Abstract
The frequency of A3669G single nucleotide polymorphism (SNP) of human glucocorticoid receptor has been reported increased in polycythemia vera. We investigated the frequency of A3669G SNP and its impact on disease phenotype and progression in 499 patients with primary myelofibrosis (PMF). The distribution of the A3669G allele differed between PMF patients and 2 healthy control populations (odds ratio, 1.6 and 1.8). The variant allele at the homozygous state (G/G) was associated with higher white blood cell count, larger spleen index, and higher frequency of circulating CD34+ cells at diagnosis. The latter association remained significant after correction for the JAK2V617F genotype. In patients JAK2V617F mutated, the G/G genotype was associated with shorter overall survival (77.6 months vs 298 months, P = .049) and blast transformation (BT)–free survival (76.7 months vs 261 months; P = .018). The latter association remained significant after correction for the known BT risk factors, such as age, sex, white blood cell count, percentage of blasts, IPSS prognostic score, and homozygosity for JAK2V617F (hazard ratio = 3.3; P = .006). In conclusion, the glucocorticoid receptor A3669G is a susceptibility allele for PMF: it contributes to confer the phenotype of excess myeloproliferation, and it cooperates with the JAK2V617F mutation in determining BT.
Introduction
Philadelphia negative myeloproliferative neoplasms (MPNs), that is, polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF), are a group of disorders characterized by clonal expansion of hematopoietic stem cells with different phenotypes. Recurrent point mutations, such as JAK2V617F and MPLW515L, mutations in TET2, AXL1, EZH2, SF3B1, IDH1, and IDH2 genes, uniparental disomy, and DNA copy variations are common,1,2 so that the neoplastic process is thought to be initiated by acquired molecular lesions. However, the study of germline variants that could predispose to acquire the mutations or the disease itself, determine the phenotype, or influence the outcomes of MPNs is a growing research field in this area.
The most studied germline genetic variants in MPNs are on the JAK2 gene. Using a candidate gene approach, Pardanani et al identified several JAK2 single-nucleotide polymorphisms (SNPs) exhibiting high linkage disequilibrium that were significantly associated with PV and ET.3 Several independent groups reported that a particular JAK2 haplotype, designed 46/1 or GGCC, was strongly associated with MPN development.4-8 Moreover, Tefferi et al reported that nullozygosity for the haplotype produced a shorter survival in PMF.9 Consistent with these reports, a novel JAK2 locus with a mutated T allele was revealed to contribute significantly to the occurrence of JAK2V617F-positive and -negative MPNs in the Japanese population.10
Two non-JAK2 germline variants were found to be associated with MPNs. Tefferi et al found a differential distribution of CCDC26 glioma-risk alleles in IDH1-mutated myeloid malignancies including MPNs, compared with either their IDH2R140-mutated or IDH-unmutated counterparts.11 Hernández-Boluda et al documented a strong association with leukemia and primary nonmyeloid cancers of the Gln/Gln genotype in the codon 751 of the DNA repair XPD gene in ET and PV patients.12 Other polymorphisms were investigated as host-modifying factors in MPNs, such as TP53 exon 4 codon 72 in PMF13 and MDM2 SNP 309 in members of families with MPN.14 However, in these cases, the allele frequencies were not different from the general population, and the SNPs did not determine different presenting characteristics or survival or predispose to hereditary diseases.
The A3669G (rs6198) SNP in the untranslated region of human glucocorticoid receptor (GR) stabilizes the mRNA of the dominant-negative β isoform of the GR and contributes to development of erythrocytosis in PV, where it has been reported with higher frequency than in normal controls.15 High frequency of the A3669G SNP has been also reported in rheumatoid arthritis,16 in persons predisposed to central adiposity,17 and in Diamond-Blackfan anemia.18 In the present study, we investigated whether the A3669G SNP of the human GR gene is a susceptibility allele for PMF and whether its presence influences the phenotype and the outcomes of the disease.
Methods
Study population
The stored DNA from peripheral blood (PB) granulocytes of 499 patients with PMF consecutively studied at the Center for the Study of Myelofibrosis of the IRCCS Policlinico S. Matteo Foundation in Pavia from 1990 to 2011 was the primary source material of this study. The patients belong to a database of cases diagnosed with PMF according to the World Health Organization (WHO) criteria19 for whom complete clinical information is available with relatively long follow-up. The ethics committee of the IRCCS Policlinico S. Matteo Foundation had approved the written informed consent for PMF patients to donate samples for molecular research on their disease. Before sampling, all patients signed this consent form. This study was conducted in accordance with the Declaration of Helsinki.
