Abstract
The Sox4 transcription factor mediates early B-cell differentiation. Compared with normal pre-B cells, SOX4 promoter regions in Ph+ ALL cells are significantly hypomethylated. Loss and gain-of-function experiments identified Sox4 as a critical activator of PI3K/AKT and MAPK signaling in ALL cells. ChIP experiments confirmed that SOX4 binds to and transcriptionally activates promoters of multiple components within the PI3K/AKT and MAPK signaling pathways. Cre-mediated deletion of Sox4 had little effect on normal pre-B cells but compromised proliferation and viability of leukemia cells, which was rescued by BCL2L1 and constitutively active AKT and p110 PI3K. Consistent with these findings, high levels of SOX4 expression in ALL cells at the time of diagnosis predicted poor outcome in a pediatric clinical trial (COG P9906). Collectively, these studies identify SOX4 as a central mediator of oncogenic PI3K/AKT and MAPK signaling in ALL.
Key Points
Sox4 is required for survival, progression, and proliferation in pre-B acute lymphoblastic leukemia.
Sox4 activates critical survival signaling pathways and correlates with poor clinical outcome.
Introduction
The Sox4 (SRY-related HMG-box) transcription factor is expressed in early B- and T-cell development similar to LEF1.1 In the absence of Sox4, B-cell development is arrested at the pro-B to pre-B cell transition.2 In Sox4−/− mice, pro-B cells fail to proliferate in response to IL7 and to expand and differentiate past the pre-B cell receptor checkpoint.2 Interestingly, SOX4 functions as transcription factor yet closely interacts with membrane-proximal cytokine receptor signaling.3 The PDZ domain-containing adaptor protein syntenin (SDCBP) recruits the Sox4 protein directly to the cytoplasmic tail of the IL5Rα chain.3 With regard to its receptor-proximal activation, Sox4 resembles the SMAD and STAT transcription factors. In addition to its IL5Rα interaction, Sox4/syntenin associates with the cytoplasmic tails of other transmembrane receptors, including syndecans and ephrins.4,5 Because IL5Rα signaling is dispensable for early B-cell development, the receptor system that recruits syntenin-mediated activation of Sox4 in early B-cell development remains to be identified. A role for Sox4 in acute myeloid leukemia was proposed based on the identification of viral insertions, activating Sox4 expression in leukemias developing in mice carrying endogenous retroviruses.6 Overexpression of Sox4 was also found to block differentiation of myeloid progenitor cells.7 Importantly, Sox4 overexpression causes myeloid leukemia6 and thereby cooperates with haplo-insufficiency of the myeloid differentiation factor PU.1.8 In acute lymphoblastic leukemia (ALL), we found SOX4 was strongly up-regulated on tyrosine kinase inhibitor (TKI) treatment and high expression of SOX4 correlates with poor clinical outcome of patients with ALL. However, the role of Sox4 in ALL cells remains unclear. In this study, using a conditional Sox4 knockout mouse model, we showed that the deletion of Sox4 compromised proliferation and viability of leukemia cells and that SOX4 plays as a central mediator of oncogenic PI3K/AKT and MAPK signaling in ALL.
Methods
Pre-B and leukemia cell culture
The work described here involves animal experiments (approved by Children's Hospital Los Angeles Institutional Animal Care and Use Committee) and analysis of clinical data (No Human Subjects, Exempt No. 4; IRB approval under Children's Oncology Group (COG) and Eastern Cooperative Oncology Group study protocols). Primary human leukemia cells were cultured on OP9 stroma cells in α minimum essential medium without ribonucleotides and deoxyribonucleotides (MEMα; Invitrogen), supplemented with 20% FBS, 2mM l-glutamine, 1mM sodium pyruvate, 100 IU/mL penicillin and 100 mg/mL streptomycin. Human ALL cell lines were maintained in RPMI with GlutaMAX (Invitrogen) containing 20% FBS, 100 IU/mL penicillin, and 100 mg/mL streptomycin. Mouse BCR-ABL1 transformed pre-B cells were cultured in IMDM (Invitrogen) with GlutaMAX containing 10% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin, and 50μM β-mercaptoethanol. Normal mouse pre-B cells were cultured in the presence of 10 ng/mL IL7.
