Abstract
Previous studies have implicated activation-induced cytidine deaminase (AID) in B-cell translocations but have failed to identify any association between their chromosomal breakpoints and known AID target sequences. Analysis of 56 unclustered IgH-CCND1 translocations in mantle cell lymphoma across the ∼ 344-kb bcl-1 breakpoint locus demonstrates that half of the CCND1 breaks are near CpG dinucleotides. Most of these CpG breaks are at CGC motifs, and half of the remaining breaks are near WGCW, both known AID targets. These findings provide the strongest evidence to date that AID initiates chromosomal breaks in translocations that occur in human bone marrow B-cell progenitors. We also identify WGCW breaks at the MYC locus in Burkitt lymphoma translocations and murine IgH-MYC translocations, both of which arise in mature germinal center B cells. Finally, we propose a developmental model to explain the transition from CpG breaks in early human B-cell progenitors to WGCW breaks in later stage B cells.
Introduction
Most human lymphoma translocations, including the t(14;18)/IgH-BCL2 in follicular lymphoma and the t(11;14)/IgH-CCND1 in mantle cell lymphoma (MCL), appear to arise in B-cell progenitors in the bone marrow and to depend on the RAG complex for chromosome breaks at JH and DH segments in the immunoglobulin heavy chain (IgH) locus. Almost nothing is known about the chromosomal breakage process at BCL2, CCND1, or other IgH partner loci because no murine or other animal models currently exist for this type of translocation. Recently, we showed that many such breakpoints are specifically targeted to the dinucleotide sequence CpG,1,2 and we proposed a chromosome breakage model in which activation-induced cytidine deaminase (AID) creates stable T:G mismatches at methylated CpG sites, followed by RAG cutting of repair intermediates.1 Here we analyze a large set of “unclustered” t(11;14) breakpoints (ie, those that fall outside of the CCND1 major translocation cluster [MTC]), and we show that most of these breaks occur at or near known AID hotspots.
Methods
Samples were obtained from the University of Washington Hematopathology Laboratory (22 cases) and the University of Nebraska (10 cases) after approval by the University of Washington Institutional Review Board. t(11;14) breakpoints were mapped using translocation comparative genomic hybridization (translocation CGH).3 Breakpoints were amplified and Sanger sequenced using patient-specific primers (available on request). Statistical methods used to analyze proximity to CpG, and WGCW motifs were described by Tsai et al1 and in supplemental Methods and supplemental Results (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results and discussion
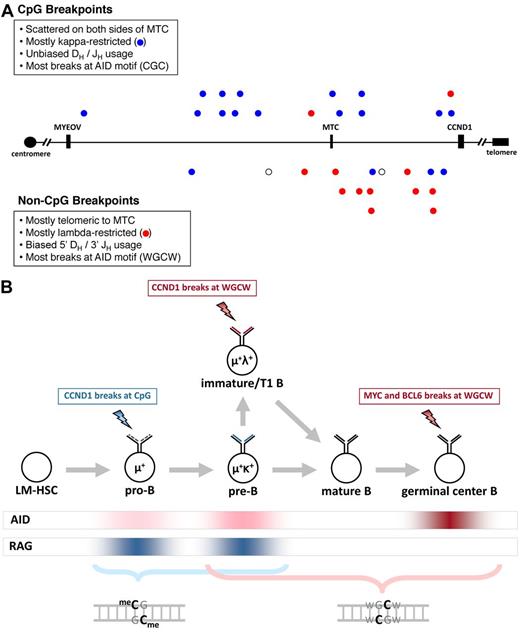
We used a modification of array-based CGH called translocation CGH3 to identify and clone 56 unclustered t(11;14) breakpoints in 32 cases of MCL. A total of 55 of the 56 breakpoints (98%) at the IgH locus on chromosome 14q32 mapped to a JH, DH, or VH gene segment and showed features consistent with RAG-dependent breakage during V(D)J recombination (see Table 1 and supplemental Tables 1-4, and supplemental Figure 2). The breakpoints on chromosome 11q13 span 329 kb of the 344-kb bcl-1 breakpoint region between CCND1 and MYEOV, the closest gene centromeric to CCND1 (Figure 1A). The most centromeric breakpoint was located 12 kb from MYEOV, 331 kb from CCND1, and > 200 kb from the most distant known breakpoint (Table 1).4,5 The most telomeric breakpoint was only 2 kb from the CCND1 gene. Previous studies7-9 have suggested that the majority of t(11;14) breakpoints are telomeric to the MTC, but 12 (37%) of our 32 cases had breakpoints centromeric to the MTC. Nevertheless, the other 20 cases (63%) had breakpoints telomeric to the MTC, significantly more than would be expected if the breakpoints were equally distributed across the entire bcl-1 region (P = .0003, binomial test).
