Abstract
The human leukocyte antigen (HLA) genes are candidate genetic susceptibility loci for childhood acute lymphoblastic leukemia (ALL). We examined the effect of HLA-DP genetic variation on risk and evaluated its potential interaction with 4 proxies for early immune modulation, including measures of infectious exposures in infancy (presence of older siblings, daycare attendance, ear infections) and breastfeeding. A total of 585 ALL cases and 848 controls were genotyped at the HLA-DPA1 and DPB1 loci. Because of potential heterogeneity in effect by race/ethnicity, we included only non-Hispanic white (47%) and Hispanic (53%) children and considered these 2 groups separately in the analysis. Logistic regression analyses showed an increased risk of ALL associated with HLA-DPB1*01:01 (odds ratio [OR] = 1.43, 95% CI, 1.01-2.04) with no heterogeneity by Hispanic ethnicity (P = .969). Analyses of DPB1 supertypes showed a marked childhood ALL association with DP1, particularly for high-hyperdiploid ALL (OR = 1.83; 95% CI, 1.20-2.78). Evidence of interaction was found between DP1 and older sibling (P = .036), and between DP1 and breastfeeding (P = .094), with both showing statistically significant DP1 associations within the lower exposure categories only. These findings support an immune mechanism in the etiology of childhood ALL involving the HLA-DPB1 gene in the context of an insufficiently modulated immune system.
Introduction
Evidence from a growing number of studies indicates that exposure to common infections early in life may play a role in the etiology of childhood leukemia.1 However, unlike leukemia in some animals2 and HTLV-associated adult T-cell leukemia,3 studies have not been able to show a direct pathologic involvement of specific infection(s) for leukemia in children.4 It has been suggested that for childhood leukemia, particularly for precursor B-cell acute lymphoblastic leukemia (ALL), a delay in exposure to common childhood infections leaves the immune network undermodulated, and subsequent exposure to infections results in an unfavorable immune response; specifically, a proliferative advantage to preleukemia cells.5 This “delayed infection” hypothesis suggests that exposure to infections or other immune-modulatory factors early in life may result in a protective effect against childhood ALL. Evidence from epidemiologic studies using proxies for early life immune modulation support this hypothesis, such as childhood social contacts increasing the likelihood of infectious transmissions and breastfeeding, but provide limited insight on mechanistic aspects. Host genetic variation within immune response genes may contribute to risk of childhood ALL and may operate in concert with early life exposures that are involved in immune modulation.
Within the MHC region on chromosome 6 reside a large number of genes involved in immunologic processes, including the highly polymorphic human leukocyte antigen (HLA) genes. The cell surface glycoproteins encoded by the classical HLA genes of the class II region, HLA-DP, -DQ, and -DR, are expressed exclusively on antigen-presenting cells (eg, macrophages, B lymphocytes, and dendritic cells) and primarily bind and present endocytosed extracellular proteins to CD4+ T lymphocytes to initiate a targeted immune response.6,7 The known polymorphisms are largely localized to exon 2, the region of the HLA class II gene that determines characteristics of the peptide-binding groove of the HLA molecule and the resulting diversity of antigens that are presented to the immune system.8 Because of their association with the peptide-binding groove, the HLA polymorphic regions have functional relevance and significance for disease susceptibility; > 50 autoimmune and infectious disease associations with HLA alleles have been identified.7,9 There are many examples of HLA associations with neoplasms, including Hodgkin lymphoma,10,11 nasopharyngeal carcinoma,12 gastric cancer,13 invasive cervical carcinoma,14 and childhood leukemia.15-18 Specifically, in the United Kingdom Childhood Cancer Study (UKCCS), Taylor et al in 2002 reported a significantly increased risk of childhood ALL associated with presence of the DPB1*02:01 allele, as well as with other rarer alleles that share antigen binding site pocket profiles with the DPB1*02:01 allotype.18 Further examination of these data as supertypes (a classification based on overlapping peptide binding specificity) showed that childhood ALL risk increased in the presence of DP2 and DP8 supertypes and decreased in the presence of the DP1 supertype.19,20
In the current analysis, we examined the associations of HLA-DPA1 and DPB1 genetic variation, both independently and jointly with 4 separate proxies indicative of early life immune modulation, including markers for infectious exposures in infancy (presence of older siblings, daycare attendance by age 6 months, and reported ear infections in infancy) and breastfeeding. We focused on non-Hispanic white and Hispanic children enrolled in the Northern California Childhood Leukemia Study (NCCLS), which together compose the majority of the study population. We previously reported an ethnic difference in risk for birth order and daycare attendance21,22 and, thus, consider these 2 groups separately in the current analysis. This study is unique in the inclusion of Hispanic children, an assessment of potential contribution of the HLA-DPA1 locus, and the evaluation of gene-environment interaction.
