast cells in multiple tissues. Runx1P1N/P1N mice fail to develop a basophil-dependent reaction, IgE-mediated chronic allergic inflammation of the skin, but respond normally when tested for IgE- and mast cell–dependent passive cutaneous anaphylaxis in vivo or IgE-dependent mast cell degranulation in vitro. These results demonstrate that Runx1P1N/P1N mice exhibit markedly impaired function of basophils, but not mast cells. Infection with the parasite Strongyloides venezuelensis and injections of IL-3, each of which induces marked basophilia in wild-type mice, also induce modest expansions of the very small populations of basophils in Runx1P1N/P1N mice. Finally, Runx1P1N/P1N mice have normal numbers of the granulocyte progenitor cells, SN-Flk2+/−, which can give rise to all granulocytes, but exhibit a > 95% reduction in basophil progenitors. The results of the present study suggest that P1-Runx1 is critical for a stage of basophil development between SN-Flk2+/− cells and basophil progenitors.
Introduction
Basophils are the least prevalent of the granulocytes, generally representing less than 1% of leukocytes in the peripheral blood. Basophil studies have been hampered by the rarity of these cells and, until recently, the lack of tools such as basophil-deficient mice with which to assess their roles in vivo. However, recent studies have unveiled evidence for several previously unrecognized roles for basophils that are distinct from those of mast cells.1-11
In addition to hampering investigations of basophil function, the small numbers of basophils and the paucity of tools for their analysis have made studies of basophil development challenging and therefore there have been few studies of this process. Arinobu et al showed that basophil lineage-restricted progenitors (BaPs) are identifiable in the BM and that the transcription factor CCAAT/enhancer-binding protein-α (C/EBPα) is important for the fate decision to develop into terminally differentiated basophils.12 Ohmori et al reported that the IL-3–STAT5 axis is important for differentiating granulocyte-monocyte progenitors to BaPs,13 and Siracusa et al showed that thymic stromal lymphopoietin (TSLP) can facilitate the development of BaPs into mature basophils.8
Despite such progress, many of the details of the basophil differentiation pathway remain to be determined. For example, it is known that IL-3–deficient,8,14,15 TSLP receptor (TSLPR)–deficient,8 and IL-3/TSLPR double-deficient8 mice have normal baseline numbers of basophils, indicating that other factors are more important in maintaining basophil levels at baseline. Moreover, C/EBPα-deficient mice die within 8 hours of birth16 and STAT5-deficient mice die in utero,17 limiting the ability to use these animals to evaluate factors that might regulate basophil development at baseline in adult mice in vivo.
Runt-related transcription factor (Runx) proteins are a family of transcription factors18,19 that have crucial roles during the development of many tissues and the immune system. Each of the 3 kinds of Runx proteins, Runx1, Runx2, and Runx3,19,20 has distinct roles in development, with Runx1 being required for hematopoiesis,18 Runx2 for osteogenesis,21,22 and Runx3 for neurogenesis, thymopoiesis, and the control of gastric epithelial-cell proliferation.23-25 Although a constitutive deficiency in Runx1 is embryonically lethal, studies of conditional Runx1-knockout mice have indicated that Runx1 can regulate the differentiation of hematopoietic stem cells (HSCs), B lymphocytes, natural killer T (NKT) cells, and T lymphocytes.18,26-30 Mx-Cre Runx1-knockout mice, which have an inducible Runx1 inactivation system, exhibit normal numbers of HSCs, a normal myeloid-cell (neutrophil) compartment, a severe reduction in megakaryocyte differentiation and platelet formation, and defects in B and T lymphocytes.31 All 3 Runx genes can be transcribed from the distal (P1) or proximal (P2) promoters,32 and P1- and P2-derived Runx1 variants differ in their N-terminal end sequences. It has been reported previously that variation in the expression of P1- versus P2-Runx1 can be regulated developmentally, but it remains to be elucidated how such Runx1 variants influence the development of different types of immune cells.33
We report herein evidence indicating that P1-derived Runx1 is important for basophil development in mice at baseline. P1-Runx1–deficient mice have a drastic reduction (more than 90%) in basophils but normal numbers of the other granulocytes (neutrophils and eosinophils) and normal numbers of mast cells in multiple anatomic sites. The results of the present study strongly suggest that, in mice, P1-Runx1 is an important regulator of the differentiation of basophils, but not other granulocytes, and plays a nonredundant role in basophil, but not mast cell, development.
Methods
Mice
Runx1P1N/P1N mice, which have been described previously,34 were backcrossed onto a C57BL/6 background (8-10 generations, 6-12 weeks of age). We mated Runx1P1N/+ mice and Runx1P1N/+ mice in our animal facility to obtain Runx1P1N/P1N mice and littermate Runx1+/+ wild-type (WT) control mice. All animal care and experimentation was conducted according to the guidelines of RIKEN, Stanford University, and the National Institutes of Health with the specific approval of the institutional animal care and use committee of Stanford University.