Colony forming assay
The methylcellulose colony-forming assays were performed with 10 000 BCR-ABL1–transformed mouse pre-B ALL cells. Cells were resuspended in mouse MethoCult medium (StemCell Technologies) and cultured on 3-cm diameter dishes, with an extra water supply dish to prevent evaporation. After 7 to 22 days, colony numbers were counted.
Quantitative RT-PCR
Total RNA from cells was extracted using RNeasy isolation kit from QIAGEN. cDNA was generated using a poly(dT) oligonucleotide and the SuperScript III Reverse Transcriptase (Invitrogen). Quantitative real-time PCR was performed with the SYBRGreenER mix (Invitrogen) and the ABI7900HT real-time PCR system (Applied Biosystems) according to standard PCR conditions. Primers for quantitative RT-PCR are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Single-locus quantitative ChIP and ChIP-on-chip analysis
ChIP assays were performed with modifications as described.9 Briefly, 1 × 107 human pre-B cells were cross-linked with 1% formaldehyde. After sonication by a Q700 (Qsonica), immunoprecipitations were performed using 5 μg Sox4 or control IgG antibody. Complexes were washed with low and high salt buffers, eluted, and reverse-crosslinked, and the DNA was precipitated. Immunoprecipitated DNA sequences were analyzed by qPCR (Antibodies and primer sequences used for qChIP analyses are listed in supplemental Tables 2C and 1, respectively). TEAD2 and SPI1 were used as a negative control, whereas LCK and an up-stream regulatory of SPI1 were used as a positive control.8,10 SOX4 ChIP-on-Chip was performed in the prostate cancer cell line (LNCaP) in triplicates for SOX4-YFP and in duplicates for YFP empty vector control (GSE11874).11 Briefly, the Sox4 ChIP and input from each sample were Cy3/Cy5 fluorescent labeled and cohybridized to NimbleGen 25K human promoter array set (2 microarrays that tile 4000 bp upstream and 750 bp downstream of the transcription start site of a gene promoter on average). Raw hybridization data were Z-score normalized, and ratios of ChIP to input DNA were determined for each sample. In the enrichment plots, the ratios of ChIP versus Input in the gene promoter regions were shown for the 3 replicates of Sox4 and for the 2 replicates of control, respectively, and the horizontal lines represent the 95th percentile of the ratios of ChIP to input for Sox4 and control samples, respectively.
Western blotting
Cells were lysed in CelLytic buffer (Sigma-Aldrich) supplemented with 1% protease inhibitor cocktail (Pierce), 1% phosphatase inhibitor cocktail (Calbiochem), and 1mM PMSF. Protein samples were loaded on NuPAGE (Invitrogen) 4% to 12% Bis-Tris gradient gels and transferred on polyvinylidene difluoride membranes (Invitrogen). For the detection of mouse and human proteins by Western blot, primary antibodies were used together with the WesternBreeze immunodetection system (Invitrogen). The antibodies used for Western blotting are listed in supplemental Table 2B.
Flow cytometry
Approximately 1 million cells/sample were resuspended in PBS blocked using Fc blocker for 10 minutes on ice, followed by staining with the appropriate dilution of the antibodies for 15 minutes on ice. Cells were washed and resuspended in PBS with PI (0.2 μg/mL) as a dead cell marker. The antibodies used for flow cytometry are listed in supplemental Table 2. For annexin V staining, annexin V binding buffer (BD Bioscience) was used instead of PBS and 7AAD (BD Bioscience) instead of PI. PE labeled annexin V was purchased from BD Bioscience. For BrdU staining, the BrdU Flow Kit was purchased from BD Bioscience and performed according to the manufacturer's protocol.