Methods
Study participants
The NCCLS is an ongoing case-control study designed to investigate the etiology of pediatric leukemias. Beginning in 1995, newly diagnosed childhood leukemia cases were ascertained at the time of diagnosis from major pediatric hospitals located in a 17-county San Francisco Bay Area study region, which was expanded in 1999 to 35 counties in Northern and Central California. Comparison with the California Cancer Registry (1997-2003) showed that the NCCLS case ascertainment protocol has captured ∼ 95% of children diagnosed with leukemia in the participating study hospitals. When considering both participating and nonparticipating hospitals within the study region, cases ascertained through the NCCLS represent ∼ 76% of all diagnosed cases. For each eligible case, statewide birth records maintained by the Center for Health Statistics of the California Department of Public Health were used to generate a list of randomly selected controls that matched the case on child's date of birth, sex, Hispanic status (a biologic parent who is Hispanic), and maternal race. Information obtained through the birth certificates and commercially available searching tools were used to trace and enroll 1 or 2 matched controls for each case.
Cases and controls were considered eligible if they were under 15 years of age at date of diagnosis for cases (or corresponding reference date for controls), resided in the study region at the date of diagnosis, had a parent or guardian who spoke either English or Spanish, and had no prior history of malignancy. Approximately 85% of eligible cases consented to participate. Among all eligible controls contacted, 86% consented to participate.23 The overall participation for the control subjects was 59% (the number of enrolled controls divided by the total number of control searches, excluding the known and presumed ineligibles). A detailed description of control selection in the NCCLS is reported elsewhere.23,24 A previous evaluation showed that participating controls in the NCCLS are representative of the sampled population with respect to parental age, parental education, and mother's reproductive history.24
The current analysis included non-Hispanic white and Hispanic childhood ALL case and control subjects recruited between 1995 and 2008 (study phases 1-3), the 2 largest racial/ethnic groups that together compose ∼ 85% of enrolled subjects. The other race/ethnicity groups were not considered because of the small number of subjects. Children were classified as Hispanic if at least 1 biologic parent self-identified as Hispanic. The non-Hispanic white group was composed of children with both biologic parents self-identifying as non-Hispanic white. Children younger than 1 year at diagnosis/reference date were excluded because of growing evidence that these leukemias may be etiologically distinct compared with leukemia diagnosed at later ages.25 This resulted in 669 ALL cases and 977 controls 1 year of age and older who were non-Hispanic white or Hispanic. Among these, only subjects with a DNA sample available at the start of this current project were included resulting in 590 cases (88.2%) and 854 controls (87.4%).
The study protocol was approved by the Institutional Review Boards of University of California, Berkeley and all collaborating institutions, and written informed consent was obtained from all participating subjects. This study was conducted in accordance with the Declaration of Helsinki.
