Abstract
Recent genomic studies have outlined a landscape of recurrent alterations within some subtypes of lymphoid cancer. Yet, the timing and cellular contexts during which these alterations occur (ie, the molecular ontogeny) remain poorly understood. Lymphoid malignancies offer an exceptional opportunity to delineate the ontogeny of somatic alterations, as lymphocyte differentiation absolutely requires the introduction of indelible genetic rearrangements at antigen receptor loci during specific stages of maturation. We review competing models of lymphomagenesis and highlight evolving evidence that somatic alterations in uncommitted hematopoietic progenitors contribute to some mature lymphoid neoplasms. These progenitors could serve as reservoirs for further clonal evolution and thereby contribute to therapeutic resistance, tumor relapse, and the development of second hematologic malignancies. Defining the pathways that are dysregulated within early progenitors and the ontogeny of subsequent alterations that contribute to lymphoid transformation could establish novel therapeutic targets across a variety of hematologic malignancies and even guide avenues for future preventive strategies.
Introduction
B-cell and T-cell lymphomas compose > 90% of lymphoid malignancies worldwide and 4% of new cancers each year.1 Recent studies have catalogued hundreds of genetic and transcriptional alterations across large panels of lymphomas, with extensive representation of the 2 most common subtypes, follicular lymphoma (FL) and diffuse large B-cell lymphoma (DLBCL).2-11 Within a short period, the large majority of lymphoma-associated alterations will probably be known, leaving 2 great challenges. The first is to clarify which alterations functionally contribute to the lymphoma phenotype. The second challenge is to define the clonal architecture of individual lymphomas, including the cellular contexts in which each alteration develops (ie, the molecular ontogeny).
New insights from both hematologic and epithelial tumors indicate that malignancies can harbor extensive clonal heterogeneity across multiple stages of differentiation.12-17 However, the investigation of ontogeny in nonlymphoid cancers requires the use of surrogates for differentiation stage, such as immunophenotype, which are inherently variable (eg, through epigenetic plasticity). In contrast, lymphocyte differentiation absolutely requires the introduction of indelible genetic alterations at antigen receptor loci. Like all hematopoietic cells, lymphocytes derive from hematopoietic stem cells (HSCs) through a process of lineage-specific differentiation that is orchestrated by master regulators, including transcription factors and microRNA. Both the RAG recombinase and activation-induced deaminase (AID) introduce characteristic rearrangements at antigen receptor loci, as well as off-target damage at oncogenes and tumor suppressor genes that contribute to lymphoid transformation.18,19 Thus, lymphoid cancers offer an exceptional opportunity to delineate the stages of differentiation at which individual alterations develop.
We review recent insights into lymphomagenesis, with a particular focus on the involvement of hematopoietic precursors with multilineage potential. Although still a nascent field, we build on evidence from select lymphoid tumor subtypes to speculate whether common processes underlie lymphomagenesis and how these processes may affect phenotypes, such as relapse, therapeutic resistance, and second hematologic malignancies.
Diverse genetic alterations in human lymphomas
As outlined in the Introduction, a series of studies have recently reported hundreds of single nucleotide variants, insertions/deletions, amplifications, and chromosomal rearrangements in primarily B-lineage lymphomas and lymphoma cell lines.2-11 For example, Morin et al sequenced the exome or whole genome of 14 lymphomas and identified between 20 and 135 coding single nucleotide variants per tumor.7 Many of the described alterations represent passenger events. However, subsets are recurrent (ie, identified in more than 1 case), cluster within specific subsets of lymphoma, and can plausibly be linked to malignant phenotypes (Table 1). For the purposes of this Perspective, functionally relevant alterations can be roughly divided into 4 large classes:
Modifiers of proliferation and apoptosis. In addition to well-known alterations (eg, IGH-MYC, IGH-CCND1, deletions of PTEN and CDKN2A/CDKN2B), more recent studies have identified both gain-of-function and loss-of-function alterations that primarily dysregulate NF-κB and/or JAK-STAT signaling, including mutations of TNFAIP3,4 CARD11,5 MYD88,20 CD79A/CD79B,3 PRDM1, STAT6,7 and JAK2.9,21 Less common mutations in genes, such as KRAS and BRAF, have also been reported.22 Additional alterations that suppress apoptosis (eg, IGH-BCL2, loss of TP53 and miR-15a/miR-1623 ) but may not directly drive proliferation are common in human lymphomas.
