Abstract
Biologic factors that predict the survival of patients with a diffuse large B-cell lymphoma, such as cell of origin and stromal signatures, have been discovered by gene expression profiling. We attempted to simulate these gene expression profiling findings and create a new biologic prognostic model based on immunohistochemistry. We studied 199 patients (125 in the training set, 74 in the validation set) with de novo diffuse large B-cell lymphoma treated with rituximab and CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) or CHOP-like therapies, and immunohistochemical stains were performed on paraffin-embedded tissue microarrays. In the model, 1 point was awarded for each adverse prognostic factor: nongerminal center B cell–like subtype, SPARC (secreted protein, acidic, and rich in cysteine) < 5%, and microvascular density quartile 4. The model using these 3 biologic markers was highly predictive of overall survival and event-free survival in multivariate analysis after adjusting for the International Prognostic Index in both the training and validation sets. This new model delineates 2 groups of patients, 1 with a low biologic score (0-1) and good survival and the other with a high score (2-3) and poor survival. This new biologic prognostic model could be used with the International Prognostic Index to stratify patients for novel or risk-adapted therapies.
Introduction
Diffuse large B-cell lymphoma (DLBCL) comprises 30%-40% of all non-Hodgkin lymphoma (NHL) cases in the developed world and consists of a heterogeneous group of tumors both morphologically and clinically.1 The addition of rituximab to the standard chemotherapy protocol of CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) has significantly improved the survival of patients with DLBCL.2-7 One of the most important clinical predictors of survival in these patients is the International Prognostic Index (IPI).8 Although some authors have suggested that the IPI has lost predictive power in the rituximab era, it remains a valuable tool for risk stratification of DLBCL patients.6,9
Gene expression profiling (GEP) can also stratify DLBLC patients into 2 biologic prognostic groups, the germinal center B cell–like (GCB) and activated B cell–like (ABC) subtypes.10-12 Because genome-wide GEP requires frozen tumor tissue, various immunohistochemical algorithms have been developed for paraffin-embedded tissue to reproduce the GEP findings and predict the cell of origin and survival in DLBCL. One of the most widely accepted methods is the Hans algorithm which uses antibodies against CD10, BCL6, and MUM1.13 Several other algorithms have recently been proposed, including the Choi algorithm which has proven to be a sensitive and specific predictor of cell of origin and survival.14,15 Recently, the cellular composition of the tumor microenvironment has also been shown to be a powerful predictor of survival in patients with DLBCL. Lenz et al performed GEP on a large series of DLBCL patients treated with rituximab (R)–CHOP and defined 2 important stromal signatures, stromal-1 and stromal-2.16 The stromal-1 signature reflects extracellular matrix deposition and histiocyte infiltration, and portends a good prognosis. Meyer et al recently attempted to reproduce the stromal-1 signature using an antibody against SPARC (secreted protein, acidic, and rich in cysteine) to evaluate expression in stromal cells and histiocytes in the tumor microenvironment.17 They showed that patients with SPARC positivity in the tumor stroma had a significantly longer survival than those without significant SPARC expression. The stromal-2 signature largely reflects angiogenesis and blood vessel density in the tumor stroma, and portends a poor prognosis. Cardesa-Salzmann et al recently attempted to reproduce the stromal-2 signature by measuring microvessel density (MVD) in DLBCL, and found high MVD to be an adverse prognostic factor.18
Therefore, we attempted to simulate the GEP findings16 and create a biologic prognostic model (BPM) based on immunohistochemistry that incorporates the cell of origin and surrogates for the stromal-1 and stromal-2 signatures. This model would potentially facilitate risk stratification of DLBCL patients for whom only paraffin-embedded tissue was available for study.