Abs, flow cytometry, and cell culture
The Abs used for cytometry were from BD Pharmingen, eBiosciences, or BioLegend. For analysis of lineage cells, we used mIgE-biotin (R35-72), CD49b-Alexa Fluor 488 (DX5), Gr-1–FITC (RB6-8C5), Siglec-F–PE (E50-2440), NK-1.1-APC (PK136), B220-APC (RA3-6B2), CD11c-FITC (HL3), c-Kit–APC (2B8), FcϵRIα-PE (MAR-1), CD3-FITC (145-2C11), CD4-FITC (L3T4), CD8-APC (53-6.7), and CD11b-FITC (M1/70). Surface staining was performed for 15-20 minutes with the corresponding mixture of fluorescently labeled Abs. Data were acquired on a FACSCalibur flow cytometer or FACSAria II cell sorter (BD Biosciences) and analyzed with FlowJo Version 8.8.6 software (TreeStar). The cell sorting technique used has been described previously.35 Briefly, BM cells were depleted for the lineage markers CD3 (145-2C11), CD4 (L3T4), CD5 (53-7.3), CD8 (53-6.7), B220 (RA3-6B2), Gr-1 (RB6-8C5), CD11b (M1/70), and Ter119 (Ter-119) by MACS LD columns with anti–rat IgG microbeads (Miltenyi Biotec). SN progenitors were sorted on a FACSAria II cell sorter using the labeled mAbs Pacific Blue–conjugated CD3 (145-2C11), CD4 (L3T4), CD8 (53-6.7), CD11b (M1/70), Ter119 (Ter-119), Gr-1 (RB6-8C5), Sca-1–PE/Cy5.5 (D7), β7-integrin–PE (M293), c-Kit–APC-eFlour780 (2B8), CD150-PE/Cy5 (TC15-12F12.2), Ly6C-FITC, FcϵRIα-FITC (MAR-1), CD71-FITC (RI7217), CD41-FITC (MWReg30), CD27-APC (LG.3A10), and Flk2-biotin (A2F10). BaPs were sorted on a FACSAria II using the following labeled mAbs: FITC-conjugated CD4 (L3T4), CD8 (53-6.7), Gr-1 (RB6-8C5), CD11b, B220, CD11c, FcϵRIα-PE (MAR-1), CD34-eFlour660 (RAM34), and c-Kit–APC-eFlour780 (2B8). Basophil mast cell bipotential progenitors (BMCPs) were sorted on a FACSAria II using the following labeled mAbs: Pacific Blue–conjugated CD3 (145-2C11), CD4 (L3T4), CD8 (53-6.7), CD11b (M1/70), Ter119 (Ter-119), Gr-1 (RB6-8C5), β7-integrin–PE (M293), c-Kit–APC (2B8), and PE-Cy7–FcγR.93 Single cells were sorted using a FACSAria II into 96-well round-bottom plates containing growth medium (IMDM) supplemented with 20% FCS and IL-3 (30 ng/mL), IL-5 (20 ng/mL), IL-6 (10 ng/mL), GM-CSF (20 ng/mL), and SCF (20 ng/mL). All cytokines were purchased from PeproTech. After 7 days in culture at 37°C, half of each well was removed from culture and the remaining half was supplemented with fresh medium and growth factors. The half that was removed was split into 2 parts: half was analyzed by flow cytometry on a LSRFortessa (BD Biosciences) and the other half was used for cytospin followed by anti–mMCP-8 staining36 and May-Grunwald-Giemsa staining as described previously35,36 ; we performed those analyses again after an additional 4 days of culture. For some BMCP cultures, in addition to the culture medium described above (containing 5 cytokines), we used medium containing 10 cytokines, namely, IMDM supplemented with 20% FCS with SCF (20 ng/mL), IL-3 (20 ng/mL), IL-5 (50 ng/mL), IL-6 (20 ng/mL), IL-7 (20 ng/mL), IL-9 (50 ng/mL), IL-11 (10 ng/mL), GM-CSF (10 ng/mL), erythropoietin (2 units/mL), and thrombopoietin (10 ng/mL; R&D Systems), as described by Arinobu et al.12
Semiquantitative RT-PCR analysis
Total RNA was prepared from total BM cells and then subjected to first-strand cDNA synthesis with RT using oligo-dT primers. Semiquantitative PCR was performed with 3-fold serially diluted cDNA templates. The primers were described previously.36
IgE-mediated chronic allergic skin inflammation
IgE-mediated chronic allergic skin inflammation was elicited as described previously.37 Briefly, mice were passively sensitized with IgE by an IV injection of 300 μg of trinitrophenol (TNP)–specific IgE (IGELb4).38 The next day, 10 μg of TNP11-conjugated ovalbumin (OVA; Biosearch Technologies) in 10 μL of PBS was injected intradermally into the left ear pinna of the mice under light anesthesia, and an equal amount of OVA was injected into the right ear pinna using a microsyringe. Ear thickness was measured with a dial thickness gauge (G1-A; Oazki) at the indicated time points. The difference in ear thickness was calculated at each time point.
Passive cutaneous anaphylaxis
Mice were sensitized passively with an intradermal injection of 2 μg of DNP-specific IgE (SPE-7; Sigma-Aldrich) in 20 μL of PBS into the right ear pinna. As a control, the same volume of PBS was injected into the left ear pinna. The mice were challenged 24 hours later with an IV injection of 250 μg of DNP30-BSA (LSL) plus 1.25 mg of Evans blue dye (Sigma-Aldrich) in 250 μL of PBS. Thirty minutes after antigen challenge, the mice were euthanized, and the Evans blue dye was extracted from each dissected ear pinna in 500 μL of acetone/water (7:3) at 37°C overnight. The Evans blue in the extracts was measured with a spectrophotometer at 620 nm and calculated based on the standard.
BMCMC degranulation assay
For the BM-derived cultured mast cell (BMCMC) assay, cells were sensitized with 1 μg/mL an anti-DNP IgE mAb (SPE-7 or ϵ-2639 ) for 12 hours at 37°C. After sensitization, the cells were washed twice with Tyrode buffer (10mM HEPES, pH 7.4, 130mM NaCl, 5mM KCl, 1.4mM CaCl2, 1mM MgCl2, and 5.6mM glucose), suspended in the same buffer containing 0.1% BSA, and stimulated with polyvalent dinitrophenyl-human serum albumin (DNP23-HAS; Biosearch Technologies) at 0, 6.25, 12.5, 25, 50, and 100 ng/mL for 30 minutes. For the β-hexosaminidase reaction, 50 μL of supernatant or cell lysate and 100 μL of 1.3 mg/mL p-nitrophenyl-N-acetyl-D-glucosamide (in 0.1M citrate, pH 4.5) were added to each well of a 96-well plate, and the color was developed for 60 minutes at 37°C. The enzyme reaction was then stopped by adding 150 μL of 0.2M glycine-NaOH, pH 10.2, and the absorbance at 405 nm was measured in a microplate reader (Bio-Rad). Cells were lysed with Tyrode buffer containing 1% Triton X-100 and the β-hexosaminidase activity was measured. The percentage of β-hexosaminidase released was calculated using the following formula: release (%) = supernatant/(supernatant + cell lysate) × 100.