Modifiers of differentiation. Both gain-of-function and loss-of-function alterations have been reported that may either arrest differentiation at a specific stage or drive differentiation to a specific lineage. Many of the affected genes encode transcription factors, including BCL6, NOTCH1, FOXO1, PAX5, IRF4, BTG1, ETS1, and MEF2B7-9,11,22,24 or otherwise affect transcription factor function.25
Epigenetic modifiers. These include factors involved in adding and removing post-translational modifications to DNA and histones (eg, MLL2,7 EZH2,6 KDM2, EP300,8 CREBBP,8 JMJD2C,21 and TET2 and DNMT3A26 ) as well as mediators of RNA splicing and nuclear export.24,27
Modifiers of the microenvironment and immune regulation. Both gain-of-function and loss-of-functional alterations can modify stromal interactions or suppress immune responses to lymphoma cells, including mutations of B2M, CD58, and CIITA,10 TNFRSF9 and TNFRSF14,28 PDL1, PDL2, and EPHA7.9,29
These 4 classes extensively overlap; for example, loss of the soluble tumor suppressor EPHA7 in follicular lymphoma promotes proliferation through up-regulation of MAP kinase and STAT3 signaling.29 In addition, many alterations in both coding genes and noncoding RNA do not fit easily within any of the 4 categories. Nonetheless, a more complete picture of the complement of alterations necessary for lymphoid transformation is coming into view. Yet the timing of these alterations relative to each other and to the stages of normal lymphocyte differentiation remains almost completely obscure.
Models of lymphomagenesis
The World Health Organization Classification of Tumors of Hematopoietic and Lymphoid Tissues includes > 80 subtypes of lymphoid cancer, many of which are defined by their histologic, immunophenotypic, and transcriptional resemblance to a particular stage of lymphoid differentiation (Figure 1).30 This has led to the hypothesis that each lymphoma originates within a lymphocyte at the differentiation stage that corresponds most closely to the lymphoma phenotype. Cells at most stages of lymphocyte differentiation do not have inherent self-renewal capacity. Thus, alterations that confer self-renewal and other malignant phenotypes must be introduced either before or after lymphoid lineage commitment.
B-cell maturation and corresponding lymphoma phenotypes. The stage of maturation is noted under each cell, and the corresponding malignant lymphoma is noted above the cell. The shaded brown region represents the germinal center. The dotted arrow indicates the possible role of a memory B–like cell in multiple myeloma,43 as outlined in the text. ABC indicates activated B cell–like; GCB, germinal center B cell–like; and SLL, small lymphocytic lymphoma.
B-cell maturation and corresponding lymphoma phenotypes. The stage of maturation is noted under each cell, and the corresponding malignant lymphoma is noted above the cell. The shaded brown region represents the germinal center. The dotted arrow indicates the possible role of a memory B–like cell in multiple myeloma,43 as outlined in the text. ABC indicates activated B cell–like; GCB, germinal center B cell–like; and SLL, small lymphocytic lymphoma.
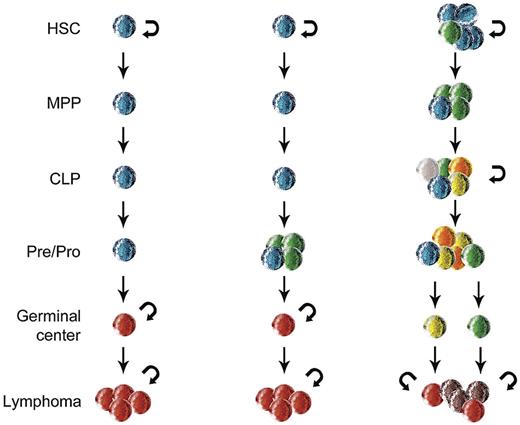
Figure 2 outlines 3 models of lymphoid transformation that differ based on the timing and cellular contexts of tumor-promoting alterations. In the first model, a lymphocyte acquires all of the alterations at a single stage of maturation. In the second model, the cell acquires initial alterations at early stages of lymphoid differentiation that may drive clonal expansion or resistance to apoptosis. These cells develop additional alterations at later stages of differentiation that complement the earlier lesions. In the third model, the initial alterations occur within uncommitted progenitors that either have inherent self-renewal capacity or obtain that capacity through the alterations. Like cells in the second model, the altered progenitors in the third model can acquire additional alterations during lymphoid differentiation. At some stages, the cells may retain partial or complete self-renewal capacity and thus form reservoirs that contribute to clonal diversification.
Three models of lymphoma ontogeny. In the first model (left), a normal lymphocyte acquires the full complement of alterations at a single stage of maturation that corresponds to the lymphoma phenotype. In the second model (center), 1 or more initiating events occur during early lymphoid maturation and the remaining alterations develop at a stage corresponding to the lymphoma phenotype. In the third model (right), the initial alterations are acquired within hematopoietic stem cells that acquire additional alterations at multiple stages of differentiation. At some stages, the cells have self-renewal capacity (indicated by semicircular arrows) that promotes further clonal heterogeneity, as indicated by cells of multiple colors. MPP indicates multipotent progenitor; and CLP, common lymphoid progenitor.