Methods
Patients
Two hundred thirty-five patients with de novo DLBCL treated with rituximab and CHOP or CHOP-like therapies were studied by the Leukemia and Lymphoma Molecular Profiling Project (LLMPP) consortium, and a training set of 125 patients was derived from this cohort based on the patients having complete immunohistochemical data (see “Construction of the prognostic model”). The validation set consisted of 74 patients from the Nebraska Lymphoma Study Group who were also treated with rituximab (R) and CHOP or CHOP-like therapies. The total of 199 patients from the training and validation sets was treated as follows: R-CHOP (166 patients, 83%); R-CNOP (cyclophosphamide, mitoxantrone, vincristine and prednisone; 31 patients, 16%); and R-ESHAP (etoposide, methylpredisolone, cytarabine, and cisplatin; 2 patients, 1%). The following sites participated in the study: Nebraska Lymphoma Study Group, Omaha, Nebraska (60 cases); British Columbia Cancer Agency, Vancouver, British Columbia (51 cases); Norwegian Radium Hospital, Oslo, Norway (43 cases); University of Barcelona, Barcelona, Spain (30 cases); Cleveland Clinic Foundation, Cleveland, Ohio (20 cases); Oregon Health Sciences Center, Portland, Oregon (14 cases); University of Arizona, Tucson, Arizona (12 cases); and the University of Würzburg, Würzburg, Germany (5 cases). This study was approved by the institutional review boards of the respective institutions, and all patients gave written informed consent in accordance with the Declaration of Helsinki.
The data used in this study originated from 3 previously published studies15,17,18 that examined the same cohort of patients. The classification of DLBCL into GCB and non-GCB subtypes was done by Meyer et al,15 and expression of SPARC in the tumor microenvironment was evaluated in a separate article by Meyer et al.17 Cardesa-Salzmann et al evaluated the MVD in the same group of patients.18
Tissue microarray immunohistochemistry
Hematoxylin and eosin–stained sections from a representative formalin-fixed, paraffin-embedded tissue block were used to define diagnostic tumor areas, and 1 to 3 representative tissue cores, 0.6-1.0 mm in diameter, were obtained from each case and inserted into recipient paraffin blocks in a grid pattern using a tissue arrayer (Beecher Instruments). Paraffin-embedded sections 5-μm thick were then cut and stained according to previously described procedures with antibodies against GCET1, CD10, BCL6, MUM1, FOXP1, CD31, and SPARC (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).15,17,18 The immunoperoxidase stains, except for CD31, were performed on either a Benchmark XT (Ventana) or Autostainer Plus (DAKO). The cutoffs for tumor positivity were 80% for GCET1, MUM1, and FOXP1, and 30% for CD10 and BCL6 as defined by the Choi algorithm.14 The scoring of these antibodies was estimated visually in 10% increments by 2 authors (A.M.P. and P.N.M.), and the DLBCL cases were classified as either GCB or non-GCB subtype.15 The number of stromal cells and histiocytes in the tumor microenvironment that expressed SPARC was also estimated by 2 authors (A.M.P. and P.N.M.) in each case and graded in 5% increments. The number of positive cells ranged from 0%-30%, and 2 groups were defined: SPARC < 5% and SPARC ≥ 5%.17 The agreement between the 2 scoring authors was 90%. Any discrepancies were resolved by joint review over a double-headed microscope. Immunoperoxidase staining for CD31 was performed according to a previously described method.18 The CD31 stains were performed on an automated Bond Max immunostainer (Vision BioSystems).