Rescue assays, PI3K, AKT, and BCL2L1
BCR-ABL1 transformed murine Sox4fl/fl pre-B ALL cells were transduced with inducible Cre-puromycin or an empty vector control. Deletion of Sox4 in Sox4fl/fl pre-B ALL cells was induced with 4-hydroxy-tamoxifen (4-OHT) for 2 days and cells were transduced with the retroviral vector to express a molecule of interest, myr-AKT, myr-p110α subunit of PI3K or BCL2L1 (Bcl-xL) tagged to GFP. Percentage of viable cells was measured on day 2. The percentage of viable transduced cells normalized to EV control viability measurement (set as 100%).
In vivo leukemia cell transplantation
BCR-ABL1 transformed murine Sox4fl/fl pre-B ALL cells were transduced with inducible Cre-puromycin or an empty vector control. Puromycin-resistant cells were selected 2 days after transduction and 1 × 106 leukemia cells were injected into sublethally irradiated (250 cGy) NOD/SCID mice. Seven mice per group were injected via tail vein injection. When a mouse became terminally sick, it was killed, and bone marrow and spleen were collected for flow cytometry analysis.
Patient outcome and gene expression microarray data
Patient outcome and RMA normalized gene expression microarray data were obtained from the National Cancer Institute TARGET Data Matrix (http://target.nci.nih.gov/dataMatrix/TARGET_DataMatrix.html) of the COG Clinical Trial P9906 (GSE11877)12 and from the German Berlin-Frankfurt-Münster (BFM) Clinical Trial ALL-REZ BFM 2002 (GSE4698).13 The end point of the clinical data are overall survival (OS) and risk stratification, which were determined by a combination of 3 prognostic factors: intermediate risk, S2; and high risk, S3/S4.
Survival analysis
Kaplan-Meier survival analysis was used to estimate overall survival (OS). Basically, patients with B-ALL (COG clinical trial, P9906, n = 207) were segregated into 2 groups according whether they had more than or less than the median expression level of the SOX4 probe-set. Log rank test was used to compare survival differences between patient groups. R package “survival” Version 2.35-8 was used for the survival analysis and Cox proportional hazards regression model in R package for the multivariate analysis (R Development Core Team, 2009).
Results
Sox4 function in BCR-ABL1 ALL
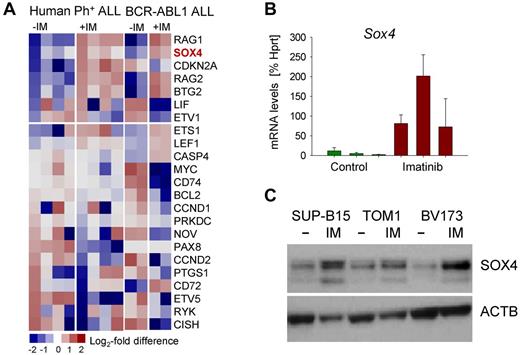
Tyrosine kinase inhibitor (TKI) treatment of Ph+ ALL cells results in strong up-regulation of BCL6, which then enables Ph+ ALL cells to survive TKI treatment in a state of dormancy and quiescence.14,15 For this reason, we examined other TKI-induced genes for a potential role in leukemia cell survival and TKI-resistance. SOX4 gene expression was strongly up-regulated in human Ph+ and murine BCR-ABL1 driven pre-B ALL by TKI-treatment as verified by qRT-PCR and Western blot (Figure 1). To test Sox4 function in normal pre-B cells and a mouse-model for Ph+ ALL, we obtained a conditional knockout mouse model for Sox4 and isolated bone marrow B cell precursors from Sox4fl/fl mice16 for culture in the presence of IL7 (pre-B cells) or transformation with BCR-ABL1 (model for Ph+ ALL). To test the function of Sox4 in a genetic model, we induced Sox4-deletion by 4-OHT inducible activation of Cre for 2 days (supplemental Figure 1). Because previous work demonstrated that loss of Sox4 results in a profound block at the pre-B cell checkpoint,2 we also studied Sox4 function in normal Sox4fl/fl pre-B cells. Loss of Sox4 in