HLA-DPA1/DPB1 genotyping
DNA specimens from buccal cytobrushes were obtained from case and control children either at the hospital (for some cases) or during the in-home personal interview and were extracted using an automated DNA extraction system (AutoGen) and FlexiGene reagents from QIAGEN. Whole genome amplification of buccal cell DNA was performed using GenomePlex reagents (Rubicon Genomics) according to the manufacturer's protocol. Whole genome amplification products were cleaned with a Montage PCR9 filter plate (Millipore). When buccal cytobrush DNA was inadequate or not available (26.6% of subjects), DNA was isolated from dried bloodspots collected at birth and archived at −20°C by the Genetic Diseases Screening Program of the California Department of Public Health. After extraction using the QIAamp DNA Mini Kit (QIAGEN), these DNA samples were whole-genome amplified using REPLI-g reagents (QIAGEN). Regardless of source, DNA specimens were quantified using human-specific Alu-PCR to confirm a minimum level of amplifiable human DNA.26
HLA-DPA1 and DPB1 genotyping was conducted on 1444 unique samples, in addition to 10 sets of Center d'Etude du Polymorphisme Humain family trios and duplicates for 10% of study samples for quality control. We used immobilized sequence-specific oligonucleotide probe strips that were designed to type both HLA-DPA1 and DPB1 loci on the same strip (Roche Molecular Systems). There were 21 probes for DPA1 and 48 probes for DPB1 on these strips. Exon 2 of the DPA1 and DPB1 loci were amplified in separate PCR reactions using biotinylated primer pairs specific for each locus. Subsequent to amplification, 35 μL of each PCR product was heat denatured at 95°C for 5 minutes and allowed to hybridize to the immobilized sequence-specific oligonucleotide probe strip membranes using an automated hybridization/wash instrument, the SLT Profiblot (Tecan Systems). The developed probe patterns on the strips were then scanned, and the scanned images were analyzed using a computer pattern matching program, StripScan Version 5.7.8 (Roche Molecular Systems) with the ImMunoGeneTics/HLA reference alignment Version HLADB-2.10.0-July 2005 (http://www.ebi.ac.uk/imgt/hla/) to determine the genotypes. A resource cataloging all common and well-documented HLA alleles formulated by the American Society for Histocompatibility and Immunogenetics was used to help resolve ambiguities to the lowest value common allele in the genotype determination process.27
No Mendelian errors based on the HLA-DPA1 and DPB1 genotypes were observed for the Center d'Etude du Polymorphisme Humain family trios, and the genotypes between duplicate samples were 100% concordant. Of the 1444 unique study samples genotyped, 11 samples (0.8%) failed to provide an interpretable strip probe hybridization pattern for genotype determination. This resulted in a total of 585 ALL cases (265 non-Hispanic white and 320 Hispanic) and 848 controls (408 non-Hispanic white and 440 Hispanic) available for the analysis. For a large subset of the ALL cases (87%), data on hyperdiploidy and TEL-AML1 chromosomal translocation were available as described in detail previously.28 Subtypes of the cases included 291 common ALL (cALL, defined as CD10+ and CD19+ ALL age 2-5 years), 172 high-hyperdiploid (51-67 chromosomes), 89 positive for the TEL-AML1 chromosomal translocation, and 70 ALL with normal karyotypes.
Data collection
The proxies for early life immune-modulatory exposures considered in this current analysis have been described in detail in previous NCCLS publications.22 Data on child's social contacts inside and outside the home (birth order and daycare attendance), common infections during the first year of life, and breastfeeding were obtained using a standardized questionnaire administered in-person with the parents/guardians of each child.
The child's birth order was determined based on a reproductive history of the biologic mother. Information on the child's social contacts outside the home was obtained through a history of daycare and preschool attendance before the reference date (date of diagnosis for cases and corresponding date for matched controls) or before the age of 6 years, whichever occurred first. For each daycare and/or preschool the child attended, information on age at attendance, duration of attendance, hours per week of attendance, and number of other children present was obtained. The calculation of a quantitative summary measure, called total child-hours of exposure, has been previously described in detail.21 Briefly, child-hours at each daycare facility were calculated as follows: (number of months attending the daycare) × (mean hours per week at this daycare) × (number of other children at this daycare) × (4.35 weeks per month). For each child, the child-hours in each daycare setting were summed to obtain the total child-hours of exposure.
Respondents were asked for a history of common infectious illnesses the child had during the first year of life, including severe diarrhea/vomiting, ear infection, persistent cough, mouth and eye infection, influenza, and unspecified “other infection.” Questions were asked with an emphasis on the timing of exposure, specifically during the age of < 3 months, 3-5 months, and/or 6-12 months.
HLA-DPB1 supertype classification
Although each HLA class II allele encodes a distinct molecule, it has been shown that many of these molecules have overlapping structural and functional features through shared peptide-binding pockets and thus can be clustered into distinct functional groups referred to as supertypes. Twenty-three of the 28 observed DPB1 alleles can be clustered into 1 of 6 supertypes described previously by Taylor et al.19 The 6 supertypes were defined by 3 dimorphic amino acid residues in pockets 6, 4, and 1 at positions β11, β69, and β84, respectively, which have been shown to play significant roles in the binding characteristics of the peptide-binding groove.19,29,30 Three of the supertypes have a positively charged lysine (K) at β69, whereas the other 3 have a negatively charged glutamic acid (E). This is accompanied by either a glycine (G) or leucine (L) at β11 and a G or an aspartic acid (D) at β84. The 6 supertypes have been referred to as DP1 (GKD), DP2 (GEG), DP3 (LKD), DP4 (GKG), DP6 (LED), and DP8 (GED).