Three models of lymphoma ontogeny. In the first model (left), a normal lymphocyte acquires the full complement of alterations at a single stage of maturation that corresponds to the lymphoma phenotype. In the second model (center), 1 or more initiating events occur during early lymphoid maturation and the remaining alterations develop at a stage corresponding to the lymphoma phenotype. In the third model (right), the initial alterations are acquired within hematopoietic stem cells that acquire additional alterations at multiple stages of differentiation. At some stages, the cells have self-renewal capacity (indicated by semicircular arrows) that promotes further clonal heterogeneity, as indicated by cells of multiple colors. MPP indicates multipotent progenitor; and CLP, common lymphoid progenitor.
Sporadic Burkitt lymphoma (BL) may represent an example of the first model. BL is thought to originate from centroblasts within the germinal center.31 The lymphoma cells maintain many morphologic and immunophenotypic features of nonmalignant centroblasts. The genetic lesions within cases of sporadic BL primarily occur within the germinal center,32 including translocations of MYC that involve aberrant class-switch recombination and off-target somatic hypermutation in both BCL6 and MYC.33,34 Importantly, enforced expression of Myc in murine B cells is sufficient to generate lymphomas with self-renewal capacity that extends to the bulk of tumor cells.35 Eμ-Myc pre-B/B-cell lymphomas can propagate within congenic recipients after transplantation of fewer than 10 cells.35 Although the expression of stem cell markers (Sca-1, AA4.1) varies across the lymphoma cells, it does not distinguish between cells that possess or lack tumor-propagating potential.35
In contrast with sporadic BL, endemic BL may represent an example of the second model, as the IGH-MYC rearrangements in these lymphomas characteristically involve RAG-mediated breaks at the IGH locus. Similarly, translocations that fuse IGH to CCND1 in mantle cell lymphoma (MCL), BCL2 in follicular lymphoma, and MALT1 in marginal zone lymphomas all involve the combined activity of RAG and AID within pro/pre-B cells.18,36,37 Multiple studies have identified AID mRNA and/or protein expression within progenitor B cells concurrent with expression of RAG.38-42 According to a process outlined by the Lieber laboratory, methylated cytidines at CpG dinucleotides are converted by AID to thymidines, which results in a T:G mismatch.18 These mismatches can either be nicked by RAG or converted to breaks through base excision repair. The resulting translocations are present within all cells of each lymphoma and directly contribute to lymphoma pathogenesis. On that basis, the IGH rearrangements could represent the initial alteration within a previously normal maturing lymphocyte, as outlined in the second model in Figure 2.
Independent of the timing of alterations during lymphoid transformation, hierarchical relationships may also exist such that subclones at a differentiation stage with inherent self-renewal potential propagate a bulk tumor population that expresses a different immunophenotype. For example, Matsui et al reported that CD138−CD20+CD27+ memory B cells compose a subpopulation within multiple myeloma cell lines and primary samples (Figure 1).43 These cells share immunoglobulin rearrangements with the bulk tumor population, give rise to clonogenic multiple myeloma in vitro, and engraft into Nod.Scid mice in vivo.43 The same group reported that the Hodgkin Reed-Sternberg (HRS) cell lines L428 and KM-H2 contain rare CD27+ B cells capable of generating both the CD27+ subset and the CD27− bulk tumor cell populations.44 CD27+ B cells were detectable in the blood of patients with Hodgkin lymphoma and shared immunoglobulin rearrangements with lymph node-resident HRS cells. Thus, in both multiple myeloma and Hodgkin lymphoma, a subpopulation of immunophenotypically distinct B cells could represent tumor-propagating cells.44
Involvement of uncommitted progenitors in lymphomagenesis
Studies of both common and uncommon lymphoma subtypes indicate that at least some alterations within mature lymphoid neoplasms can develop in uncommitted hematopoietic progenitors. HSCs are long-lived, which affords the opportunity to acquire mutations over time. Thus, the requirement for multiple oncogenic events to accrue within HSCs is consistent with the later age of onset among the common lymphoid tumor subtypes (ie, FL, MCL, and chronic lymphocytic leukemia (CLL)/small lymphocytic lymphoma. Evidence supporting the involvement of uncommitted progenitors in mature lymphoid neoplasms includes:
Stem cell leukemia/lymphomas