normal pre-B cells had only mild effects on viability and the ability of pre-B cells to differentiate (Figure 2A top). Withdrawal of IL7 from normal pre-B cells strongly induces differentiation and Ig light chain gene rearrangement.17 To examine whether Sox4 has a measurable effect on early B-cell differentiation, we overexpressed Sox4 in the presence and absence of IL7. However, gain of Sox4 function did neither affect IL7-responsiveness of normal pro and pre-B cells, nor the ability of large cycling pre-B cells (Fraction C' according to Hardy nomenclature)18 to proceed to small resting pre-B-cell stages (Fraction D) and to subsequently rearrange Vκ-Jκ light chain gene rearrangements (supplemental Figure 2). Compared with normal pre-B cells, Sox4 ablation in BCR-ABL1 driven pre-B ALL had a severe effect on viability and apoptosis (Figure 2A bottom). Furthermore, Cre-mediated deletion of Sox4 in B cell precursor ALL (supplemental Figure 1) resulted in G0/G1 arrest of BCR-ABL1 ALL cells (Figure 2B). To elucidate the function of Sox4 on colony forming ability, we studied the effect of Sox4 deletion in methylcellulose serial replating assays. Absence of Sox4 leads to loss of self-renewal capacity and failure to form colonies in 3 serial replatings (Figure 2C).
Up-regulation of SOX4 in response to TKI treatment in Ph+ ALL. Ph+ ALL (BV173, SUP-B15, TOM1, and NALM1; supplemental Table 6) and murine pre-B ALL were treated with the tyrosine kinase inhibitor (TKI) Imatinib for 16 hours (IM; A). Based on gene expression analyses we identified SOX4 as a gene that is highly up-regulated. BCR-ABL1–transformed mouse pre-B ALL cells were treated with or without Imatinib (IM; 2μM) for 16 hours and mRNA levels of SOX4 were measured by quantitative RT-PCR (B). In panel C, 3 human Ph+ ALL cell lines were treated with or without Imatinib (IM; 2μM) for 16 hours and SOX4 protein levels were assayed by Western blot using β-actin as loading control.
Up-regulation of SOX4 in response to TKI treatment in Ph+ ALL. Ph+ ALL (BV173, SUP-B15, TOM1, and NALM1; supplemental Table 6) and murine pre-B ALL were treated with the tyrosine kinase inhibitor (TKI) Imatinib for 16 hours (IM; A). Based on gene expression analyses we identified SOX4 as a gene that is highly up-regulated. BCR-ABL1–transformed mouse pre-B ALL cells were treated with or without Imatinib (IM; 2μM) for 16 hours and mRNA levels of SOX4 were measured by quantitative RT-PCR (B). In panel C, 3 human Ph+ ALL cell lines were treated with or without Imatinib (IM; 2μM) for 16 hours and SOX4 protein levels were assayed by Western blot using β-actin as loading control.
Sox4 is required for pre-B ALL cell survival and proliferation. Effects of inducible activation of Cre or empty vector (EV) control in Sox4fl/fl pre-B ALL cells was studied on viability/apoptosis (A), cell cycle regulation (B), and colony formation in methylcellulose (C). In panel A, Sox4fl/fl was deleted in IL7-dependent normal pre-B cells (top) and BCR-ABL1–transformed ALL cells and viability changes were measured in both after 2 days of induction of Cre. In panel B, cells were stained with BrdU and 7AAD after 1 day of Sox4 deletion and percentages of cells in G0/1, S and G2/M phases of the cell cycle are indicated (n = 3; P = .008 for S phase) and shown in a bar graph (bottom). In panel C images for colonies in a serial replating assay are shown and quantitative analysis (right; n = 3).