Blood vessel density measurement
The MVD was quantified by analyzing digitalized images of the CD31-stained tissue microarray cores with Olympus Cell B Basic Imaging Software. Microvessel areas were defined as vascular areas delineated by CD31-positive staining, and the MVD was calculated as the sum of all microvessel areas (μm2) divided by the total area of the core analyzed (μm2). The MVD values were then grouped in quartiles (first quartile: 0-0.00635; second quartile: 0.00635-0.01047; third quartile: 0.01047-0.01509; fourth quartile: 0.01509-0.08270). The TMAs were independently scored by 2 observers (T.M.C.-S. and L.C.), and discrepancies were resolved by joint review over a double-headed microscope.18
Construction of the prognostic model
Of the 235 cases, only 125 cases with complete immunohistochemical data were used to form the training set. These 125 cases were compared with the other 110 cases not used in the analysis to assess whether there were any clinical differences between the 2 groups, and none were found (supplemental Table 2). Univariate and multivariate analyses were then performed on the 125 patients. Because all 3 parameters (Choi algorithm, SPARC, and MVD) were previously shown to be significant predictors of survival,15,17,18 they were reevaluated in this cohort and used to tally a BPM score. One point was awarded for each adverse prognostic marker: non-GBC subtype, SPARC < 5%, and MVD quartile 4. This BPM was then tested in multivariate analysis after adjusting for the IPI. The BPM was also validated in another multivariate analysis of 74 other patients (validation set) treated with rituximab and CHOP or CHOP-like therapies by the Nebraska Lymphoma Study Group. The immunohistochemical studies of the validation set were scored using tissue microarrays as previously done in the training set. The clinical characteristics of patients in the training and validation sets were also compared for any significant differences.
Statistical analysis
The Kaplan-Meier method was used to estimate overall (OS) and event-free survival (EFS) distributions. Overall survival was defined as the time from initial diagnosis to death from any cause or last contact. Event-free survival was defined as the time from initial diagnosis to the date of progression, relapse, death, or last contact. Patients who were alive and relapse-free at the time of last follow-up were treated as censored. The log-rank test was used to compare the survival distributions. The association between the BPM and certain clinical characteristics including age, sex, performance score, clinical stage, elevated serum lactate dehydrogenase (LDH), number of extranodal sites, IPI score, and response to initial therapy was evaluated by using the χ2 test or the Fisher exact test for small samples. Comparison of clinical characteristics between the training and the validation sets was evaluated using χ2 tests. Cox regression analysis was used to compare prognostic factors in multivariate analysis after adjusting for the IPI. Model fit of the Cox regression models was examined using various residual plots, and it was determined that there was no serious lack of fit of the models.
Validation of the BPM was assessed using discrimination and calibration information. Discrimination of the model was measured with the concordance index where, in a randomly selected pair of patients, if the patient with the worse outcome (ie, dies or has the event first) has the worse predicted outcome by the model, then the result is said to be concordant. A random prediction would result in a concordance index of 0.5 and perfect prediction results in an index of 1.0. The concordance index with a 95% confidence interval (CI) is calculated for the both the training and validation sets for OS and EFS using the SAS macro SurvCsTD by the method of Kremers.19 Calibration of the model was assessed by comparing the predicted probability of survival from the training set to the observed probability of survival from the validation set along with 95% CIs. The survival probabilities were compared for both OS and EFS at 1-, 2-, 3-, and 5-year time points for the combined groups using the biologic prognostic model score. The analysis was conducted with SAS software (SAS Institute Inc). P values less than .05 were considered statistically significant.
Results
The training set included 125 patients with complete immunohistochemical data, including 70 (56%) men and 55 (44%) women (supplemental Table 2). The median age of the patients was 62.6 years (range, 22-92 years), and 57 patients (46%) were younger than 60 years of age. At the time of analysis, 83 patients were alive and 42 had died. The median follow-up of the surviving patients is 3.8 years (range, 0.3-11.3 years).