Statistical analysis
Python for Population Genetics Version 0.7.0 (Pypop) software was used to test HLA-DPA1 and DPB1 genotype frequencies for deviation from Hardy-Weinberg equilibrium in controls, to estimate HLA-DPA1-DPB1 haplotype frequencies from genotype data of unrelated persons using the expectation-maximization algorithm, and to evaluate the patterns of linkage disequilibrium between the 2 loci.31 Comparison of HLA-DPA1 and DPB1 allele frequencies between cases and controls was performed using the Fisher exact test. A logistic regression model adjusting for child's age and sex was used to calculate odds ratios (ORs) and 95% CIs associated with carrier status of specific HLA-DPA1 and DPB1 alleles (referred to as phenotype) and supertypes. Analyses were conducted among non-Hispanic white and Hispanic children separately; and if no evidence of heterogeneity between the 2 groups based on the χ2 test of homogeneity (P > .05) was present, a pooled analysis was performed with adjustment for race/ethnicity (non-Hispanic white, Hispanic white, and Hispanic other).
The multivariable evaluation of the 4 proxy exposure measures (older sibling, daycare child-hours, ear infection, and breastfeeding) have been reported in previous NCCLS publications,22 and those results served as the basis for the focus on the specific variables chosen for evaluation in the joint analysis. The joint effect between the DP1 supertype and each of the 4 proxy measures was evaluated separately using logistic regression models that included an interaction term representing the product of the supertype (presence or absence) and exposure measure, and additionally adjusting for the other 3 proxy measures, child's age and sex, maternal age, maternal education, annual household income, and study phase. The risk estimate for the interaction term is defined as the ratio of the joint effect of the exposure variable and DP supertype (ORGE) and the product of the individual effects [ORinteraction = ORGE / (ORGE' × ORG'E)]. An ORinteraction significantly different from 1.0 is an indication of multiplicative interaction. P values for the interaction term of < .10 were considered statistically significant.
Results
For both non-Hispanic white and Hispanic children, compared with controls, cases tended to have less annual household income and mothers with less formal education (Table 1). Case-control difference for maternal age was observed only in non-Hispanic white children with mothers of cases appearing younger.
A total of 6 DPA1 alleles were observed in non-Hispanic white and Hispanic subjects. DPA1*01:03 was the most common with a frequency of about 0.80 (Table 2). In contrast to DPA1, DPB1 was considerably more polymorphic. A total of 28 different alleles were observed in the non-Hispanic white and Hispanic populations combined (Table 2). However, only 5 alleles were present at 5% or higher, including DPB1*01:01, *02:01, *03:01, *04:01, and *04:02. Observed genotype frequencies for DPA1 and DPB1 did not differ from what was expected based on Hardy-Weinberg proportions in non-Hispanic white and Hispanic controls.
Strong linkage disequilibrium was present between the 2 loci (D′ = 0.92, Wn = 0.73, P < .001), with only 4 common haplotypes (> 5%) observed (DPA1*01:03-DPB1*02:01, DPA1*01:03-DPB1*03:01, DPA1*01:03- DPB1*04:01, DPA1*01:03- DPB1*04:02). Cases and controls had similar frequencies of common DPA1-DPB1 haplotypes.
In the logistic regression analysis evaluating DPA1 and DPB1 phenotypes and childhood ALL risk, no evidence of heterogeneity in effect between non-Hispanic white and Hispanic children was observed (Table 3). Similarly elevated risk estimates were observed for DPB1*01:01 carriers in both groups, and the pooled analysis showed a statistically significant increased risk of childhood ALL associated with carriers of the allele (OR = 1.43; 95% CI, 1.01-2.04; Table 3).