Several genetically defined diseases manifest as both lymphoma and myeloid leukemias, suggesting that the initiating lesion occurs within a cell with multilineage potential. Most notable among these diseases are the myeloid and lymphoid neoplasms associated with eosinophilia and abnormalities of PDGFRA, PDGFRB, or FGFR1, which typically present with some combination of myeloproliferative neoplasm with eosinophilia, T-cell lymphoblastic lymphoma and progression to acute leukemia.30 Among the acute leukemias observed with 8p11 (FGFR1) rearrangements, two-thirds have myeloid or bilineage phenotypes and the remaining one-third are purely lymphoid.45 In a murine bone marrow transplant model, expression of ZFN198-FGFR1 results in myeloproliferation and T-cell lymphoblastic lymphoma. Mice transplanted with ZFN198-FGFR1-expressing bone marrow have a 30-fold expansion of Lin−Sca+Kit+ cells, suggesting that early HSCs are targeted by the transgene.46 The T-cell lymphoblastic lymphomas harbor homozygous deletions involving Tcrα and Tcrδ that most probably occur during V(D)J recombination. These deletions preclude functional T-cell receptor (TCR) signaling, which results in arrest at the CD4+/CD8+ stage.46 Nonmalignant T cells that lack a functional TCR undergo clonal deletion, but the ZFN198-FGFR1–expressing cells are presumably protected from apoptosis by the prosurvival effects of aberrant FGFR1 signaling.46
B-lineage priming in chronic lymphocytic leukemia
Kikushige et al isolated CD34+CD38− HSCs and CD34+CD38−CD90+ long-term repopulating HSCs from patients with CLL and normal controls.47 On transplantation into immunodeficient mice, the cells from patients with CLL generated a higher percentage of pro-B cells than those from normal controls.47 Indeed, mice transplanted with HSCs from patients with CLL develop CD5+ B cells that are either monoclonal or oligoclonal but harbor different immunoglobulin rearrangements than the original CLLs. Thus, the CD5+ B cells appear to reflect specific priming of the HSCs toward the CLL phenotype and not contamination with CLL cells.47 This priming is associated with the expression of early B-cell transcription factors (IKZF1, TCF3, and IRF8).47 The authors speculated that the propensity to progress into CLL develops within HSCs and that the progeny of these HSCs acquire additional alterations during B-cell development, including translocations and mutations involving off-target activity of RAG and AID (as outlined in model 3 in Figure 2).47 Cytogenetic abnormalities identified in the original CLLs were not present in the patient's HSCs or the B cells that developed after xenogeneic transplantation.47 However, extensive sequencing and epigenetic profiling were not performed, so the specific alterations within the HSCs that underlie lineage priming remain unknown.
If early progenitors can be primed toward B-cell transformation, it would follow that multiple phenotypes of B-cell lymphoma would develop in some patients. Indeed, Sanchez et al reported 53 cases of B-cell malignancies with 2 or more clonal immunophenotypic populations.48 Three or more distinct immunoglobulin rearrangements (IGH, IGκ, and/or IGλ) could be recovered from all 44 cases with material available, including 24 (45%) with surface IGκ in one population and surface IGλ in the other.48 Overall, 4.8% of 477 consecutive B-cell malignancies had 2 or more B-cell clones, including cases of CLL, hairy cell leukemia, DLBCL, MCL, and marginal zone lymphoma. In nearly one-half of cases that included CLL with an additional clone, the second clone did not phenotypically resemble CLL.48 Thus, whatever alterations occurred before clonal divergence presumably contributed to both CLL and the additional lymphoid phenotype. The same phenomenon may underlie composite lymphomas, in which 2 histologically distinct lymphomas are admixed within the same specimen and typically share the same immunoglobulin gene rearrangement.49 In contrast, 13 of the 53 cases in the series by Sanchez et al with multiple clones had identical immunophenotypes, except that one subpopulation expressed IGκ and the other expressed IGλ.48 In these cases, the tumor phenotype was presumably determined by the alterations that existed before light chain rearrangement (ie, at the pre-B cell stage or earlier).
Mutations of epigenetic modifier genes in cells with multilineage potential
Quivoron et al identified mutations of TET2 in 2% of B-cell lymphomas and 11.9% of T-cell lymphomas.50 One patient with B-cell lymphoma subsequently developed acute myelogenous leukemia (AML). Both the lymphoma and the AML harbored the same biallelic TET2 mutations, but myeloid colonies from the patient's CD34+ cells contained 0, 1, or 2 TET2 mutations (Figure 3A).50 Thus, clonal evolution of the TET2 mutations occurred within cells capable of multilineage differentiation and generated both B-cell lymphoma and AML.
Mutations within CD34+ cells in lymphoma and AML. (A) CD34+ cells from a patient with B-cell lymphoma and AML carried 0, 1, or 2 mutations in TET2. (B) CD34+ cells from a patient with T-cell lymphoma that harbored biallelic TET2 mutations carried only 1 mutation in TET2 and either 3 or 4 additional mutations that were identified in the lymphoma.
Mutations within CD34+ cells in lymphoma and AML. (A) CD34+ cells from a patient with B-cell lymphoma and AML carried 0, 1, or 2 mutations in TET2. (B) CD34+ cells from a patient with T-cell lymphoma that harbored biallelic TET2 mutations carried only 1 mutation in TET2 and either 3 or 4 additional mutations that were identified in the lymphoma.