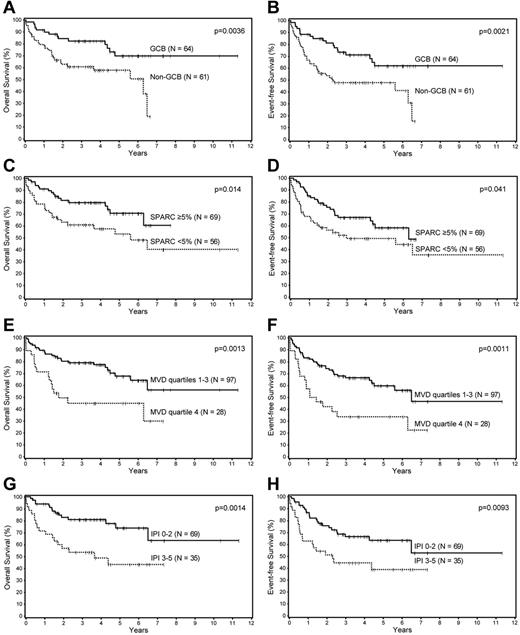
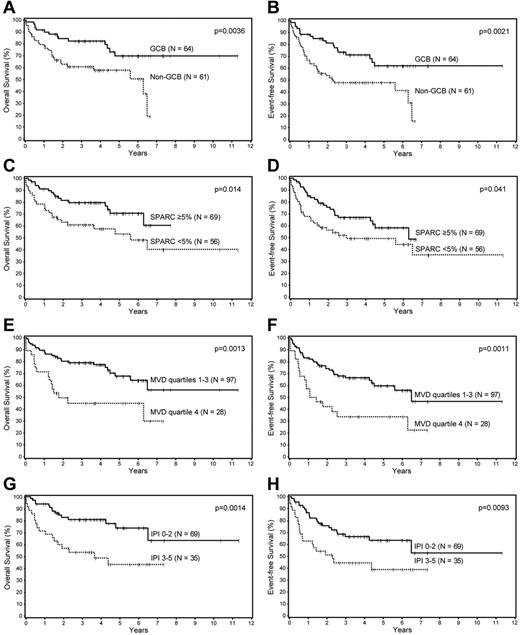
Univariate analysis of the prognostic factors in this cohort showed that the Choi algorithm, SPARC expression, MVD, and the IPI were all significant predictors of OS and EFS (Table 1; Figure 1). Multivariate analysis showed that SPARC, MVD, and the IPI were independent predictors of OS, whereas the Choi algorithm was a borderline predictor (P = .056). The Choi algorithm, MVD, and the IPI were also independent predictors of the EFS, whereas SPARC was a borderline predictor (P = .068; Table 2).
Survival analyses of patients with DLBCL. Overall and event-free survival of patients with DLBCL based on the (A-B) Choi algorithm, (C-D) SPARC expression, (E-F) microvascular density, and the (G-H) IPI.
Survival analyses of patients with DLBCL. Overall and event-free survival of patients with DLBCL based on the (A-B) Choi algorithm, (C-D) SPARC expression, (E-F) microvascular density, and the (G-H) IPI.
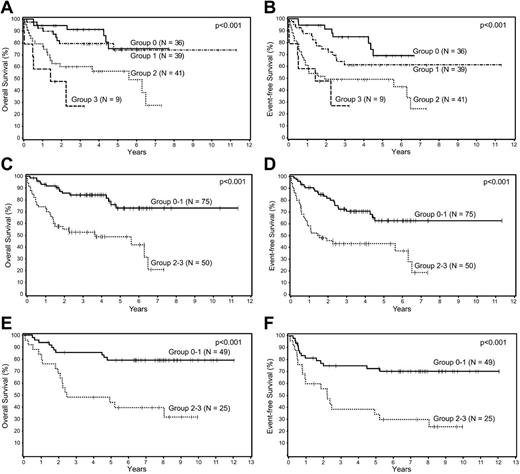
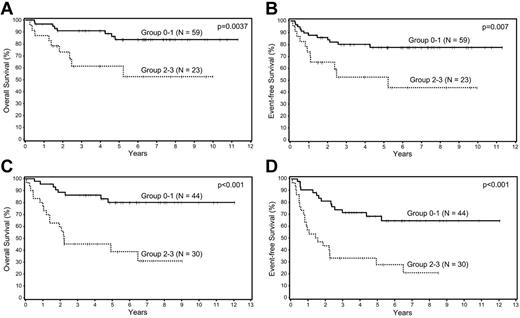
Because the Choi algorithm, SPARC, and MVD were all shown to be independent predictors of survival, they were used to create a BPM score by awarding 1 point for each adverse prognostic marker; 4 survival curves were generated corresponding to scores of 0, 1, 2, and 3 (Figure 2A-B). The curves were compared to assess the differences in survival between the groups and there was no significant difference in OS or EFS between the 2 groups with low (0, 1) scores (P = .52 and .13, respectively). The same was true for the 2 groups with high (2, 3) scores (P = .14 and .41, respectively). Based on these results, 2 prognostic groups were formed, group 0-1 (n = 75) and group 2-3 (n = 50), and compared to assess whether there were any clinical differences between these 2 groups (Table 3). The only significant difference between the 2 groups was a higher proportion of patients with advanced-stage disease in group 2-3. The new BPM using the 3 factors was highly predictive of OS (P = .0001) and EFS (P < .0001) in multivariate analysis after adjusting for the IPI (Table 4), and patients with low (0-1) BPM scores had significantly better survival than those with the high (2-3) scores (Figure 2C-D).