The cohort consisted of 291 males (58.3%), and had a median age of 51 years (range, 6-82 years). According with the WHO classification, 118 patients (23.6%) had a diagnosis of prefibrotic myelofibrosis.19 The IPSS risk20 distribution was 60.8% low, 17.4% intermediate 1, 13.7% intermediate 2, and 8.1% high, which reflects a more benign disease than commonly reported.21 JAK2V617F genotyping was done in 481 patients and 295 harbored a JAK2V617F mutation giving a frequency of the mutation of 61.3%; 176 had a heterozygous mutation (36.5%), whereas 119 (24.7%) had a homozygous mutation. Karyotype was obtained in 136 patients and was normal in 97 (71.3%) and abnormal in 39 (28.7%). After a median follow-up of 51 months, 72 patients (14.4%) had died, producing a median survival of 292 months (24.3 years), and 88 patients (17.4%) had documented blast transformation (BT), with a median time to BT of 256 months (21.3 years).
Control populations
To compare allele frequencies of the GR A3669G SNP, we used 2 control populations. One was derived from healthy Italian subjects belonging to the bone marrow donor registry whose samples were made anonymous for the purpose of the study (n = 111). The second population was derived from the Wellcome Trust Case Control Consortium (WTCCC),22 consisting of 2837 controls from England, Scotland, and Wales. The analyzed control samples from the WTCCC were filtered according to the quality control criteria previously reported.22
JAK2 V617F mutation and GR A3669G SNP determination
DNA was isolated from blood granulocytes, obtained by density gradient centrifugation, using the QIAamp DNA Blood Mini Kit (QIAGEN). JAK2V617F mutant allele burden was measured by quantitative RT-PCR, with the CFX96 real-time PCR detection system. The JAK2V617F allele burden was calculated by comparison with standard curves obtained with DNA from a PV patient with 100% allele burden and a normal subject (positive and negative controls, respectively).
The presence of the A3669G SNP (resulting in a TCATT instead of a TCGTT) was determined by high resolution melting (HRM) analysis and then confirmed by direct sequencing. We designed the following primers on GR DNA sequence (NCBI Reference Sequence: NG_009062.1) resulting in a product length of 102 bp; forward primer: 5′CAGACTGTAAAACCTTGTGTGGA3′; reverse primer: 5′CTGCCAATTCGGTACAAATC3′. HRM data were analyzed using Precision Melt Analysis software (Bio-Rad). PCR reactions were carried out in duplicate; in a final reaction volume of 15 μL, we used 30 ng of genomic DNA, 7.5 μL of SsoFast EvaGreen Supermix (Bio-Rad) and 2.5μM of each of the primer. Products were heated to 95°C for 30 seconds and cooled to 60°C to allow heteroduplex formation then, the HRM step consisted of an initial denaturation step at 95° for 2 minutes, followed by a 45-cycle program (denaturation at 95°C for 5 seconds and annealing at 60°C for 5 seconds). HRM data were collected over the range from 65°C to 90°C in 0.2°C increments. Curves for each duplicate were checked on the shape and peak height to meet reproducibility.
To confirm HRM results, samples were subjected to direct sequencing using the same primers used in HRM analysis. To better corroborate the genotypic status of our samples, we also performed a direct sequencing using the following primers that amplified a longer fragment16 : forward primer: 5′AGTGTCTTTTTACCT ACGCA3′; reverse primer: 5′ATGTTTCTCCATATTTGGCA3′. Between 30 and 40 ng of genomic DNA was used as template for each PCR. PCR amplification was done in a Veriti 96-well Thermal Cycler and PCR product was analyzed by agarose gel (2.2%) electrophoresis, finally visualized by Sybr-safe staining. For sequencing, the expected fragment of 173 bp has been purified by QIAquick Gel Extraction Kit (QIAGEN) and ∼ 40 ng of purified DNA has been used in cycle Sequencing reaction, done by BigDye Terminator Version 1.1 Cycle Sequencing Kit (Applied Biosystems). Treated with DyeEx 2.0 Spin Kit (QIAGEN), products were subjected to sequencing analysis by using an automatic sequencer (ABI PRISM 377DNA Sequencer, Applied Biosystems-PerkinElmer Life and Analytical Sciences).