In another case, the patient's T-cell lymphoma harbored biallelic TET2 mutations, but only 1 of the mutations was present within myeloid colonies derived from the patient's CD34+ cells (Figure 3B). Thus, 1 mutation was present within an early progenitor, whereas the other was acquired during clonal differentiation.50 Exome sequencing identified 7 mutations in other genes within the T-cell lymphoma. Among these, mutations of PLZF and CRIM1 were present in 8 of 8 CD34+ cells that also had mutant TET2. In contrast, a mutation of ZNF774 was present in only 6 of 8 cells that also had mutant TET2, indicating that the ZNF774 mutation was acquired later than the others (Figure 3B).50 None of the 3 mutations were observed in 65 CD34+ cell-derived colonies that had wild-type TET2.50
The same group recently reported the co-occurrence of TET2 and DNMT3A mutations in T-cell lymphomas.26 Two patients with T-cell lymphomas that harbored both TET2 and DNMT3A mutations also had myelodysplastic syndrome (MDS). In one of these 2 patients, the MDS harbored the same TET2 and DNMT3A mutations found in the patient's T-cell lymphoma. In the other case, the MDS harbored only the DNMT3A mutation but not the TET2 mutation.26 Thus, both patients presumably had DNMT3A-mutated cells with multilineage potential. In one case, those precursors also harbored a TET2 mutation, but in the other case the precursors did not.26 These findings demonstrate that molecular ontogeny can differ across patients, with the same mutations acquired at either early or late stages of differentiation. The finding that TET2 and DNMT3A are recurrently mutated within CD34+ cells is also consistent with the previously described notion that epigenetic dysregulation within precursor cells creates a premalignant state conducive to the acquisition of transforming alterations.51,52
Mutations associated with follicular lymphoma within HSCs
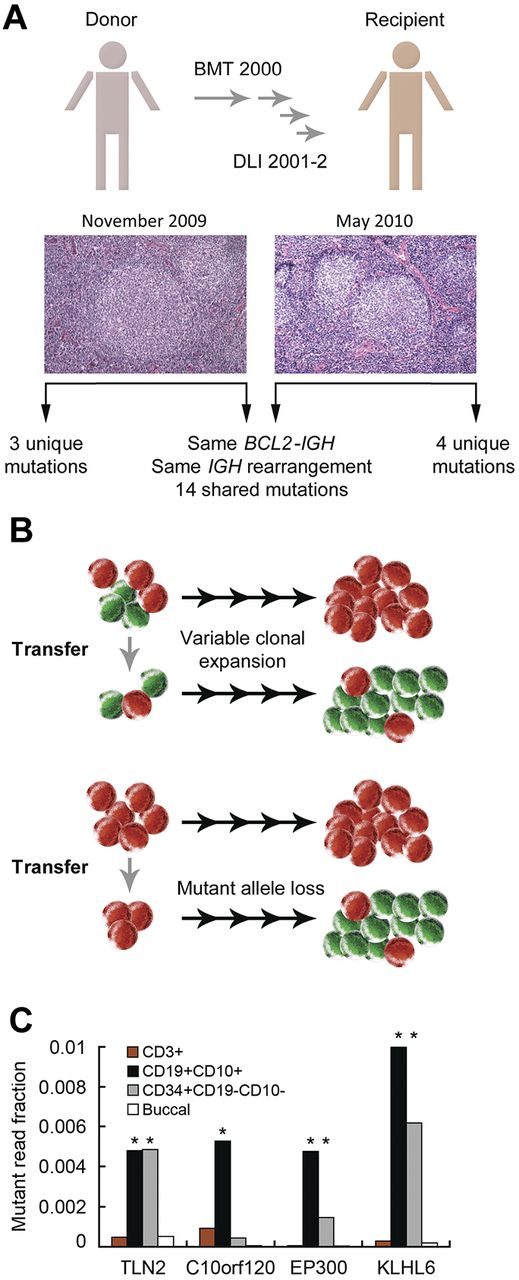
To explore lymphoma ontogeny, we used a donor-recipient pair who both developed FL 7 years after allogeneic bone marrow transplantation and donor lymphocyte infusions (DLIs; Figure 4A).53 Both FLs harbored the same BCL2-IGH and IGH V-DJ rearrangements,53 indicating that they derived from the same B-lineage committed precursor.18 The BCL2-IGH rearrangement was present in 1 in 2000 cells in the DLI and thus occurred at least 7 years before the clinical onset of either FL.53
Molecular ontogeny in donor-derived FL. (A) Bone marrow transplantation (BMT) and DLIs were performed for chronic myelogenous leukemia. FL developed concurrently in the recipient and the donor. Sequencing of both FLs identified shared and disparate mutations. (B) Models for subclonal loss of the CTSS mutation from the recipient's FL. (C) Ultra-deep sequencing of sorted populations from the DLI. DNA from the donor's buccal swab (buccal) was used for germline material. *P < .01 versus with buccal for the same mutation.
Molecular ontogeny in donor-derived FL. (A) Bone marrow transplantation (BMT) and DLIs were performed for chronic myelogenous leukemia. FL developed concurrently in the recipient and the donor. Sequencing of both FLs identified shared and disparate mutations. (B) Models for subclonal loss of the CTSS mutation from the recipient's FL. (C) Ultra-deep sequencing of sorted populations from the DLI. DNA from the donor's buccal swab (buccal) was used for germline material. *P < .01 versus with buccal for the same mutation.