Survival analyses of patients with DLBCL according to biologic prognostic model score. Overall and event-free survival of patients with DLBCL in the training set divided into 4 groups based on the (A-B) biologic prognostic model score, and into 2 groups based on combined scores in the (C-D) training set, and the (E-F) validation set.
Survival analyses of patients with DLBCL according to biologic prognostic model score. Overall and event-free survival of patients with DLBCL in the training set divided into 4 groups based on the (A-B) biologic prognostic model score, and into 2 groups based on combined scores in the (C-D) training set, and the (E-F) validation set.
The validation set included 74 patients with complete immunohistochemical data. There were 42 (57%) men and 32 (43%) women, and the median age was 63 years (range, 19-89 years). At the time of analysis, 48 patients were alive and 26 had died. The median follow-up of the surviving patients is 7.9 years (range, 1.4-12 years). There were no significant differences in the clinical characteristics of the training and the validation sets. Similar to the training set, the BMP was highly predictive of OS (P = .0002) and EFS (P = .0008) in multivariate analysis after adjusting for the IPI (Table 4), and patients with low (0-1) BPM scores had significantly better survival than those with the high (2-3) scores (Figure 2E-F).
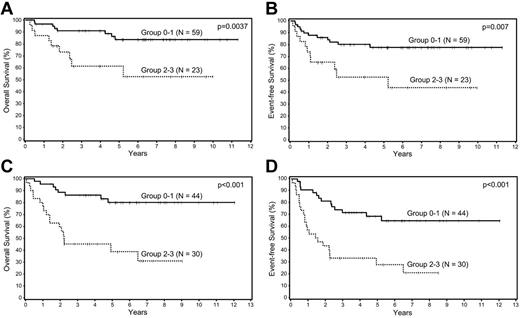
Patients in the training and validation sets were combined into 1 cohort and then divided into 3 prognostic groups according to IPI scores: low risk (0-1), intermediate risk (2-3), and high risk (4-5). The BPM was highly predictive of OS and EFS in both the low-risk and intermediate-risk IPI groups (Figure 3). This survival analysis was not done for the high-risk IPI group because of the small number of patients (N = 15).
Survival analyses of patients with DLBCL according to biologic prognostic model score in the IPI groups. Overall and event-free survival in the (A-B) low and (C-D) intermediate IPI groups.
Survival analyses of patients with DLBCL according to biologic prognostic model score in the IPI groups. Overall and event-free survival in the (A-B) low and (C-D) intermediate IPI groups.
The biologic prognostic model validation showed good concordance in both the training and validation datasets. The concordance index for the training set was 0.75 (95% CI: 0.67-0.83) for OS and 0.72 (95% CI: 0.65-0.79) for EFS. The concordance index for the validation set was 0.73 (95% CI: 0.62-0.84) for OS and 0.69 (95% CI: 0.59-0.78) for EFS. Calibration of the data showed excellent agreement between the predicted probability and the observed probability for OS in the 0-1 risk group in the validation set (supplemental Table 3). There was also good agreement between the predicted and observed probability for OS in the 2-3 risk group, with the 95% confidence intervals for the observed probability containing the predicted probabilities. The EFS data also showed good agreement between predicted and observed probabilities for each group at each time point.