Data analysis and statistics
All statistical analyses considered clinical and laboratory parameters from PMF cohort obtained at diagnosis. The Pearson χ test with 1 degree of freedom was used to compare allele and genotypes frequencies between cases and controls, using the PLINK toolset.23 Deviations from the Hardy-Weinberg equilibrium within cases and controls and in the combined cohorts were tested by the Fisher exact test as implemented in PLINK. The population attributable risk, corresponding to the minor allele was estimated as previously reported.23 Logistic regression was used to assess the ability of the patients' SNP genotypes to predict hematologic and biologic relevant parameters. We examined the following covariates: age, sex, hemoglobin, white blood cell (WBC) count, percentage of blasts in PB, CD34+ cells in PB, IPSS risk categories,20 and JAK2V617F mutational status. Association of genotypes with PMF outcomes was assessed by overall survival (OS) and blast transformation (BT)–free survival. Survival differences, estimated by Kaplan-Meier analysis for patients with different genotypes, were assessed using a log-rank test. Times to an event or death were measured as the time between diagnosis and the event of interest. For censored cases, it represented the time from the diagnosis to date last known alive without an event. The hazard ratio with 95% CI for genetic variants was estimated by Cox regression analysis. Cox regression was also used to estimate multivariable hazard ratio for the variant that remained significant after the replication step and after adjustment for multiple comparisons. The common prognostic factors were included in the model. Because of low number of chromosomal analyses, we did not consider karyotype information in the prediction analysis of disease outcomes. P < .05 was defined the threshold for defining statistically significant associations. Logistic regression, survival analyses, and Cox regression analyses were performed with STATISTICA 9.0 software (StatSoft).
Results
GR A3669G variant distribution between patients and healthy controls
Genotypes and allele frequencies of the GR A3669G SNP in our population of patients with PMF and the 2 control groups are listed in Table 1. GR variant, as tagged by the G allele, occurred significantly more frequently in PMF patients (262 of 998 alleles; 26.2%) compared with 111 healthy volunteers from Italy genotyped at this locus (38 of 222 alleles, 18.2%; odds ratio [OR], 1.62; 95% CI, 1.12-2.34; P = .009), and 2837 healthy donors from United Kingdom genotyped by the WTCCC (917 of 5674 alleles, 16%; OR, 1.84; 95% CI, 1.57-2.15; P < .001). The A/G plus G/G genotype frequency was significantly higher in patients with PMF with respect to local controls (P = .028) and WTCCC controls (P < .001).
To determine whether the minor G allele of the GR A3669G SNP favored the acquisition of JAK2V617F, we analyzed the frequency of GR SNP genotypes and of G allele in patients stratified for the JAK2V617F genotype. Compared with WTCCC healthy controls, A3669G SNP was enriched in both V617F-positive and -negative patients (OR, 1.84 and 1.91, respectively), and the G allele distribution between V617F-positive and -negative PMF patients was not significantly different (26.2% vs 27.0%; P = .8; Table 2). When the analysis was restricted to patients with JAK2V617F allele burden > 50% (homozygous genotype) or < 50% (heterozygous genotype), the frequency of subjects who were homozygous for the mutant allele (G/G) was significantly higher in patients with the V617F homozygous than with the heterozygous genotype (11.6% vs 4.5%; P = .024), and there was a marginally significant increased frequency of the G allele in JAK2V617F homozygous compared with heterozygous genotypes (30.3% vs 23.3%; P = .069).
The GR A3669G genotype frequencies were in Hardy-Weinberg equilibrium both in the PMF patients and healthy controls (P > .05). However, to exclude that G allele homozygosity was generated in vivo by somatic recombination,24 we tested the GR A3669G genotype in nonclonal cells of patients with PMF. We analyzed 12 patients with GG homozygous GR A3669G SNP in the granulocytes; the analysis of buccal mucosa cells revealed that all subjects were germline homozygous carriers of the G polymorphic allele.
GR A3669G polymorphism, clinical and biologic features, and JAK2V617F genotype
Compared with patients with A/A genotype of the GR A3669G SNP, those with G/G genotype had higher WBC count (mean, 8.8 × 109/L vs 12.2 × 109/L; P = .046), and those with A/G or G/G genotype had higher spleen index (151 cm2 vs 172 cm2; P = .040; Table 3). Compared with patients with A/A or A/G genotypes, patients with G/G genotype had higher CD34-positive cells frequency in PB (155 × 106/L vs 46 × 106/L; P = .03).