Previous work has shown more than 90% of the DPB1 alleles can be clustered into 6 supertypes based on similarities in functional (peptide binding) properties.19 The most common supertype, DP4, was present in a large majority of non-Hispanic white (79.2%) and Hispanic (82.5%) control children. The DP1 supertype, which includes DPB1 alleles *01:01, *05:01, and *50:01, showed elevated ORs and a statistically significant increased risk associated with the high-hyperdiploid ALL subtype (OR = 1.83; 95% CI, 1.20-2.78; P = .005; Table 4).
For the evaluation of interaction, we focused on the associated DP1 supertype and 4 proxies for early life immune modulation, including breastfeeding, history of ear infections during the first year of life, and birth order and daycare attendance as indicators for the likelihood of infectious transmissions. This analysis was conducted within a subset of the subjects, including those with complete data for all variables (non-Hispanic whites: 231 cases and 364 controls; Hispanics: 276 cases and 398 controls). Within this subset of subjects, similar to previous NCCLS reports,21,22 having an older sibling (OR = 0.57; 95% CI, 0.40-0.82) and greater daycare child-hours (≥ 2000 vs < 2000: OR = 0.46; 95% CI, 0.25-0.85) was associated with a statistically significant reduced risk of childhood ALL only in non-Hispanic whites and not Hispanic children (Table 5). In contrast, the risk estimate for history of ear infection before 6 months of age was the same in both race/ethnicity groups and the pooled analysis showed a statistically significant reduced risk of childhood ALL (vs no ear infection in the first year: OR = 0.51; 95% CI, 0.31-0.83). Breastfeeding did not show a statistically significant association with childhood ALL risk in either group (Table 5). Based on these main effect results, interaction analyses focused on non-Hispanic whites for the evaluation of older sibling and daycare child-hours and both non-Hispanic white and Hispanic children combined for ear infection and breastfeeding.
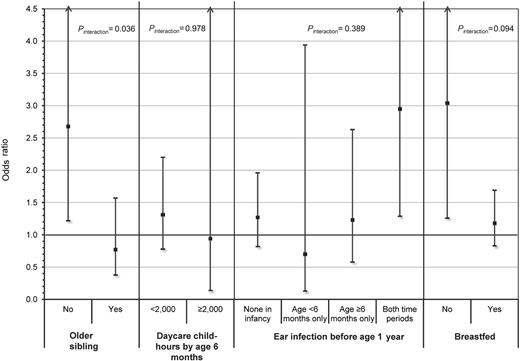
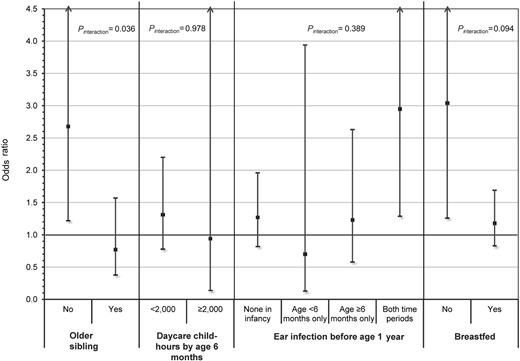
The risk of childhood ALL associated with DP1 supertype differed between children without an older sibling (OR = 2.68; 95% CI, 1.22-5.91) and with an older sibling (OR = 0.77; 95% CI, 0.38-1.57) and showed statistically significant evidence of interaction (ORinteraction = 0.33; 95% CI, 0.12-0.93; P = .036) in non-Hispanic white children (Figure 1). A consistent finding was observed for breastfeeding (ORinteraction = 0.47; 95% CI, 0.20-1.14; P = .094) where the DP1 supertype was associated with an increased risk of ALL among children who did not breastfeed (OR = 3.04; 95% CI, 1.26-7.30), whereas the DP1 supertype was not associated with risk among children who did breastfeed (OR = 1.18; 95% CI, 0.83-1.69). In contrast, daycare child-hours and ear infections in infancy showed little evidence of multiplicative interaction with the DP1 supertype in the risk of childhood ALL (Figure 1), but these analyses had limited statistical power. However, the point estimates were in the direction consistent with the results observed for the older sibling and breastfeeding proxy measures.