Discussion
Diffuse large B-cell lymphoma is a biologically heterogeneous disease which is reflected in the varied response to therapy and survival of individual patients. The IPI is the most widely used prognosticator in DLBCL, yet patients with the same IPI score may have quite different responses to treatment and survival outcomes. Prediction of the outcome of patients with DLBCL is important for multiple reasons including the selection of the best initial therapy, enrollment in clinical trials, and evaluation of novel therapeutic agents.20
Gene expression profiling studies have identified 2 biologic and prognostic subtypes of DLBCL based on the cell of origin, the GCB and ABC subtypes.10-12 More recently, GEP has also defined 2 prognostic signatures in the tumor microenvironment of DLBCL, stromal-1 and stromal-2.16 The stromal-1 signature predicts a favorable outcome and reflects extracellular matrix deposition and histiocyte infiltration. In contrast, the stromal-2 signature carries a poor prognosis and reflects angiogenesis and blood vessel density in the tumor stroma. We attempted to reproduce these GEP findings using paraffin-embedded tissue from DLBCL patients treated with standard immunochemotherapy.
In this study, we used data from 3 previously published studies of a cohort of DLBCL patients treated with rituximab and CHOP or CHOP-like regimens.15,17,18 Meyer et al15 classified the cases of DLBCL into GCB and non-GCB subtypes using the Choi algorithm.14 In a separate study, Meyer et al examined the expression of SPARC in the tumor microenvironment in an attempt to simulate a stromal-1 signature.17 SPARC is a glycoprotein expressed in a variety of normal cells, including a specific population of macrophages associated with tissue injury or certain tumors.21 They found that high SPARC expression by cells in the microenvironment of DLBCL, specifically macrophages and fibroblasts, portends a good prognosis.17 Concurrently, Cardesa-Salzmann et al attempted to simulate the stromal-2 signature in the same cohort of patients by measuring the MVD.18 They used an antibody against CD31 (PECAM1) to stain the tumor vessels because this was one of the genes included in the stromal-2 signature originally described by Lenz et al.16 This was the first study in the rituximab era to look at MVD in DLBCL, with high MVD predicting a poor clinical outcome.18
One hundred twenty-five patients from this cohort with complete immunohistochemical data were used in our study to form the training set of patients. Univariate analysis of these patients showed that the cell of origin, SPARC expression, MVD, and the IPI were all significant predictors of survival, and multivariate analysis demonstrated that all 4 factors were also independent predictors of survival. Based on these results, we constructed a BPM using the 3 factors derived from immunohistochemistry done on paraffin-embedded tissue. One point was awarded for each adverse prognostic factor (non-GCB subtype, SPARC < 5%, and MVD quartile 4) and survival curves were generated for scores of 0, 1, 2, and 3. Because the curves for scores of 0 and 1 overlapped, and the curves for scores of 2 and 3 also overlapped, they were grouped together, respectively, and 2 prognostic groups were formed: group 0-1 and group 2-3. The BPM was then tested in a multivariate analysis and was found to be highly predictive of both OS and EFS after adjusting for the IPI. We found that patients with a low BPM score (0-1) had a significantly better survival than those with a high BPM score (2-3). The BPM was then validated by multivariate analysis of a separate set of 74 patients, and was again predictive of OS and EFS after adjusting for the IPI. Furthermore, the BPM was highly predictive of OS and EFS in the low-risk (0-1) and intermediate-risk (2-3) IPI groups when the training and validation sets were combined, identifying patients with more aggressive disease within each of these clinical groups.