Because excess myeloproliferation in PMF is known to be also associated with the JAK2V617G genotype,25 we explored how the A3669G SNP and the acquired somatic JAK2V617F mutation contribute to determine the disease phenotype in a bivariate analysis.
We documented by stepwise multiple regression analysis that JAK2V617F mutational status was the only significant determinant of higher WBC count (P < .001) and spleen index (P < .001), whereas homozygous G/G genotype of the A3669G SNP resulted in the only significant determinant of higher frequency of CD34+ cells in PB (P = .008).
GR polymorphism and disease outcomes
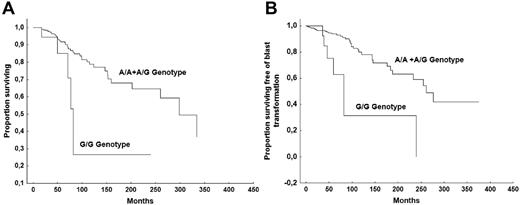
To test whether the GR A3669G variant might affect disease outcomes, we analyzed the risk of incurring into severe anemia (hemoglobin < 10 g/dL), large splenomegaly (spleen extending > 10 cm from the left costal margin), thrombocytopenia (< 150 × 109/L platelet count), leucopenia (< 4 × 109/L WBC count), death for any cause, and BT in patients with different GR A3669G genotypes. None of the analyzed outcomes was significantly different among the 3 genotypes of the SNP. However, when the analysis was restricted to patients bearing the JAK2V617F mutation (n = 295), the OS and BT-free survivals were significantly shorter in patients who were homozygous for the mutant allele G/G (n = 21) with respect to those heterozygous (A/G) or homozygous for the wild-type allele (A/A; n = 274; median survival, 77.6 months vs 298 months, P = .049; and 76.7 months vs 261 months, P < .018, respectively; Figure 1). In multivariable analyses, including age, sex, hemoglobin, WBC count, IPSS prognostic groups, percentage of blasts in PB, CD34+ cell frequency in PB, and the JAK2V617F homozygous genotype, the homozygosity for the minor GR A3669G allele G/G was not identified as an independent factor for unfavorable OS, but it was identified as an unfavorable factor for BT (Table 4). Patients who bore JAK2V617F mutation and who were either homozygous for the wild-type allele (A/A) or heterozygous (A/G; n = 273) developed 36 events (13.2%), with an event rate of 0.47 per 100 person-years, whereas patients with a G/G genotype (n = 21) had 7 events (33.3%) with an event rate of 13.6 per 100 person-years.
Comparison of survival. OS (A) and BT-free survival (B) in patients who are JAK2V617F mutated and who bear the G/G genotype of the glucocorticoid receptor A3669G polymorphism versus those who bear the A/G or A/A genotype.
Comparison of survival. OS (A) and BT-free survival (B) in patients who are JAK2V617F mutated and who bear the G/G genotype of the glucocorticoid receptor A3669G polymorphism versus those who bear the A/G or A/A genotype.
Discussion
Gene polymorphisms have been recently studied to reveal associations of germline variants with occurrence, phenotypes, and outcomes of MPNs.3-14 In this study, we focused on the A3669G (rs6198) polymorphism of the GR gene in PMF because its frequency has been reported to be increased in PV patients, and the variant has been documented to be able to induce an excess proliferative signaling in hematopoietic cells.15 With a large cohort of patients with WHO-classified PMF, in this study we provide evidence that the A3669G SNP represents a host predisposition factor for PMF. With respect to the risk for the development of JAK2V617F-associated MPNs conferred by the JAK2 haplotype 46/1, the GR SNP is a minor risk factor. Indeed, in our PMF population, the ORs with respect to healthy populations were 1.6 and 1.8, in contrast to 3.7 of JAK2 46/1, and the population attributable risk estimated over the set of 499 PMF patients versus the combined set of 2948 controls was 9.1% in contrast to 28%.4 The overall strength of association between PMF and the polymorphic variant resulted independent from the JAK2V617F mutation. Nonetheless, we documented that patients who were homozygous for the G variant were highly represented in the population with the V617F homozygous genotype. These results may be interpreted according to the hypothesis that GR A6639G polymorphism produces a host genetic fertile ground for the development of PMF and that, at the homozygous state, the minor G allele contributes to uniparental disomy of chromosome 9 that gives rise to homozygous JAK2V617F mutants in PMF.