Exome sequencing of both FLs identified 14 somatic nonsynonymous mutations that were shared between the lymphomas as well as 4 mutations solely within the recipient's FL and 3 mutations solely within the donor's FL (Figure 4A).53 Based on ultra-deep sequencing (median read coverage ∼ 400 000), 13 of the 14 shared mutations were enriched within the DLI.53 All 4 mutations unique to the recipient's FL and a mutation in RFTN1 identified only in the donor's FL were not enriched in the DLI or the other FL,53 consistent with subsequent acquisition during clonal divergence. In addition, a mutation in cathepsin S (CTSS) was initially detected only in the donor's FL, but deep sequencing recovered the mutation in the DLI and in 4.7% of reads from the recipient's FL.53 Thus, either the transplanted BCL2-IGH–rearranged cells were a polyclonal population that included a subset harboring the CTSS mutation or the CTSS mutation was lost in the majority of FL cells within the recipient (Figure 4B).
As expected, the BCL2-IGH rearrangement was only detectable within the CD19+ population in the DLI.53 To clarify whether other mutations identified in both FLs were present in CD19− cells, we performed ultra-deep sequencing of flow-sorted populations from the DLI (median depth, 235 590). Three mutations (TLN2 T588M, EP300 V1148_F1149del, KLHL6 K485_T486insK) but not C10orf120 Q181H were enriched within the CD34+CD10−CD19− population (Figure 4C), which contains multipotent progenitors and HSCs. The mutated genes included EP300, which is altered in 40% of FLs and DLBCLs,7,8 and KLHL6, which is recurrently altered in CLL and multiple lymphoma subtypes.7,24 Thus, alterations that presumably contribute to FL pathogenesis can occur both within CD34+ cells (and thus before the BCL2-IGH rearrangement) and also over a protracted period of clonal evolution.
Network compatibility in FL ontogeny
In our study of donor-derived FL, an ARID1A R1276* mutation was present in the recipient's FL but not within the DLI.53 Somatic mutations in ARID1A occur commonly in a wide variety of cancer types, suggesting that ARID1A is a tumor suppressor.54 Although the donor's FL lacked the ARID1A mutation, it harbored a deletion at the ARID1A locus.53 Therefore, the 2 lymphomas underwent convergent evolution within separate hosts. Mutations in ARID1A have not been reported from extensive sequencing studies of FL, so loss in both of our cases suggests a specific selection for ARID1A haploinsufficiency within the context of the multiple preexisting mutations. Such selection could reflect a concept known as network compatibility,55 which describes the requirement that functional alterations acquired during carcinogenesis both (1) contribute to the malignant phenotype and (2) do not introduce detrimental synthetic interactions. Tumor-related alterations can initiate compensatory changes within affected cells to mitigate deleterious effects from the alteration (ie, functional buffering). As a consequence, any subsequent alterations must also be compatible with preexisting buffering effects.
Up to 25% of healthy persons harbor circulating BCL2-IGH–rearranged cells,56 the majority of which have already traversed the germinal center.57 Yet, FL is diagnosed in only 1 in 24 000 persons in the United States annually. The conventional explanation for this discrepancy is that cells harboring BCL2-IGH rearrangements must subsequently acquire the additional alterations necessary to become fully malignant. An alternative explanation is that alterations necessary for FL development must occur before the BCL2-IGH rearrangement, possibly to foster network compatibility with aberrant BCL2 expression. The finding that mutations of MLL2 are found in nearly 90% of FL cases7 further challenges the notion that BCL2-IGH rearrangement must be the initiating event in FL.
Finally, Eμ-BCL2 mice aberrantly express BCL2 within B cells but do not develop lymphoid neoplasms. In contrast, vavP-BCL2 mice, which express BCL2 throughout the hematopoietic compartment, develop an FL-like disease that appears to depend on T-cell expansion.58 Yet, none of the mutations that we recovered from CD34+ cells was detectable in CD3+ T cells (Figure 4C). This would suggest that the complement of alterations within CD34+ cells in our cases supported B-lineage priming (as observed in CLL47 ) and/or was not compatible with the transcriptional changes inherent to T-cell maturation (ie, network incompatibility).