Sox4 is required for pre-B ALL cell survival and proliferation. Effects of inducible activation of Cre or empty vector (EV) control in Sox4fl/fl pre-B ALL cells was studied on viability/apoptosis (A), cell cycle regulation (B), and colony formation in methylcellulose (C). In panel A, Sox4fl/fl was deleted in IL7-dependent normal pre-B cells (top) and BCR-ABL1–transformed ALL cells and viability changes were measured in both after 2 days of induction of Cre. In panel B, cells were stained with BrdU and 7AAD after 1 day of Sox4 deletion and percentages of cells in G0/1, S and G2/M phases of the cell cycle are indicated (n = 3; P = .008 for S phase) and shown in a bar graph (bottom). In panel C images for colonies in a serial replating assay are shown and quantitative analysis (right; n = 3).
Mechanism of Sox4-mediated survival signaling in ALL
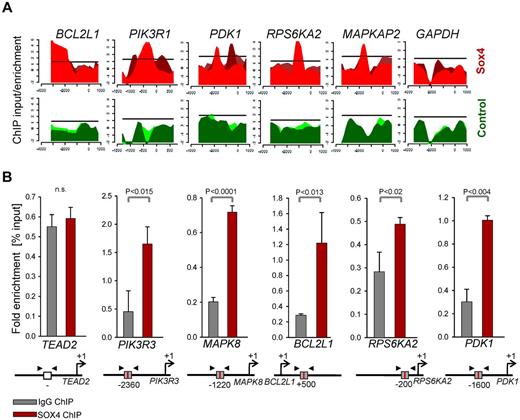
To study the mechanism of Sox4 regulation in pre-B ALL, we performed a loss-of-function gene expression array experiment (Figure 3A). Deletion of Sox4 reduced mRNA levels of multiple genes that play a known role in survival signaling in Ph+ ALL cells including Bcl2l1 (Bcl-xL), molecules involved in the MAPK pathway Mapk8, Map2k6, Rps6ka2, and PI3K/AKT-related molecules Pik3r2, Pik3r3, and Mtor (Figure 3A-C). Interestingly, gain-of-function based on Sox4 overexpression had the opposite effect: overexpression of Sox4 resulted in increased mRNA levels of Bcl-xL (Bcl2l1; Figure 3C), Mapk8, and PI3K/AKT (Pik3r2, Pik3r3; supplemental Figure 3). Because Sox4 functions as a transcription factor and carries a Sry-related HMG box DNA-binding domain,1 we identified Sox4 target genes via promoter binding. ChIP-on-chip analysis in human prostate cancer cells (GS11874) revealed that SOX4 binds directly to promoters of BCL2L1 (BCL-XL), genes of the MAPK pathway and multiple genes that are involved in PI3K/AKT signaling (Figure 4A; supplemental Figure 4). Single-locus quantitative ChIP confirmed binding of SOX4 to PIK3R3, MAPK8, BCL2L1, RPS6KA2, and PDK1 promoters in human Ph+ ALL cells via its putative DNA binding motif WWCAAWG1,10,19 (Figure 4B, supplemental Figure 5). Transcriptional regulation of components within the PI3K/AKT pathway is indeed functionally relevant because Cre-mediated deletion of Sox4 caused multiple dephosphorylation events (supplemental Figure 6), including AKTS473, SRCY416, and RPS6S235/6 (Figure 5A). Although a global analysis of phospho-tyrosine protein changes (using the 4G10 antibody; supplemental Figure 6) revealed some increases of tyrosine-phosphorylation, in the majority of cases, deletion of Sox4 caused loss of tyrosine-phosphorylation (supplemental Figure 6). Besides its function as transcriptional regulator, some of the dephosphorylation events may also reflect the ability of Sox4 to function as cytoplasmic activator of signaling as described previously.3-5 In addition, inducible deletion of Sox4 resulted in strong up-regulation of both Arf and p53 protein (Figure 5B). Consistent with previous findings,15 TKI-treatment (Imatinib) resulted in down-regulation of Arf expression (Figure 5C). Negative regulation of Arf was dependent on Sox4: loss of Sox4 prevented the down-regulation of Arf expression after imatinib treatment (Figure 5C). Loss of viability on Sox4 deletion in the leukemia cells was rescued by transduction with BCL2L1 (Bcl-xL; Figure 5D-E) and transiently rescued by constitutively active (myristoylated; CA) mutants of AKTCA and the p110 catalytic subunit of PI3K (p110αCA; supplemental Figure 7). These findings demonstrate that Sox4 mediates critical survival signals in Ph+ ALL via activation of BCL2L1 and PI3K/AKT.