Histologic analysis
Ear, back skin, and stomach specimens were fixed with 10% formalin and embedded in paraffin. Then, 4-μm sections were stained with 0.1% Toluidine blue for histologic examination of mast cells. Mast cells were quantified according to area (per square millimeter) for ear and back skin and per linear millimeter of tissue for glandular stomach and forestomach. Images were captured with an Olympus BX60 microscope using a Retiga-2000R QImaging camera run by Image-Pro Plus Version 6.3 software (Media Cybernetics).
ELISA
BMCMCs from WT or Runx1P1N/P1N mice were sensitized with an anti-DNP IgE mAb39 overnight and then stimulated with 10 ng/mL of DNP23-HSA (Biosearch Technologies) for 16 hours. ELISA for IL-6 was performed using an ELISA kit from BD Biosciences.
Nematode infection
WT or Runx1P1N/P1N mice were infected with 10 000 Strongyloides venezuelensis L3 larvae. BM and spleen were analyzed 8 days after infection.
Treatment with cytokines in vivo
WT or Runx1P1N/P1N mice were treated with daily IP injections of IL-3 (200 ng/d; PeproTech) for 7 consecutive days, TSLP (400 ng/d; R&D Systems) for 5 consecutive days, or vehicle (PBS) for 7 or 5 consecutive days. Basophils in the BM and spleen were analyzed the day after the 7th day (for IL-3 vs PBS) or 5th day (for TSLP vs PBS) injection. The IL-3 complex (IL-3 10 μg plus anti–IL-3 Ab 10 μg; MP2-8F8; BD Biosciences) was prepared as described previously13 and mice were analyzed 3 days after a single IV injection.
Results
Basophils are severely reduced in Runx1P1N/P1N mice
To investigate the roles of the P1-Runx1 variant protein in vivo, we recently established mice in which the N-terminal sequences for P1-Runx1 were replaced with neor gene (Runx1P1N allele), resulting in the absence of both P1-Runx1 transcripts and protein.34 We had demonstrated previously a requirement for P1-Runx1 in lymphoid tissue inducer cell differentiation,40 and found that Runx1P1N/P1N mice have severe reductions in NKT cells, mild T-cell deficits, and an increase in Lin−c-Kit+Sca-1+ HSCs.40 However, there have been no previous reports describing the myeloid cell compartment in these mice. When we analyzed myeloid cells in Runx1P1N/P1N mice, we found that they have a severe reduction in basophils. Compared with corresponding WT mice, Runx1P1N/P1N mice have a greater than 90% reduction of basophils in the BM, spleen, and blood (Figure 1A-B). To examine this phenotype using a different approach, we performed RT-PCR for Mcpt8, which encodes the basophil-associated marker, mouse mast cell protease 8.36 Under the RT-PCR conditions used, Mcpt8 mRNA was not detectable in total BM cells of Runx1P1N/P1N mice, but was readily detected in corresponding samples from WT mice (Figure 1C). These results provided additional evidence of the drastic reduction in basophils in Runx1P1N/P1N mice.
Runx1P1N/P1N mice have markedly reduced numbers of basophils. (A) BM, spleen, and blood were isolated from WT and Runx1P1N/P1N mice and stained with anti-IgE and anti-DX5 mAbs. Data shown are representative of 5 independent experiments, each of which gave similar results. (B) The numbers of basophils are shown as means + SEM. ***P < .0001; no asterisks, P > .05. (C) Semiquantitative RT-PCR analysis for Mcpt8, which encodes mMCP-8, was performed using RNA prepared from total BM cells from WT or Runx1P1N/P1N mice. cDNA was diluted 3-fold. Data shown are from 1 of 3 independent experiments, each of which gave similar results.
Runx1P1N/P1N mice have markedly reduced numbers of basophils. (A) BM, spleen, and blood were isolated from WT and Runx1P1N/P1N mice and stained with anti-IgE and anti-DX5 mAbs. Data shown are representative of 5 independent experiments, each of which gave similar results. (B) The numbers of basophils are shown as means + SEM. ***P < .0001; no asterisks, P > .05. (C) Semiquantitative RT-PCR analysis for Mcpt8, which encodes mMCP-8, was performed using RNA prepared from total BM cells from WT or Runx1P1N/P1N mice. cDNA was diluted 3-fold. Data shown are from 1 of 3 independent experiments, each of which gave similar results.
Normal numbers of eosinophils, neutrophils, and mast cells in Runx1P1N/P1N mice
There are 3 types of granulocytes: neutrophils, eosinophils, and basophils. Because Runx1P1N/P1N mice virtually lack basophils, we analyzed numbers of the other granulocytes in the mutant mice. Neutrophils (Gr-1highSiglec-F−) and eosinophils (Gr-1lowSiglec-F+) were detected by flow cytometry at normal numbers in both the BM and spleen of Runx1P1N/P1N mice compared with WT mice (Figure 2A-B). In addition to these granulocytes, numbers of monocytes (Gr-1lowSiglec-F−), NK cells (NK1.1+CD3−), total T cells (CD3+), B cells (B220+), and dendritic cells (CD11c+) were not significantly different in Runx1P1N/P1N mice compared with WT mice in either the BM or spleen (Figure 2A-B). As we reported previously,40 NKT cells (NK1.1+CD3+) were reduced in both the BM and spleen (Figure 2A-B). These data indicate that, among granulocyte populations, basophils are uniquely deficient in Runx1P1N/P1N mice.
Phenotypic analysis of other granulocytes and leukocytes in Runx1P1N/P1N mice. (A) Representative flow cytometric plots of neutrophils (Gr-1high SiglecF−), eosinophils (Gr-1int SiglecF+), monocytes (Gr-1int SiglecF−), NK cells (NK1.1+CD3−), NKT cells (NK1.1+CD3+), B cells (B220+), conventional dendritic cells (DCs; CD11c+B220−), plasmacytoid dendritic cells (CD11c+B220+), and T cells (CD3+), and their cell counts (B) from BM and spleens from WT or Runx1P1N/P1N mice. Data shown are from 1 of 3 independent experiments, each of which gave similar results. Data in panel B show means + SEM.