Multiple individual biomarkers, as well as prognostic models, have been evaluated in patients with DLBCL using different techniques. Some of the models are based on mRNA expression by GEP or by RT-PCR technology, and provide a quantitative measurement of gene expression.20 These techniques are highly accurate and predictive of outcome, but are impractical for routine use because they are expensive, not generally available, and usually require fresh or cryopreserved tumor tissue. However, Malumbres et al22 have created a paraffin-based, 6-gene prognostic model in R-CHOP–treated DLBCL patients using RT-PCR. The genes they used included LMO2, BCL6, FN1, CCND2, SCYA3, and BCL2, and their model divided patients into low- and high-risk groups based on a mortality predictor score that was independent of the IPI. Another study analyzed the expression of 36 genes with the ArrayPlate assay in CHOP- and R-CHOP–treated DLBCL patients using formalin-fixed tissue.23 The best prognostic model in their study was a 2-variable model using the MYC and HLA-DR genes. This model was also shown to be complementary to the IPI in stratifying patients with regard to prognosis. Recently, Alizadeh et al proposed a 2-gene model based on the expression of LMO2 and the tumor microenvironment marker TNFRSF9 in patients with DLBCL.24 This study used the quantitative real-time PCR and was performed on formalin-fixed tissue. The 2-gene model was an independent predictor of survival in multivariate analysis. However, none of the technologies used in these models are simple to use or commercially available. In contrast, our method using immunohistochemistry performed on paraffin-embedded tissue offers another approach to evaluate the prognosis of DLBCL patients. Immunostaining is inexpensive, simple to perform, and routine in most clinical laboratories. However, each of these new BPMs will need to be validated in prospective trials in the future. One potential challenge for our model is the use of multiple antibodies with different cutoffs which could lead to increased variability in interobserver scoring. However, with the trend toward personalized therapy of hematolymphoid neoplasms, incorporation of multiple markers to prognostic panels is to be expected.
Our BPM based on tissue immunohistochemistry is the first model in the rituximab era to combine the cell-of-origin and stromal signatures into 1 prognostic score. Although the reproducibility of immunohistochemical stains and algorithms has been questioned in the literature, in our experience, the Choi algorithm and SPARC immunohistochemical stains are easy to score and highly reproducible.25,26 Furthermore, we have previously demonstrated the reproducibility of the Choi algorithm.14,15 Although microvessel density assessment by image analysis is not currently available in most laboratories, as with all new technologies in medicine, it is critical to evaluate the potential contribution of image analysis in advancing our diagnostic and prognostic capabilities. As image analysis becomes more widely available, pathologists would have the option of performing or outsourcing this ancillary study, as is routinely done with other laboratory tests. Moreover, with the imminent onset of virtual microscopy imaging, access to these analytical tools will inevitably become more widespread in academic and tertiary care institutions. Therefore, in the future, our biologic prognostic model could be used in conjunction with the IPI to stratify patients in clinical trials for novel or risk-adapted therapies.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Cancer Institute grant U01-CA-114778.
National Institutes of Health
Authorship
Contribution: A.M.P. performed immunohistochemical stains, analyzed the results, and wrote the manuscript; T.M.C.-S. and P.N.M. performed immunohistochemical stains and analyzed the results; L.C. and E.C. analyzed the results and contributed cases to the study; L. M. Smith performed the statistical analysis; K.F., T.C.G., J.D., R.D.G., L.R., E.S.J., G.O., A.R., R.M.B., R.T., J. R.C., L. M. Staudt, J.M.C., L.H.S., J.M.V., A.L.-G., and W.C.C. contributed cases and data to the study; D.D.W. designed the study, analyzed the results, and wrote the manuscript; and all coauthors reviewed and contributed to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dennis D. Weisenburger, MD, Department of Pathology and Microbiology, 983135 Nebraska Medical Center, Omaha, NE 68198-3135; e-mail: dweisenb@unmc.edu.
References
Author notes
T.M.C.-S. and P.N.M. have equally contributed to the study.