We found that the GR A3669G variant has a role in determining the disease phenotype. In particular, minor allele homozygous patients (G/G) had at diagnosis higher WBC count, larger splenomegaly, and higher number of CD34+ cells in PB, all characteristics representative of excess myeloproliferation. Because there is evidence that JAK2V617F mutation, in particular at the homozygous state, is responsible for hyperproliferation in PMF,25 we endeavored to dissect the contribution of the 2 genetic backgrounds to the disease phenotype. At multivariable analysis, only excess mobilization of CD34+ cells in PB resulted to independently associate with the G/G genotype, indicating the complex interplay of the 2 genetic determinants on disease presentation.
We further documented an unfavorable influence of the G/G genotype on OS and risk of BT. For both outcomes, the influence of G variant was exploited only in the presence of JAK2V617F mutation. At the multivariable analysis, the influence of G/G genotype on survival was nullified by the influence of known prognostic markers of the disease, such as older age, anemia, and high percentage of blasts in PB, whereas an independent role of G/G genotype in favoring BT was consistently evidenced. In the category of patients with the JAK2V617F mutation, G/G genotype produced a 3.3 times higher risk of BT than the A/A or A/G counterpart.
Biologic plausibility supports the minor allele of the GR A3669G SNP as a determinant of the hyperproliferative phenotype and as a predisposing factor for BT in patients with PMF. Varricchio et al have suggested that the polymorphic allele stabilizes GRβ mRNA, thus preventing the creation of complexes between STAT-5 and GRα with the consequent overexpression of the transcription factor NFE2,15 a crucial factor for erythromyeloid lineage differentiation.26-28 However, GR regulates the expression of a high number of genes, some of them involved in the cell's response to environmental factors as the stem cell derived factor-1 receptor CXC chemokine receptor 4.29 The blunting of the GR activity by overexpression of the β-chain could cooperate with the epigenetic silencing in lowering the CXC chemokine receptor 4 expression on CD34+ cells, a mechanism responsible for hemopoietic stem cell mobilization.30,31 However, although our results would suggest that A3669G has a functional role in myeloproliferation, other potential functional variants in linkage disequilibrium with A3669G may be present. Thus, further mechanistic studies are needed to clarify the precise role of A3669G in the susceptibility of PMF.
The results of this study deserve to be discussed first from the perspective of PMF epidemiology. The GR A3669G variant is differently distributed among different populations, with an allele frequency ranging from 4% in Sub-Saharan Africans and 20% in Europeans.32 Thus, one could hypothesize that the prevalence diversity of the host predisposing polymorphic allele could cause prevalence diversity of PMF in different ethnic groups and could determine differences in disease phenotype among them. If this would be demonstrated, it would justify the great differences of presentations among different series of PMF patients, and different results on the role JAK2V617F mutation exerts on BT as we have recently pointed out.21 Moreover, the GR A3669G allele may be hypothesized to be a candidate predisposition variant for the study of familiar cases of MPN. Our results finally indicate GR A3669G variant as a new predisposition marker for BT that can be prospectively used in association with the known predictors of BT in the prognostic assessment of PMF patients. In particular, the assay of this polymorphism would focus on the assessment of BT risk in patients bearing the JAK2V617F mutation: patients identified with the highest risk of BT could benefit from close surveillance and individualized therapeutic approaches.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC, Milano) Special Program Molecular Clinical Oncology 5X1000 to AIRC-Gruppo Italiano Malattie Mieloproliferative and AIRC (code 10413). A detailed description of the AIRC-Gruppo Italiano Malattie Mieloproliferative project is available at http://www.progettoagimm.it. This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data are available from www.wtcc.org.uk. Funding for the project was provided by the Wellcome Trust (award 076113).
Authorship
Contribution: G.B. designed the study, collected and registered cases on the database, analyzed the data, and wrote the manuscript; V.P. contributed to the GR A3669G SNP determination; V.R. designed the study, collected cases, and wrote the manuscript; L.V. contributed to the JAK2 V617F mutation detection by allele-specific PCR; A.C. performed statistical analysis; A.M. performed genetic statistical analysis; P.C. performed the quantitative PCR assay for JAK2 V61F mutation; R.C. and M. Massa designed the study and wrote the manuscript; G.V. contributed with CD34+ cell assay in PB; A.R.M. designed the study and interpreted data; and M. Martinetti provided samples of local healthy controls.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Giovanni Barosi, Unità di Epidemiologia Clinica e Centro per lo Studio della Mielofibrosi, Fondazione IRCCS Policlinico S. Matteo, Viale Golgi 19, 27100 Pavia, Italy; e-mail: barosig@smatteo.pv.it.