Phenotypic analysis of other granulocytes and leukocytes in Runx1P1N/P1N mice. (A) Representative flow cytometric plots of neutrophils (Gr-1high SiglecF−), eosinophils (Gr-1int SiglecF+), monocytes (Gr-1int SiglecF−), NK cells (NK1.1+CD3−), NKT cells (NK1.1+CD3+), B cells (B220+), conventional dendritic cells (DCs; CD11c+B220−), plasmacytoid dendritic cells (CD11c+B220+), and T cells (CD3+), and their cell counts (B) from BM and spleens from WT or Runx1P1N/P1N mice. Data shown are from 1 of 3 independent experiments, each of which gave similar results. Data in panel B show means + SEM.
Basophils are often compared with mast cells because they share certain features such as the expression of the high-affinity IgE receptor (FcϵRIα) and the ability to secrete, after the appropriate stimulation, a similar (although distinct) spectrum of mediators, including histamine, lipid mediators, and cytokines.2,41 To examine whether there is also a deficit in mast cells in these mutant mice, we quantified numbers of mast cells in several tissues. Compared with normal WT mice, Runx1P1N/P1N mice exhibited no differences in the numbers of mast cells in the peritoneal cavity (Figure 3A), ear or back skin, glandular stomach, or forestomach (Figure 3B). These findings reveal that, unlike basophils, the mast cell populations analyzed are not dependent on P1-Runx1 to achieve normal numbers at baseline.
Runx1P1N/P1N mice have normal numbers of mast cells in multiple anatomic sites. (A) Cells from peritoneal lavage fluid were stained with anti-mIgE and anti–c-Kit mAbs. Data shown are from 1 of 5 independent experiments, each of which gave similar results. The numbers of peritoneal mast cells are shown as means + SD. ns indicates not significant (P > .05). (B) Toluidine blue staining for mast cells (some indicated by solid arrows) in 4-mm-thick paraffin sections of ear pinnae from WT (top) and Runx1P1N/P1N mice (bottom). The numbers of mast cells in the ear pinnae, back skin, or stomach are shown as means + SD. ns indicates not significant (P > .05).
Runx1P1N/P1N mice have normal numbers of mast cells in multiple anatomic sites. (A) Cells from peritoneal lavage fluid were stained with anti-mIgE and anti–c-Kit mAbs. Data shown are from 1 of 5 independent experiments, each of which gave similar results. The numbers of peritoneal mast cells are shown as means + SD. ns indicates not significant (P > .05). (B) Toluidine blue staining for mast cells (some indicated by solid arrows) in 4-mm-thick paraffin sections of ear pinnae from WT (top) and Runx1P1N/P1N mice (bottom). The numbers of mast cells in the ear pinnae, back skin, or stomach are shown as means + SD. ns indicates not significant (P > .05).
Basophil, but not mast cell, function is abolished in Runx1P1N/P1N mice
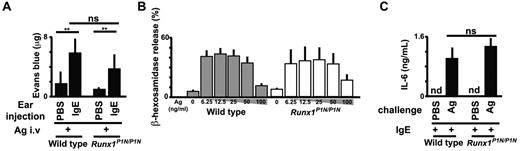
Although Runx1P1N/P1N mice have normal numbers of mast cells (as shown in Figure 3), we wished to examine the function of mast cells in Runx1P1N/P1N mice. It is well known that the development of IgE-dependent passive cutaneous anaphylaxis requires mast cells.42 We injected the ear pinnae of WT mice and Runx1P1N/P1N mice with a DNP-specific IgE mAb or with PBS as a control, and then challenged them intravenously the next day with antigen (DNP-BSA) plus Evans blue. Thirty minutes after antigen challenge, the mice were killed, the ears were dissected, and Evans blue was extracted. There were no significant differences in the amount of extracted dye at IgE- or PBS-injected sites between WT and Runx1P1N/P1N mice (Figure 4A).
Runx1P1N/P1N mice have normal mast-cell functions. (A) Analysis of passive cutaneous anaphylaxis reactions in WT and Runx1P1N/P1N mice that received intradermal injections of IgE anti-DNP into the right ear pinnae and of saline into the left ear pinnae (control; none). After sensitization, mice were challenged intravenously with DNP-BSA. Data show means + SD of the extravasation of Evans blue into the ears. (B) Degranulation of WT and Runx1P1N/P1N BMCMCs, assessed as the release of β-hexosaminidase. BMCMCs were sensitized with anti-DNP IgE and stimulated with the indicated concentrations of DNP-HSA (0, 6.25, 12.5, 25, 50, and 100 ng/mL). Data show the means + SD. (C) ELISA of IL-6 in BMCMCs from WT and Runx1P1N/P1N mice sensitized with anti-DNP IgE and stimulated with DNP-HSA (10 ng/mL). nd indicates not detected. ***P < .0001; **P < .001; no asterisks, P > .05 relative to the corresponding WT mice. Data are from 1 of 3 independent experiments, each of which gave similar results.
Runx1P1N/P1N mice have normal mast-cell functions. (A) Analysis of passive cutaneous anaphylaxis reactions in WT and Runx1P1N/P1N mice that received intradermal injections of IgE anti-DNP into the right ear pinnae and of saline into the left ear pinnae (control; none). After sensitization, mice were challenged intravenously with DNP-BSA. Data show means + SD of the extravasation of Evans blue into the ears. (B) Degranulation of WT and Runx1P1N/P1N BMCMCs, assessed as the release of β-hexosaminidase. BMCMCs were sensitized with anti-DNP IgE and stimulated with the indicated concentrations of DNP-HSA (0, 6.25, 12.5, 25, 50, and 100 ng/mL). Data show the means + SD. (C) ELISA of IL-6 in BMCMCs from WT and Runx1P1N/P1N mice sensitized with anti-DNP IgE and stimulated with DNP-HSA (10 ng/mL). nd indicates not detected. ***P < .0001; **P < .001; no asterisks, P > .05 relative to the corresponding WT mice. Data are from 1 of 3 independent experiments, each of which gave similar results.