Summary plot showing the risk of childhood ALL associated with carriers of the DP1 supertype stratified by levels of select proxy exposure measures for early life immune modulation. ORs and 95% CIs represent the risk of childhood ALL associated with carriers of the DP1 supertype and were calculated using multivariable logistic regression adjusting for age, sex, maternal age, maternal education, annual household income, phase of study enrolled, and all other proxy measures presented. Two-way interactions between DP1 supertype carrier status and each of the 4 proxy measures separately were evaluated using a similar model, which additionally included an interaction term representing the product of the DP1 supertype and proxy exposure measure. Results for the 2 social contact variables, older sibling and daycare child-hours, were based on an analysis conducted in non-Hispanic white children only. The other 2 measures, ear infection and breastfeeding, were analyzed among all subjects because no evidence of heterogeneity was observed by race/ethnicity. Two-sided P values (Pinteraction) of < .10 were considered evidence of a significantly different effect of the DP1 supertype on ALL risk between levels of the proxy exposure measure.
Summary plot showing the risk of childhood ALL associated with carriers of the DP1 supertype stratified by levels of select proxy exposure measures for early life immune modulation. ORs and 95% CIs represent the risk of childhood ALL associated with carriers of the DP1 supertype and were calculated using multivariable logistic regression adjusting for age, sex, maternal age, maternal education, annual household income, phase of study enrolled, and all other proxy measures presented. Two-way interactions between DP1 supertype carrier status and each of the 4 proxy measures separately were evaluated using a similar model, which additionally included an interaction term representing the product of the DP1 supertype and proxy exposure measure. Results for the 2 social contact variables, older sibling and daycare child-hours, were based on an analysis conducted in non-Hispanic white children only. The other 2 measures, ear infection and breastfeeding, were analyzed among all subjects because no evidence of heterogeneity was observed by race/ethnicity. Two-sided P values (Pinteraction) of < .10 were considered evidence of a significantly different effect of the DP1 supertype on ALL risk between levels of the proxy exposure measure.
Discussion
Exposure to common infections and the role of immune-related processes have both emerged as strong candidate risk factors for childhood ALL.5 In this current study, we examined the HLA-DP association in a large non-Hispanic white and Hispanic population of childhood ALL cases and controls in California. The DP1 supertype was associated with childhood ALL; this effect was most prominent in the high-hyperdiploid ALL subtype. Furthermore, we observed a statistically significant interaction between the DP1 supertype and 2 separate measures used as proxies for early life immune-modulatory exposures, having an older sibling and breastfeeding, in the risk of childhood ALL.
In a previous study by Taylor et al, DPB1*02:01 and rarer alleles that share functional properties with the DPB1*02:01 allotypic protein were associated with risk of cALL.18 In addition, DPB1*04:02 was associated with an increased risk, whereas DPB1*01:01 showed a reduced risk. Further investigation of HLA-DPB1 alleles as supertypes indicated a reduced risk associated with DP1 (GKD),19 which appeared to be driven by the DPB1*01:01 allele.20 This effect was most marked for TEL-AML1 and high hyperdiploid-positive ALL.20 A DP1 association in childhood ALL, particularly in high-hyperdiploid ALL, is also present in the NCCLS; however, the risk estimate was in the opposite direction (ie, increased risk) from that of the UKCCS study,20 and the other previously reported DPB1 associations were not observed.
In interpreting this discrepancy, one observation to note is the marked differences in allele frequencies for DPB1*01:01 and DPB1*02:01 between the UKCCS controls and NCCLS non-Hispanic white controls. The DPB1*01:01 allele appears to be less common in the NCCLS non-Hispanic white controls (4.5%) compared with the UKCCS controls (8.0%) but is similar to those reported in other United States white populations (4.5%-6.0%).32-34 Higher frequencies for DPB1*01:01 (more consistent with the UKCCS observations) have been reported in studies of whites residing in Europe, including populations of Germany (8.0%), France (6.6%), and Great Britain (7.5%).35-37 Similarly, the allele frequency of DPB1*02:01, one of the alleles driving the DP2 association observed in the UKCCS, appears to be lower among UKCCS controls (6.0%) versus other studies conducted in United States and European whites (10.0%-15.6%).32-37 Variation in DPB1 allele frequencies between populations is an indication of the potential diversity in underlying genetic structure and that the populations may have evolved under different selective pressures. In adaptive immunity, the HLA class II molecule selectively binds to the antigen and together with the T-cell receptor, forms a specialized complex that activates a cascade of intercellular signals that are specifically designed to elicit a productive immune response.38 The involvement of the HLA molecule as a functional unit in disease susceptibility is therefore dependent on the presence of the immunodominant antigenic peptide.17,39 Accordingly, heterogeneity by geographic region in the prevalence of certain infectious agents or other relevant exposures may potentially contribute to the variability observed.
