Abstract
Explorative genome-wide next-generation sequencing of leukaemias and lymphomas has revealed a wide spectrum of acquired mutations and considerable tumour heterogeneity that might be responsible for disease initiation, resistance to treatments and relapse. There is, therefore, a clinical need to identify these genetic abnormalities in a diagnostic setting.
Here, we present the development and validation of a targeted next generation mutation analysis tool. To compare the distribution pattern of genetic abnormalities in chronic lymphocytic leukemia (CLL), we performed targeted deep sequencing on CLL samples using a TruSeq custom designed targeted amplicon assay (TSCA, Illumina). We reveal differential mutation distribution patterns depending on clinical CLL subgroups.
The TSCA panel was designed to amplify 21 genes (table 1) with known or suspected links to either the development of CLL or as response predictors, including TP53, SF3B1 (Puente, Nature, 2011; Quesada et al, 2012) and NOTCH1 (Rossi, Blood, 2012). Where genes have known mutational hotspots in CLL, only these regions were included in our panel, for example exons 5–8 of TP53. For genes such as MAP2K1, where mutations are distributed throughout the coding region, every exon was targeted. In total, we were able to design an amplicon panel able to cover 99% of our desired 36,035bp target region.
List of genes included in CLL custom amplicon panel
| ASXL1 | ATM | CHD2 | DDX3X | FBXW7 | HMCN1 | IRF4 |
| KLHL6 | LRP1B | MAP2K1 | MAPK1 | MED12 | NOTCH1 | PCLO |
| POT1 | SAMHD1 | SF3B1 | TP53 | XPO1 | ZFPM2 | ZMYM3 |
| ASXL1 | ATM | CHD2 | DDX3X | FBXW7 | HMCN1 | IRF4 |
| KLHL6 | LRP1B | MAP2K1 | MAPK1 | MED12 | NOTCH1 | PCLO |
| POT1 | SAMHD1 | SF3B1 | TP53 | XPO1 | ZFPM2 | ZMYM3 |
In order to validate our approach, we used samples previously subjected to whole genome sequencing as controls. Of the 13 individual mutations in the control cohort, we were successfully able to detect 10 (77%) with our custom assay to an average depth of 1380x. A 19bp deletion in TP53 failed to be picked up by the variant calling software, and 2 point mutations in ATM were not detected due to the targeted nature of the assay. There was a single false positive mutation across all samples in ZFPM2, caused by a sequencing error in a homopolymer region.
The sample group consisted of 45 representative CLL cases, split into two cohorts. The first cohort consisted of 11 cases that have yet to receive any treatment, whilst the second cohort comprised 34 relapsed/refractory cases. Analysis of further samples is in progress.
We performed library preparation according to the manufacturers instructions. Each sample was dual indexed with two 8bp “barcodes” prior to equimolar pooling, and the final pooled library was processed on an Illumina MiSeq instrument using the TruSeq 2×150bp paired end sequencing protocol. The run produced 1.6Gb of passed filter sequence data, with 92.8% of above the quality threshold of Q30. The average depth of coverage across all samples was 849x.
Primary analysis of the sequencing data was performed using the cloud based data analysis package from Illumina, which carried out the alignment and variant calling. A conservative quality score threshold of >99 was set, with all variants above this carried forward for further analysis.
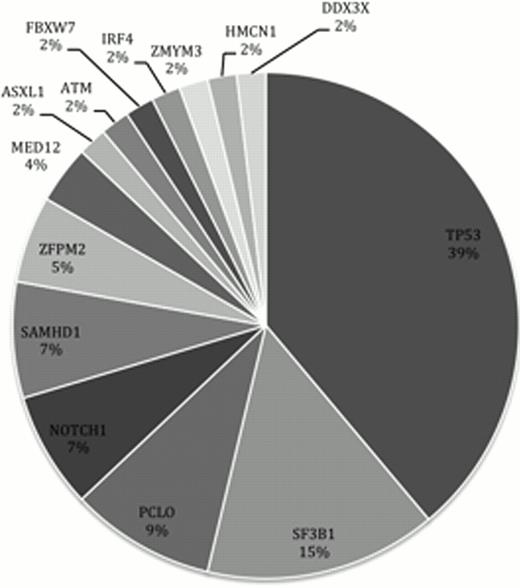
Our custom amplicon panel detected mutations in 35 of the samples, comprising 8 indels and 45 point mutations. Of the 54 mutations, 40 were missense, 8 were frame-shifts, 1 was a nonsense mutation and 5 are predicted to have functional effects on splicing domains. The most frequently mutated gene was TP53, followed by SF3B1, PCLO and NOTCH1 (figure 1).
Frequency of genes with somatic mutations in our CLL cohort.
Frequency of genes with somatic mutations in our CLL cohort.
Importantly, there was good correlation between mutation allele frequencies from whole genome sequencing, targeted deep sequencing and TSCA, demonstrating that the high sensitivity of large-scale genome sequencers can be reliably applied in a diagnostic setting.
We describe mutation hotspots and mutation distribution patterns and link them to clinical behaviour. For example: SF3B1 mutations occurred in 15% of patients and were linked to reduced progression free survival.
In conclusion, our technique allows for rapid mutation detection of the most frequently mutated genes in CLL. Further refinements in amplicon design and variant calling will lead to added precision. TSCA design and validation for other haematological diseases is in progress.
No relevant conflicts of interest to declare.

This icon denotes a clinically relevant abstract
Author notes
Asterisk with author names denotes non-ASH members.