We also tested mast cells from WT or Runx1P1N/P1N mice in vitro. We found no differences in the numbers or rate of development of BM-derived cultured mast cells (BMCMCs; > 99% c-Kit+FcϵRIα+ by flow cytometry) from WT versus Runx1P1N/P1N mouse BM cells maintained as usual in IL-3–containing medium (data not shown). BMCMCs were sensitized with a DNP-specific IgE mAb overnight, then washed, and stimulated with DNP-HSA. Degranulation was quantified by measuring β-hexosaminidase release. BMCMCs from WT versus Runx1P1N/P1N mice exhibited similar levels of degranulation (Figure 4B) and IL-6 production (Figure 4C) after challenge with IgE and specific antigen. These results detected no abnormality in IgE-dependent function in Runx1P1N/P1N mast cells.
Although it is well known that IgE-mediated immediate type reactions are mast cell–dependent, Mukai et al reported that a type of IgE-mediated chronic skin reaction (IgE-dependent chronic allergic inflammation of the skin [IgE-CAI]) is dependent on basophils but not mast cells.37 We therefore tested whether Runx1P1N/P1N mice exhibited attenuation or absence of this basophil-dependent biologic response. WT mice and Runx1P1N/P1N mice were sensitized intravenously with a TNP-specific IgE mAb and challenged intradermally the next day with the corresponding antigen (TNP-OVA) or the control carrier protein (OVA). We found that the tissue swelling associated with the IgE-CAI response was essentially eliminated in Runx1P1N/P1N mice (Figure 5A). Histologic analysis of TNP-OVA–challenged ear pinnae on day 4 showed marked infiltrates of leukocytes, including basophils (cells stained with anti–mMCP-8 Ab, which were observed in high numbers in the specimens from WT but not Runx1P1N/P1N mice; Figure 5B). Flow cytometric analysis confirmed that there were few infiltrating myeloid cells in the TNP-OVA–challenged ear pinnae of Runx1P1N/P1N vs WT mice (supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In addition, levels of mRNA for IL-4 and mMCP-8 were up-regulated in the TNP-OVA–challenged ear pinnae of WT but not Runx1P1N/P1N mice (supplemental Figure 1C). These results confirm previously reported results43 indicating that basophils play a pivotal role in eliciting myeloid cell infiltration of the dermis in IgE-CAI responses, and show that the basophil deficiency observed in Runx1P1N/P1N mice is sufficient to result in a marked reduction in the basophil-dependent IgE-CAI response.
Runx1P1N/P1N mice have markedly reduced basophil- and IgE-dependent chronic allergic inflammation. (A) WT (■) or Runx1P1N/P1N mice (○) were sensitized passively by an IV injection of TNP-specific IgE 1 day before being challenged with an intradermal injection of TNP-OVA into the left ear pinna and OVA into the right ear pinna as a control. Ear swelling at each time point is shown (means + SEM, n = 3 each). (B) Immunohistochemical staining with an anti-mMCP8 Ab (DAB substrate) to visualize basophils (some indicated with solid arrowheads) and Giemsa counterstaining (4-μm-thick, paraffin-embedded sections) to demonstrate leukocytes in ear pinnae from WT or Runx1P1N/P1N mice 4 days after challenge with OVA or TNP-OVA. Scale bars indicate 100 μm (top panel) or 25 μm (bottom panel). Data shown are from 1 of 2 independent experiments, each of which gave similar results. ***P < .0001; **P < .001; no asterisk, P > .05 relative to the corresponding WT mice.
Runx1P1N/P1N mice have markedly reduced basophil- and IgE-dependent chronic allergic inflammation. (A) WT (■) or Runx1P1N/P1N mice (○) were sensitized passively by an IV injection of TNP-specific IgE 1 day before being challenged with an intradermal injection of TNP-OVA into the left ear pinna and OVA into the right ear pinna as a control. Ear swelling at each time point is shown (means + SEM, n = 3 each). (B) Immunohistochemical staining with an anti-mMCP8 Ab (DAB substrate) to visualize basophils (some indicated with solid arrowheads) and Giemsa counterstaining (4-μm-thick, paraffin-embedded sections) to demonstrate leukocytes in ear pinnae from WT or Runx1P1N/P1N mice 4 days after challenge with OVA or TNP-OVA. Scale bars indicate 100 μm (top panel) or 25 μm (bottom panel). Data shown are from 1 of 2 independent experiments, each of which gave similar results. ***P < .0001; **P < .001; no asterisk, P > .05 relative to the corresponding WT mice.
Taken together, our results show that Runx1P1N/P1N mice exhibit a marked deficiency in a basophil-dependent immune response (as well as a marked deficiency in basophil numbers) but appear to exhibit normal levels of the IgE-dependent mast cell functions analyzed.
Nematode infection or IL-3 injection fail to induce marked basophilia in Runx1P1N/P1N mice
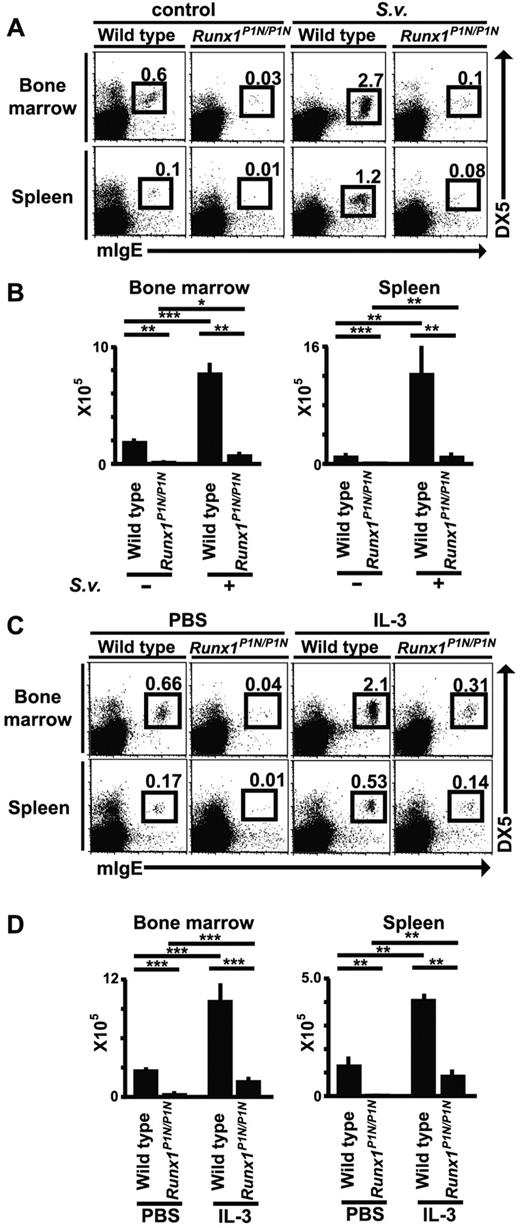
It has been reported that basophil numbers expand during infection with certain nematodes.14,15,44 To investigate this in Runx1P1N/P1N mice, WT or mutant mice were infected by subcutaneous inoculation with 10 000 S venezuelensis third-stage infective larvae. Eight days after S venezuelensis infection, we analyzed the number of basophils (mIgE+DX5+c-Kit−) in these mice by flow cytometry. In WT mice, we observed an approximately 4-fold increase in BM basophils, and a more than 10-fold increase in spleen basophils (Figure 6A-B). In Runx1P1N/P1N mice, basophils exhibited similar increases in response to S venezuelensis infection as observed in WT mice, namely, approximately 4.5-fold in the BM and approximately 8-fold in the spleen, but these expanded populations of basophils in S venezuelensis–infected Runx1P1N/P1N mice were still less than the corresponding basal levels in the uninfected WT mice.