Studies have shown DPB1*01:01, the principal allele contributing to the DP1 supertype, to be associated with an increased risk of other disease outcomes as well. Earlier case-control studies have found increased risks of both dermatitis herpetiformis and celiac disease associated with DPB1*01:01 in white populations, including those residing in the United Kingdom.40,41 More recently, a large study found that, along with other HLA class II alleles, DPB1*01:01 is associated with an increased susceptibility to sarcoidosis in a United States black population.42
The evaluation of interaction between DP1 supertypes and proxy measures in the risk of childhood ALL showed that the increased risk associated with DP1 is only found in children without an older sibling. A complementary finding was also seen for interaction with breastfeeding, where an increased risk of the DP1 supertype was found only among non-breastfed children. The dichotomous variable indicating presence or absence of an older sibling was used as a proxy measure of exposure to common infections early in life, based on the assumption that contact with older siblings would expose the index child to infections at a very young age. In the context of the “delayed infection” hypothesis, this exposure would, in turn, contribute to immune modulation, a critical component of normal immunologic development.5,43 The immune-modulatory effects of breastfeeding are also well documented.44 Findings of the current analysis, demonstrated by interactions with 2 separate proxy measures, suggest that T-cell recognition of the DP1-peptide complex in the context of an insufficiently modulated immune system may trigger an unfavorable immune response that contributes to leukemogenesis.
Several lines of evidence underscore the importance of immune-modulatory exposures early in life and the effects of exposures on the subsequent development of certain health outcomes, particularly allergic disorders and autoimmune diseases.45 The neonatal immune system is significantly down-regulated and proceeds along a series of postnatal developmental stages, most of which are mediated by environmental exposures during the first year of life. Among the notable characteristics of the neonatal immune system are the presence of high amounts of regulatory T cells and the down-regulated CD4+ T helper 1 (Th1) and T helper 2 (Th2) activities that display a strong skewing toward Th2 responses.46 A critical element of immune maturation is to increase the functional capacity of Th1, which is necessary for effective cell-mediated immunity, antitumor defense, and progression toward a normal immune balance. Various endogenous conditions and environmental exposures, particularly microbial agents as well as breastfeeding, have been shown to promote Th1 capacity and contribute to the process of Th1/Th2 balance during immune maturation.46 Capacity for T regulatory cell activity is also stimulated by early life microbial exposures, which have the ability to enhance the regulatory networks that are responsible for down-regulating immune responses after infection is controlled.47 Taken together, extensive research indicates that proper immune modulation, largely through exposures to infections early in life, may have a significant impact on later life responses to immunologic stimuli.
As described in previous NCCLS publications,21,22 the main effect of birth order and daycare attendance on childhood ALL risk was observed only in non-Hispanic whites, and not in Hispanic children. Observed differences in family structure and daycare attendance patterns between these 2 populations suggested that this differential effect may be the result of heterogeneity in the degree to which these factors serve as appropriate proxies for early life exposure to common infections. Accordingly, evidence of interaction using these surrogate measures was not found in Hispanic children, but the main effect of DP1 does appear to be consistent in both non-Hispanic white and Hispanic children. From a social contacts perspective (indirect measures of infectious exposure), a more refined measure of exposure to infections among Hispanics may be the total number of people living in the household at the time of child's birth, including nonsibling children and adults. However, such measures are not available in the NCCLS. Nevertheless, support for a role of infections in the Hispanic population also exists. History of ear infections by the age of 6 months, a direct measure of exposure, was found to be associated with a reduced risk in both Hispanics and non-Hispanic whites.22 With respect to interaction with DP1 in Hispanics, an effect was found when considering history of breastfeeding as the immune-modulatory exposure. The lack of strong evidence for DP1 interactions with daycare child-hours and ear infection by age 6 months may be the result of limited statistical power. Stratified analyses did show, however, risk estimates pointing in the directions consistent with DP1 interaction effects found for older sibling and breastfeeding.