Clonal evolution through dedifferentiation and lineage trans-differentiation
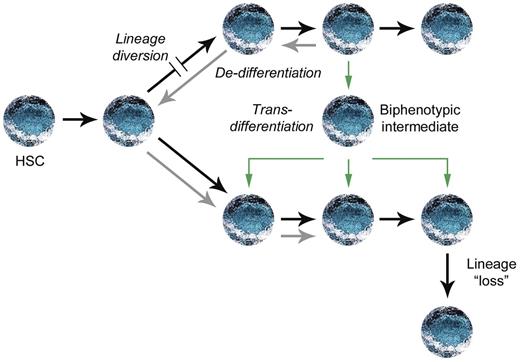
A subset of myeloid malignancies appears to derive from precursors with rearranged antigen receptor loci. Common lymphoid progenitors and pro-T cells can undergo D-J rearrangements at TCRβ and IGH as well as rearrangements of TCRδ and TCRγ before myeloid differentiation, so these are not proof of aberrant differentiation.59 Instead, cells with multilineage potential may initiate antigen receptor rearrangements and then acquire an alteration that diverts the lineage toward myeloid differentiation (Figure 5). AML with translocations of MLL may exemplify this sequence of events; these leukemias have TCR and/or IGH D-J rearrangements in one-third of cases compared with only 2% of AML with germline MLL.60
Possible pathways for aberrant lineage modification.63 A block in one lineage could result in lineage diversion to another lineage before commitment. Alternatively, committed progenitors could either undergo dedifferentiation followed by redifferentiation (gray arrows) or directly trans-differentiate through a biphenotypic intermediate. Alterations within a committed progenitor could also result in the loss of lineage-specific markers without dedifferentiation, as described in Hodgkin lymphoma.
Possible pathways for aberrant lineage modification.63 A block in one lineage could result in lineage diversion to another lineage before commitment. Alternatively, committed progenitors could either undergo dedifferentiation followed by redifferentiation (gray arrows) or directly trans-differentiate through a biphenotypic intermediate. Alterations within a committed progenitor could also result in the loss of lineage-specific markers without dedifferentiation, as described in Hodgkin lymphoma.
In contrast with MLL-translocated AML, many cases of chronic myelogenous leukemia myeloid blast crisis have IGH V-DJ and Igκ rearrangements, indicating that they derive from a committed B-cell precursor.61 To become myeloid blasts, these cells have either undergone dedifferentiation to an uncommitted stage followed by myeloid redifferentiation or have directly trans-differentiated into the myeloid lineage (Figure 5). BCR-ABL1 can drive AID expression, which initiates somatic hypermutation at immunoglobulin loci, B-lineage oncogenes, and BCR-ABL1 itself.61 Of note, BCR-ABL1 shares downstream signaling with many other gain-of-function tyrosine kinases identified in human leukemias, but the effects of these kinases on AID expression have not been described.
Ectopic expression of the B-cell transcription factor PAX5 in myeloid BCR-ABL1 leukemia cells promotes the up-regulation of AID, BLNK, and CD79A, along with surface expression of CD19, suggesting that PAX5 alone can regulate lineage switching from the myeloid to the lymphoid lineage.61 Expression of the myeloid transcription factor C/EBPα in differentiated B cells can override Pax5 and lead to reprogramming into macrophages.62 Lineage switching induced by C/EBPα involves a biphenotypic intermediate, consistent with a process of trans-differentiation, rather than dedifferentiation and redifferentiation (Figure 5).62
CD19-cre/Pax5fl/− mice, which lose Pax5 in CD19+ cells, die within 8 months of birth from aggressive lymphomas that express B220 and Igμ but lack CD19 and other B-cell markers.63 Most of these lymphomas have rearranged Igκ or Igλ and thus underwent B-lineage commitment.63 Strikingly, nonmalignant Pax5-deficient pro-B cells have in vivo self-renewal potential,64 can differentiate into myeloid cells in vivo,65 and reconstitute the T-cell compartment on transplantation into irradiated recipients.63 T cells expanded in vivo from transplanted Pax5-deficient pro-B cells carry oligoclonal heavy and light chain rearrangements and thus derive from committed B cells that underwent B-lineage commitment, dedifferentiation on loss of Pax5, and then redifferentiation into T cells.
Although loss of Pax5 can contribute to B-cell transformation, Pax5 can also serve an essential role in maintaining B-cell phenotype within established lymphoid tumors. Mice transplanted with p53-deficient bone marrow transduced with a Myc transgene develop B-cell lymphomas. When these lymphoma cells are cultured in vitro, a subset will down-regulate Pax5 as well as B-cell surface markers (CD19 and B220) and acquire myeloid morphology and surface markers (CD11b and Emr1).66 When reinjected into congenic recipients, the cells rapidly reexpress Pax5 and form B-lineage lymphomas but switch back to the myeloid phenotype on subsequent culture. Importantly, in vitro differentiation to the myeloid phenotype can be blocked by enforced expression of Pax5.66
Lineage redifferentiation from the lymphoid to the myeloid lineage may also occur in CALM-AF10–rearranged AML.67 Malignant phenotypes associated with CALM-AF10 rearrangements in humans include AML, undifferentiated leukemia, acute lymphoblastic leukemia, and T-cell lymphoma.68 In a mouse model of CALM-AF10–rearranged AML, the bulk of leukemia cells are positive for the myeloid markers Mac1 or Gr1, but the rare tumor-propagating cells express the B-cell marker B220 and lack myeloid markers.68 Both populations harbor the same Igh D-J rearrangements, and a single B220+ cell can give rise to both B220+Mac1− and B220−Mac1+ cells in vitro.68 Therefore, the lymphoid immunophenotype within the tumor-propagating cell is lost from the bulk myeloid leukemia population. A small population of B220+Mac1+ cells are also present and have intermediate tumor-propagating potential,68 supporting a process of trans-differentiation rather than dedifferentiation and redifferentiation (Figure 5).