Changes in basophil numbers after S venezuelensis infection or IL-3 injection in Runx1P1N/P1N versus WT mice. (A) WT or Runx1P1N/P1N mice were infected with 10 000 S venezuelensis larvae and basophils (mIgE+DX5+) in the BM and spleen were analyzed 8 days after infection. Data shown are from 1 of 3 independent experiments, each of which gave similar results. (B) Recombinant IL-3 (200 ng/mouse/d) was injected into WT or Runx1P1N/P1N mice for 7 consecutive days. Basophils in the BM and spleen from each mouse were stained the day after the 7th injection. Data shown in panels B and D are means + SEM.
Changes in basophil numbers after S venezuelensis infection or IL-3 injection in Runx1P1N/P1N versus WT mice. (A) WT or Runx1P1N/P1N mice were infected with 10 000 S venezuelensis larvae and basophils (mIgE+DX5+) in the BM and spleen were analyzed 8 days after infection. Data shown are from 1 of 3 independent experiments, each of which gave similar results. (B) Recombinant IL-3 (200 ng/mouse/d) was injected into WT or Runx1P1N/P1N mice for 7 consecutive days. Basophils in the BM and spleen from each mouse were stained the day after the 7th injection. Data shown in panels B and D are means + SEM.
Previous reports from our laboratory and others have shown that IL-3 is essential for the increases in basophil levels that occur after infection with the nematodes S venezuelensis14,15 and Nippostrongylus brasiliensis.15,45 To examine the responsiveness of Runx1P1N/P1N mice to IL-3 in vivo, we injected IL-3 (100 ng/d, IP injection for 7 consecutive days) into WT or Runx1P1N/P1N mice. IL-3 treatment greatly increased the numbers of basophils in WT mice by approximately 4-fold in both the BM and spleen compared with values in WT mice not treated with IL-3 (Figure 6C-D). As observed with S venezuelensis infection, although Runx1P1N/P1N mice injected with IL-3 exhibited increases in basophil numbers that were similar or even greater than those of WT mice (approximately 6-fold in the BM and more than 10-fold in the spleen), the numbers of basophils in IL-3–injected Runx1P1N/P1N mice remained at levels lower than baseline levels of basophils in vehicle-injected WT mice (Figure 6C-D).
We reported previously evidence that both IL-3 and c-Kit can contribute to resistance to a primary infection with S venezuelensis14 When we examined mast cell– and IL-3–deficient KitW/W-v, IL-3−/− mice, they exhibited a more pronounced defect in rejection of S venezuelensis during the primary infection than did either KitW/W-v or IL-3−/− mice,14 suggesting that basophils might contribute to host resistance during this infection. Runx1P1N/P1N mice cleared a primary infection with S venezuelensis significantly more slowly than did WT mice (supplemental Figure 2). Whereas we cannot rule out the possibility that other defects in Runx1P1N/P1N mice also contributed to this observation, this finding is consistent with the hypothesis that the impaired response to this infection reflects, at least in part, the drastic reduction in basophils in Runx1P1N/P1N mice.
It has been reported that TSLP injection can increase the numbers of basophils in mice.8 We found that TSLP treatment (400 ng/mouse/d IP for 5 consecutive days) increased the numbers of basophils in the BM and spleen by approximately 50%-100% in both WT mice and Runx1P1N/P1N mice (supplemental Figure 3). Moreover, as observed with S venezuelensis infection or IL-3 treatment, levels of basophils in TSLP-treated Runx1P1N/P1N mice remained lower than baseline levels of basophils in vehicle-treated WT mice.
BaPs are severely reduced in Runx1P1N/P1N mice
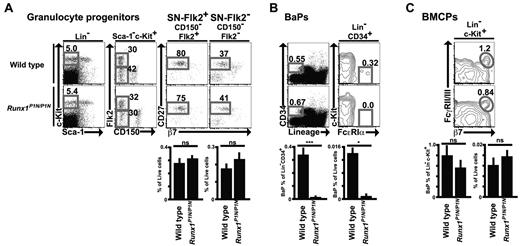
To examine whether the reduced numbers of basophils in Runx1P1N/P1N mice are associated with a deficit in BaPs, we first performed flow cytometric analysis of granulocyte progenitors in the BM. We showed previously that SN-Flk2+ (Sca-1−Lin−c-Kit+CD150− Flk2+CD27+) and SN-Flk2− (Sca-1−Lin−c-Kit+CD150−Flk2−CD27+) populations are already committed predominantly to the granulocyte fate, and that these populations can give rise to all 3 kinds of granulocytes.35 There were no significant differences between WT and Runx1P1N/P1N mice in SN-Flk2+ or SN-Flk2− (Figure 7A).