Our study had sufficient power to detect moderate to strong HLA-DP associations with ALL, but associations within certain subgroup analyses (eg, molecular subtypes) and of rare alleles and supertypes may not have been detectable with the current sample size. Assuming a phenotype frequency of ∼ 0.15 (frequency of carriers of the allele or supertype) and a type I error rate of 0.05, the current analysis had adequate power (> 70%) to detect associations for risk estimates as low as 1.4. Assuming a causal relationship, the functionally driven classification of HLA alleles into supertypes would be expected to improve chances of detecting associations. An increased probability of false-positive findings resulting from multiple testing is a possibility. However, we focused on a previously reported candidate locus, and we approached this unique analysis of gene-environment interactions within the context of a strong a priori hypothesis stemming from an infection-related premise that has been studied over the past 2 decades.1 Although multiallelic, this investigation included primarily 1 genetic locus (HLA-DPA1 and DPB1 are in strong linkage disequilibrium) and the proxy exposure measures were carefully selected based on findings from a previous publication.22 Additional confidence in the findings of the current analysis comes from observing a consistent main effect for the DP1 supertype in both non-Hispanic white and Hispanic children, and consistent evidence of interaction of DP1 observed with 2 different proxy measures related to immune modulation.
The effect of population stratification is probably minimal in the NCCLS because of the careful and detailed account of race and ethnicity obtained from the subjects and statistical adjustment. In addition, the extent of genetic admixture was assessed using a series of 80 ancestry informative markers for a subset of the cases and controls included in this study.48 Estimates of genetic ancestry (percent of European, Amerindian, and African ancestry) were determined with these ancestry informative markers, and comparison of these estimates between cases and controls showed no significant differences (data not shown).48
Potential selection biases resulting in systematic differences between cases and controls should be considered. In the NCCLS, population-based controls are selected from the statewide birth registry among all children born within the study region. A previous methodologic evaluation has shown that controls enrolled in the NCCLS are comparable with “ideal” controls who could have been enrolled under the optimal circumstances (ie, no difficulty in tracing, no refusal of participation).24 In the current analysis, cases and controls appeared to differ in maternal age in non-Hispanic whites, and annual household income in both race/ethnicities. The extent to which these differences are a result of systematic biases is unknown, but the influence of these and other factors was evaluated and addressed accordingly in the multivariable analyses. However, uncontrolled or residual confounding on risk estimates is a possibility.
Although there are numerous studies supporting the importance of early life immune modulation in childhood ALL based on proxy measures, unresolved inconsistencies still exist. The current study demonstrates that concurrent evaluation of host genetic susceptibility factors, together with early life exposures, may be important. Our findings support the conclusion that HLA-DPB1 genetic variation combined with an insufficiently modulated immune system may trigger an adverse immune response that contributes to childhood ALL risk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the clinical collaborators and participating hospitals: University of California Davis, University of California San Francisco, Children's Hospital of Central California, Lucile Packard Children's Hospital, Children's Hospital and Research Center Oakland, Kaiser Permanente Roseville, Kaiser Permanente Santa Clara, Kaiser Permanente San Francisco, and Kaiser Permanente Oakland. The authors also thank the HLA Laboratory staff at the Children's Hospital Research Center Oakland and staff at Roche Molecular Systems for their assistance with the genotyping assays, the Northern California Childhood Leukemia Study staff, the Survey Research Center, and the participating children and their families for their important contributions to this study.
This work was supported by the United States National Institute of Environmental Health Sciences (R01ES09137 and P42ES0470518), the National Cancer Institute (R03CA125823), and the United Kingdom Children with Cancer Foundation (2005/027, 2005/028, 2006/051, and 2006/052).
National Institutes of Health
Authorship
Contribution: K.Y.U., A.P.C., C.M., L.F.B., E.T., and P.A.B. conceived and designed the study; K.Y.U., A.P.C., C.M., X.M., J.L.W., J.K.W., G.V.D., P.M., H.A.E., E.T., and P.A.B. assisted in assembling the data; K.Y.U., A.P.C., C.M., S.S., L.F.B., X.M., J.L.W., J.K.W., M.T., P.B., E.T., and P.A.B. analyzed and interpreted the data; and all authors critically reviewed and edited the manuscript for intellectual content and gave final approval of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kevin Y. Urayama, University of California, Berkeley, School of Public Health, 1995 University Ave, Suite 460, Berkeley, CA 94704; e-mail: kurayama@luke.or.jp.