Distribution and features of unclustered CCND1 breakpoints and a model for CpG and WGCW breaks in IgH translocations. (A) A 400-kb segment of chromosome 11q13 (chr11:68 800 000-69 200 000 in NCBI Build 36) is shown. The MYEOV and CCND1 genes and MTC are shown for reference. CpG breakpoints are shown above the axis, and non-CpG breakpoints are shown below. Blue represents κ-restricted cases; red, λ-restricted cases; and ○, cases for which light chain restriction is unknown. (B) Our results implicate AID-initiated chromosomal breaks in the genesis of IgH-CCND1 translocations in MCL, IgH-MYC translocations in Burkitt lymphoma, and IgH-BCL6 translocations in follicular and diffuse large B-cell lymphomas (see supplemental Results). AID expression is highest in antigen-stimulated mature B cells undergoing somatic hypermutation and CSR but is expressed at lower levels in immature and transitional 1 (T1) B cells, and at even lower levels in pro-B and pre-B cells. RAG activity is associated with V(D)J recombination in pro-B cells during IgH rearrangement, in pre-B cells during light chain rearrangement, and in immature B cells during receptor editing. Lightning bolts indicate the stage at which each translocation is proposed to occur. Our model proposes that low levels of AID favor CpG breaks (blue font), whereas intermediate or high levels of AID favor WGCW breaks (red). The staggered cytosine bases on opposite strands of the CpG and WGCW motifs are highlighted to indicate the potential of this configuration to promote DSBs.
Distribution and features of unclustered CCND1 breakpoints and a model for CpG and WGCW breaks in IgH translocations. (A) A 400-kb segment of chromosome 11q13 (chr11:68 800 000-69 200 000 in NCBI Build 36) is shown. The MYEOV and CCND1 genes and MTC are shown for reference. CpG breakpoints are shown above the axis, and non-CpG breakpoints are shown below. Blue represents κ-restricted cases; red, λ-restricted cases; and ○, cases for which light chain restriction is unknown. (B) Our results implicate AID-initiated chromosomal breaks in the genesis of IgH-CCND1 translocations in MCL, IgH-MYC translocations in Burkitt lymphoma, and IgH-BCL6 translocations in follicular and diffuse large B-cell lymphomas (see supplemental Results). AID expression is highest in antigen-stimulated mature B cells undergoing somatic hypermutation and CSR but is expressed at lower levels in immature and transitional 1 (T1) B cells, and at even lower levels in pro-B and pre-B cells. RAG activity is associated with V(D)J recombination in pro-B cells during IgH rearrangement, in pre-B cells during light chain rearrangement, and in immature B cells during receptor editing. Lightning bolts indicate the stage at which each translocation is proposed to occur. Our model proposes that low levels of AID favor CpG breaks (blue font), whereas intermediate or high levels of AID favor WGCW breaks (red). The staggered cytosine bases on opposite strands of the CpG and WGCW motifs are highlighted to indicate the potential of this configuration to promote DSBs.
In 16 of 32 cases (50%), one or both CCND1 breakpoints were ≤ 4 nt from a CpG dinucleotide (referred to below as CpG breakpoints), including 10 cases (31%) with breakpoints directly at a CpG. The majority of the CpG breakpoints (10 of 16, 63%) were located at CGC (Table 1), a known AID hotspot that is distinctive for not conforming to the consensus WRC motif for AID-dependent DNA deamination.10,11 In the remaining 16 cases (50%), the CCND1 breakpoints were ≥ 10 nucleotides (nt) from the closest CpG (“non-CpG” breakpoints), including 5 cases for which der(11) breakpoint sequences were unavailable. CCND1 breakpoints were not associated with any other dinucleotide or RAG motifs (supplemental Table 5A).
The CpG and non-CpG breaks were differentially distributed across the bcl-1 breakpoint region (Figure 1A). The breakpoints in 13 (81%) of the 16 non-CpG cases were telomeric to the MTC, significantly more than the 5 of 16 (32%) expected if equally distributed across the bcl-1 region (P = .00006, binomial test). In contrast, the CpG breakpoints appear to be randomly distributed across both centromeric and telomeric regions (P = .2, binomial). Thus, the overall predominance of telomeric bcl-1 breakpoints in our series can be explained by the non-CpG breaks alone. This differential distribution of CpG and non-CpG breakpoints suggested that these 2 types of breaks are biologically distinct.
Most B-cell lymphomas show a slight preference for immunoglobulin κ over λ light chain restriction, presumably reflecting the sequential rearrangement of the κ locus before the λ locus during normal B-cell development.12,13 To explore the unusual λ restriction bias that has been observed for MCL,14 we compared light chain restriction status to CCND1 breakpoint type and location. Ten (71%) of the 14 non-CpG cases for which light chain data were available (Table 1) were λ-restricted compared with only 2 of 16 CpG cases (12%), indicating a strong association between CpG status and light chain restriction (P = .0022, 2-tailed Fisher exact test).