Impaired BaPs in Runx1P1N/P1N mice. (A-C) Representative flow cytometry plots and percentage of indicated gates of SN-Flk2+ (Sca-1−Lin−c-Kit+CD150−Flk2+CD27+) cells and SN-Flk2− (Sca-1−Lin−c-Kit+CD150−Flk2− CD27+) cells (A), and BaPs (Lin−CD34+c-Kit−FcϵRIα+) in BM from WT or Runx1P1N/P1N mice (B), and BMCPs (Lin−c-Kit+β7+FcγRII/III+) in spleen from WT or Runx1P1N/P1N mice (C). Data shown are from 1 of 3 independent experiments, each of which gave similar results.
Impaired BaPs in Runx1P1N/P1N mice. (A-C) Representative flow cytometry plots and percentage of indicated gates of SN-Flk2+ (Sca-1−Lin−c-Kit+CD150−Flk2+CD27+) cells and SN-Flk2− (Sca-1−Lin−c-Kit+CD150−Flk2− CD27+) cells (A), and BaPs (Lin−CD34+c-Kit−FcϵRIα+) in BM from WT or Runx1P1N/P1N mice (B), and BMCPs (Lin−c-Kit+β7+FcγRII/III+) in spleen from WT or Runx1P1N/P1N mice (C). Data shown are from 1 of 3 independent experiments, each of which gave similar results.
We next assessed BaPs (Lin−CD34+c-Kit−FcϵRIα+), which have been shown to differentiate predominantly into basophils.12 BaPs were severely reduced in Runx1P1N/P1N mice (Figure 7B). Ohmori et al reported that treatment with a mixture of IL-3 and anti–IL-3 (the IL-3 complex) increased the frequency of BaPs in WT mice.13 Compared with IL-3 treatment, injection of the IL-3 complex resulted in a more substantial increase in basophils (supplemental Figure 4A-B). IL-3 complex treatment also resulted in an increase of BM BaPs. As shown in supplemental Figure 4A and B, treatment with the IL-3 complex resulted in substantial increases of BaPs in both WT mice and Runx1P1N/P1N mice. As was also observed for basophils, injection of IL-3 complex into Runx1P1N/P1N mice resulted in levels of BaPs that were higher than the baseline levels of these cells in WT mice, but that were still much less than the corresponding levels of these cells in IL-3 complex–treated WT mice.
Akashi et al reported the identification of a Lin−c-Kit+β7+FcγRII/III+ BMCP in the spleen.12 However, we detected no difference in the numbers of Lin−c-Kit+β7+FcγRII/III+ cells in the spleens of WT compared with Runx1P1N/P1N mice (Figure 7C).
To examine their developmental potential, we performed single-cell cultures of SN-Flk2+ and SN-Flk2− cells. We sorted each of these progenitors to a single cell per well and, after 7 and 11 days of culture, analyzed the resulting populations by assessing their surface markers by flow cytometry and their morphology by cytospin analysis. In the case of BaPs, we sorted 100 cells per well from WT mice (we could not collect enough to analyze from Runx1P1N/P1N mice), and analyzed them 7 days after culture. Neutrophils were defined as Gr-1high cells with lobulated nuclei and no Vital Red staining of the cytoplasm, eosinophils as Gr-1int CCR3+ cells with lobulated nuclei that exhibited cytoplasmic staining with Vital Red, basophils as Gr-1−FcϵRI+c-Kit−DX5+ cells with lobulated nuclei and with cytoplasm that stained with mMCP-8 but not with Vital Red, and mast cells as Gr-1−FcϵRI+c-Kit+DX5− cells lacking Vital Red and mMCP-8 staining of the cytoplasm (supplemental Figure 5).
As shown in supplemental Figure 6, neutrophils developed from both SN-Flk2+ and SN-Flk2− cells and there were no differences between results obtained from cells derived from WT versus Runx1P1N/P1N mice. Compared with neutrophils, eosinophils developed more efficiently from SN-Flk2− than SN-Flk2+ cells, but there were also no significant differences between results obtained from cells derived from WT versus Runx1P1N/P1N mice. Basophils developed predominantly from SN-Flk2− cells in WT mice, but that potential was severely reduced in SN-Flk2− cells from Runx1P1N/P1N mice. As expected, BaPs from WT mice developed into basophils (supplemental Figure 6). In contrast, Lin−c-Kit+β7+FcγRII/III+ “BMCPs” gave rise only to mast cells whether we used our culture conditions (“5 cytokines” in supplemental Figure 7) or those used by Akashi et al12 (“10 cytokines” in supplemental Figure 7).
Together with our finding of a striking reduction in basophils in Runx1P1N/P1N mice in vivo, these in vitro results indicate that P1-Runx1 plays a role in facilitating the developmental transition from granulocyte progenitors to BaPs, and that an abnormality at this step contributes to the drastic reduction in basophils in Runx1P1N/P1N mice. Our data also suggest that Lin−c-Kit+β7+FcγRII/III+ cells do not represent the main pathway for the development of basophils in vivo.
Discussion
In the present study, we found that Runx1P1N/P1N mice, which are deficient in the P1-Runx1 transcription factor, exhibit a severe reduction in basophils at baseline (Figure 1), but have normal levels of other granulocytes and tissue mast cells (Figure 3). To our knowledge, our data are the first to identify P1-Runx1 as a transcription factor that has a nonredundant role in the development of basophils but apparently not for other granulocytes or mast cells. We reported previously that granulocyte development potential resides predominantly in SN-Flk2+ (Sca-1−Lin−c-Kit+CD150−Flk2+ CD27+) and SN-Flk2− (Sca-1−Lin−c-Kit+CD150−Flk2−CD27+) populations.35 In the present study, we found that the SN-Flk2+ and SN-Flk2− populations are present in normal numbers in Runx1P1N/P1N mice (Figure 7A), but that the number of BaPs12 is reduced profoundly in such mice (Figure 7B). These findings suggest that Runx1P1N/P1N mice have a marked restriction in the transition from granulocyte progenitors to BaPs.