In other diseases associated with both myeloid and lymphoid malignancies, it remains unclear whether the divergent phenotypes result through dedifferentiation or trans-differentiation. For example, FL, CLL, and MCL can all precede or simultaneously present with histiocytic/dendritic cell neoplasms that share the same BCL2-IGH and/or immunoglobulin rearrangements.51,69,70 Thus, the histiocytic/dendritic cells derive from a cell that underwent V(D)J recombination and then switched to the myeloid lineage. Similar to the murine models outlined in this section, the histiocytic/dendritic cell neoplasms lack expression of PAX5 but express the myeloid transcription factors C/EBPα and PU.1.69,70
Finally, epigenetic alterations may drive the loss of lineage-specific markers without true dedifferentiation (Figure 5). For example, HRS cells have rearranged and hypermutated immunoglobulin genes, indicating their derivation from mature B cells.71,72 Although HRS cell lines (L428, L591, L1236, and KMH2) lack B-cell markers and do not express immunoglobulin, they do express the B-cell transcription factors E2A, EBF1, and/or PAX5.73 The failure to express targets of these transcription factors may result from overexpression of ABF-1 and ID2, which both antagonize E2A.73 Indeed, enforced expression of ABF-1 and ID2 in non-Hodgkin lymphoma cell lines can disrupt E2A homodimers and block the expression of E2A-dependent target genes that include CD19 and CD79A.73 Thus, Hodgkin lymphoma may derive from a hypermutated B cell that undergoes loss of B-cell phenotype but not dedifferentiation.
Conclusions and testable hypotheses
The increasingly exhaustive catalog of lymphoma-associated alterations has created an opportunity to finely map molecular ontogeny across the many subtypes of lymphoma. As we have outlined, a comprehensive accounting of lymphomagenesis within an individual patient could span from HSCs through lineage commitment and include multiple pools of self-renewing precursors. Divergent and convergent clonal evolution within these cells could then drive therapeutic resistance, relapse, and second hematologic malignancies.
The complexity of these processes, as well as the preliminary evidence, suggest that marked differences in the timing and cellular context of lymphoma-associated alterations will exist across patients. Even within the same subtype of lymphoma, differences in lymphoma ontogeny may modulate phenotypes, such as relapse independently of the specific complement of alterations present within the bulk lymphoma population.
Based on the outlined principles, we propose several testable hypotheses:
As mutations are acquired in HSCs over time, lymphoid malignancies common in childhood and early adulthood (eg, Hodgkin lymphoma, BL) will generally harbor fewer tumor-promoting mutations within HSCs (akin to models 1 and 2 in Figure 2) than lymphoid diseases in older adults.
Patients who develop both lymphoid and myeloid malignancies will frequently harbor common mutations in both malignancies.
In a subset of the cases in #2, the mutations will be present in a progenitor with multilineage potential and self-renewal capacity. In a separate subset of cases, alterations within one malignancy will drive lineage switching.
Patients with lymphoid malignancies who harbor mutations in MDS/AML-associated genes within HSCs (eg, TET2) will have a higher likelihood of developing treatment-associated MDS/AML.
Lymphoid tumors with greater clonal diversity among malignant cells will have a greater likelihood of therapeutic resistance.
Similar to #5, lymphoid tumors with greater clonal diversity among premalignant cells at earlier stages of differentiation will have a greater likelihood of developing secondary malignancies.
Specific network incompatibilities (eg, between particular mutations) within differentiating lymphocytes suppress malignant transformation.
Defining the molecular ontogeny of lymphomas, especially those that are currently incurable with conventional agents, is likely to establish new targets for eradicating both malignant and premalignant cells. Effective strategies that block essential stages of lymphomagenesis would shift the focus from disease control to prevention and cure.
Acknowledgments
The authors thank Bjoern Chapuy, Jon Aster, and Michael Lieber for thoughtful comments.
D.M.W. was supported by a Stand Up To Cancer Innovative Research Grant and an American Cancer Society Research Scholar Grant.
Authorship
Contribution: O.W. and D.M.W. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for O.W. is Department of Internal Medicine III, University Hospital Grosshadern, Ludwig-Maximilians University, Munich, Germany.
Correspondence: David M. Weinstock, Dana-Farber Cancer Institute, 450 Brookline Ave, Dana 510B, Boston, MA 02215; e-mail: dweinstock@partners.org.