Non-CpG breakpoints also were significantly more likely than CpG breakpoints to involve the most 3′ JH segment (JH6) and the most 5′ DH segment (DH2-2; supplemental Tables 2 and 4). JH6 was rearranged in 8 of 14 non-CpG cases (57%) but in only 2 of 16 (11%) CpG cases (P = .019 by Fisher exact test). Similarly, DH2-2 was rearranged in 4 of 10 non-CpG cases (40%) compared with 0 of 12 CpG cases (P = .029). Three of the 4 DH2-2 cases had JH6 rearranged on the der(14) chromosome and the remaining case had a DH2-2 signal joint at the der(14) breakpoint (Table 1). Biased usage of 3′ JH segments and 5′ DH segments has been previously identified at t(14;18) breakpoints15 but not for t(11;14) breakpoints at the MTC.16 Taken together, the preferential use of λ light chain and the biased usage of 3′ JH and 5′ DH segments in non-CpG cases suggest that these rearrangements occur at a later stage of B-cell development than CpG breakpoints.
To explore whether AID is involved in generating the non-CpG breaks, we examined the proximity of these breaks to the sequence motif WGCW, where W = A or T. WGCW was recently shown to facilitate AID-dependent double-strand breaks (DSBs) during physiologic class-switch recombination (CSR) by providing 2 antiparallel AID hotspot motifs (WRC, where R = A or G).17 Strikingly, 12 of the 27 non-CpG breaks at CCND1 are within 4 nt of a WGCW motif, making it the second-most frequent break site after CpG (P = .02; supplemental Figure 4; supplemental Table 5B).
Similarly, we analyzed a large database of 156 human and 299 murine IgH-MYC breakpoints and found a striking proximity of MYC breaks to WGCW in both human Burkitt lymphomas (P < .0001) and murine plasmacytomas (P < .000001). Because of the large degree of breakpoint heterogeneity around WGCW at both CCND1 and MYC loci, we included breaks occurring within 4 bp of WGCW (see supplemental Figure 4A-C). Neither CCND1 nor MYC breakpoints were associated with other 4-nt sequence motifs having equivalent degeneracy to WGCW (P ≥ .19), including GWWC, CWWG, SATS, and STAS (S = G or C), each of which contains 2 G:C and 2 A:T base pairs and a central or peripheral palindromic dinucleotide pair (supplemental Table 5B-D).
The proximity of the CCND1 breaks to the known AID motifs CGC and WGCW provides strong and direct evidence that AID cooperates with RAG to initiate IgH-CCND1 translocations in B-cell progenitors. The palindromic nature of CpG and WGCW facilitates chromosome breakage at CCND1 by providing cytosine bases on both strands of the DNA duplex in almost direct opposition to one another (Figure 1B), thereby increasing the likelihood of a DSB.17
To explain why MYC breaks occur at WGCW but not at CpG1 whereas CCND1 breaks can occur at either CpG or WGCW, we propose the following mechanistic and developmental model for IgH translocations (Figure 1B). CpG breaks are favored at the earliest stages of B-cell development when AID is expressed at very low levels18-22 because deamination of methylated CpG gives rise to T:G mismatches that are repaired 2500-fold less efficiently than U:G mismatches at nonmethylated cytosines.1 The T:G mismatches preferentially predispose to chromosome breaks because they persist for much longer than U:G mismatches, even though it is likely that both T:G and U:G lesions arise at very low levels.
MYC breaks occur at WGCW because this motif is favored at the very high level of AID found in germinal center B cells, as indicated by the strong preference for DSBs at WGCW motifs in the IgH locus during physiologic CSR.17 We hypothesize that the non-CpG breaks in MCL might occur in immature/transitional 1 (T1) stage B cells, where AID is expressed at an intermediate level18-22 (Figure 1B), presumably in conjunction with receptor editing.22-24 This would explain their preference for WGCW motifs as well as their DH/JH bias and λ light chain preference, in contrast to the unbiased DH/JH usage and κ predominance in CpG cases (Figure 1A).
Given the strong evidence linking AID to DNA lesions within actively transcribed genes,25,26 it might seem surprising that AID is targeting CpG and WGCW sites that are so far from the CCND1 gene (Figure 1A). However, several recent genome-wide studies of AID-induced mutations and chromosomal breaks in mature murine B cells26-29 clearly show that AID lesions are not limited to genic regions. For example, Klein et al reported that 10%-20% of their AID-dependent translocation hotspots are intergenic.28 Murine models30 and other experimental systems to study chromosomal translocations in B-cell precursors are needed to better understand the mechanisms underlying these most common IgH translocations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.A.G. designed research; H.S.Y. collected data; H.A.G., A.G.T., and M.R.L. analyzed and interpreted data; Z.L. and A.G.T. performed statistical analyses; T.C.G. contributed critical reagents; and H.A.G. and M.R.L. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for A.G.T. is Department of Pathology, Stanford University, Stanford, CA.
Correspondence: Harvey A. Greisman, Department of Laboratory Medicine, University of Washington Medical Center, Box 357110, Seattle, WA 98195; e-mail: greisman@uw.edu.
References
National Institutes of Health