It is important to emphasize that whereas basophils levels are strikingly reduced in Runx1P1N/P1N mice, a few basophils can be still detected in these mice (Figure 1). Moreover, when we subjected Runx1P1N/P1N mice either to infection with the nematode S venezuelensis or to repetitive injection with IL-3, each of which results in marked expansion of basophil populations in WT mice,13-15 basophil numbers also expanded in Runx1P1N/P1N mice (Figure 6). Indeed, although basophil numbers in S venezuelensis–infected or IL-3–injected Runx1P1N/P1N mice remained lower than the corresponding baseline levels in naive WT mice, the relative increases in the numbers of BM and spleen basophils in S venezuelensis–infected or IL-3–injected Runx1P1N/P1N mice were the same as or greater than those in the identically treated WT mice (Figure 6). These findings indicate that the basophil lineage in Runx1P1N/P1N mice retains responsiveness to IL-3, but that the expansion of basophils in Runx1P1N/P1N mice injected with IL-3 or infected with a parasite that results in enhanced levels of endogenous IL-3 is subject to a marked restriction, as is the development of baseline levels of basophils in these mice.
In addition to the apparently unipotential BaPs, Arinobu et al reported that a Lin−c-Kit+β7+FcγRII/III+ bipotent progenitor of basophils and mast cells (which they named “BMCP”) can be identified by flow cytometry in the mouse spleen.12 We found that Runx1P1N/P1N and WT mice have similar numbers of Lin−c-Kit+β7+FcγRII/III+ cells in the spleen (Figure 7C). However, in the present study, these cells gave rise only to mast cells in vitro. In analyzing the cultured cells, we identified basophils by both flow cytometry (as FcϵRIα+DX5+c-Kit− cells) and by morphology (as cells with lobulated and often ring-like nuclei and exhibiting a few granules in the cytoplasm by Giemsa stain and positive staining of the cytoplasm with an Ab to mMCP-8). Mast cells were defined as FcϵRIα+DX5−c-Kit+ cells by flow cytometry and by morphology as mMCP-8− cells with many granules in the cytoplasm that stained with Giemsa stain.
It is possible that the discrepancy between our findings and those of Arinobu et al12 reflect differences in the mice analyzed and/or in aspects of the flow cytometric or culture conditions used. However, we found using flow cytometry that Runx1P1N/P1N mice and WT mice not only have similar numbers of Lin−c-Kit+β7+FcγRII/III+ “BMCPs” in the spleen (Figure 6C), but also exhibit statistically indistinguishable numbers of mast cells in the peripheral tissues analyzed (Figure 3). Our data thus indicate that Lin−c-Kit+β7+FcγRII/III+ cells may have only a limited (or no) ability to give rise to basophils. As we suggested in a prior study,35 Lin−c-Kit+β7+FcγRII/III+ cells may represent mast-cell progenitors that can give rise to a subpopulation of cells in the mast-cell lineage that have little or no surface expression of c-Kit.46 Moreover, mouse basophils can be difficult to identify based on conventional staining protocols (such as with May-Giemsa staining). We therefore recommend confirming the identity of mouse basophil populations initially identified based on testing a limited number of cell-surface markers by also searching for markers that are more specific for these cells, such as mMCP-8.
The precise mechanism by which P1-Runx1 regulates basophil differentiation remains to be elucidated. One must consider in this context at least 2 pathways of basophil development. The first is basophil differentiation in naive mice at baseline. In this pathway, both IL-3 and TSLP are dispensable.8,14,15 Alternatively, during infection with certain parasites, IL-3 is essential for basophil expansion,14,15,45 and it has been reported that injections of the IL-3 complex can result in an increase in the number of BaPs and basophils.13 We detected no significant differences in the levels of surface expression of the IL-3 receptor on basophils or SN-Flk progenitors in WT compared with Runx1P1N/P1N mice (data not shown). Moreover, whereas treatment with the IL-3 complex is not “physiologic,” such experiments revealed that IL-3 can increase numbers of both basophils and BaPs in Runx1P1N/P1N mice and in WT mice (supplemental Figure 4). These results provide further support for the conclusion that the defect(s) in basophil production in Runx1P1N/P1N mice occur despite the retention of responsiveness of cells in the basophil lineage to IL-3 and TSLP.
Based on these results, we speculate that the P1-Runx1 pathway functions in parallel with or independently of the IL-3 or TSLP-dependent pathways. Indeed, because we observed a substantial increase in basophil numbers over baseline levels in S venezuelensis–infected or IL-3–treated Runx1P1N/P1N mice (Figure 6), and because basophils were not completely absent in Runx1P1N/P1N mice in the steady state (although basophil numbers were < 10% of WT levels; Figure 1), there appears to be a P1-Runx1–independent pathway that can contribute to the development of basophils both at baseline and during IL-3–dependent (and perhaps TSLP-dependent) expansion of this cell type. It is possible that IL-3 and TSLP are not the only contributors that regulate basophil numbers and that P1-Runx1 can modulate the action of those other regulators/mechanisms of basophil development. Clearly, further studies are required to clarify the identity and interrelationships among the pathways that contribute to basophil development.
The results of the present study reveal a novel role for P1-Runx1 in basophil development and provide another example of a difference in the regulation of basophil and mast-cell development in mice. Our findings also suggest that further studies of P1-Runx1–mediated transcriptional networks may uncover additional interesting features of the developmental pathway(s) that lead(s) to the generation of these rare and enigmatic granulocytes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Chieko Tezuka, Chen Liu, and Jennifer Lilla for technical assistance; Fu-Tong Liu for providing ϵ-26 IgE; and all the members of the Galli, Karasuyama, and Taniuchi laboratories for helpful discussions.
This study was supported by grants from the National Institutes of Health (AI070813, AI023990, and CA072074 to S.J.G.) and the Japanese Ministry of Education, Culture, Sports, Science and Technology (to I.T. and K.N.).
National Institutes of Health
Authorship
Contribution: K.M., M.J.B., M.T., and K.N. performed the experiments; K.M., M.J.B., M.T., K.N., H.K., I.T., and S.J.G. designed the research and analyzed the data; K.M. and S.J.G. wrote the manuscript; and all authors reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephen J. Galli, MD, Department of Pathology, L-235, Stanford University School of Medicine, 300 Pasteur Dr, Stanford, CA 94305-5324; e-mail: sgalli@stanford.edu.