Identification of transcriptional targets of Sox4 in Ph+ ALL. We deleted Sox4 on an inducible activation of Cre in Sox4fl/fl leukemia cells (supplemental Figure 1) and studied gene expression changes after 1 day of activation of Cre or an empty vector control (EV; A). Sox4-dependent gene expression changes (GSE36543) involve the PI3K/AKT, MAPK, and critical survival pathways. Target genes were verified in a qRT-PCR (B-C; supplemental Figure 3). In panel C, BCL2L1 is verified as Sox4 target gene in BCR-ABL1 ALL cells through Cre-mediated deletion and retroviral overexpression of Sox4.
Identification of transcriptional targets of Sox4 in Ph+ ALL. We deleted Sox4 on an inducible activation of Cre in Sox4fl/fl leukemia cells (supplemental Figure 1) and studied gene expression changes after 1 day of activation of Cre or an empty vector control (EV; A). Sox4-dependent gene expression changes (GSE36543) involve the PI3K/AKT, MAPK, and critical survival pathways. Target genes were verified in a qRT-PCR (B-C; supplemental Figure 3). In panel C, BCL2L1 is verified as Sox4 target gene in BCR-ABL1 ALL cells through Cre-mediated deletion and retroviral overexpression of Sox4.
SOX4 binds to promoters of PI3K-AKT and MAPK8 pathway genes in Ph+ ALL. ChIP-on-chip data of human prostate cancer cells for SOX4 binding11 was analyzed (GSE11874) and specific binding at BCL-XL (BCL2L1), MAPK and PI3K/AKT-related loci is shown (A). Specific binding of SOX4 to PIK3R3, MAPK8, BCL2L1, RPS6KA2, and PDK1 promoters in human Ph+ ALL cells was verified by single-locus quantitative ChIP (B). The putative DNA bindinig motif (gray box) and primer (arrows) are indicated in the diagram for each of the specific gene locus.
SOX4 binds to promoters of PI3K-AKT and MAPK8 pathway genes in Ph+ ALL. ChIP-on-chip data of human prostate cancer cells for SOX4 binding11 was analyzed (GSE11874) and specific binding at BCL-XL (BCL2L1), MAPK and PI3K/AKT-related loci is shown (A). Specific binding of SOX4 to PIK3R3, MAPK8, BCL2L1, RPS6KA2, and PDK1 promoters in human Ph+ ALL cells was verified by single-locus quantitative ChIP (B). The putative DNA bindinig motif (gray box) and primer (arrows) are indicated in the diagram for each of the specific gene locus.
Sox4 regulates survival signaling via Bcl-xL and the PI3K-AKT pathway in Ph+ ALL. Effects of imatinib-treatment (IM) on phosphorylation status of survival signaling molecules (Stat5, AKT, SRC, RPS6) were compared with effects of Sox4 deletion (A). In the absence of Sox4, activation of AKT (p-AKTS473), S6 (p-S6S235/6), and SRC (p-SRCY416) were decreased. Effects of Cre-mediated deletion on protein levels of Arf (in the presence and absence of imatinib-treatment) and p53 were studied by Western blot in panels B and C. In panel D, Sox4fl/fl ALL cells were transduced with an expression vector for Bcl-xL on induction of Cre-mediated deletion of Sox4. The effects of Bcl-xL leukemia cell survival (D) and quantitative analysis (E; P = .004) are shown.
Sox4 regulates survival signaling via Bcl-xL and the PI3K-AKT pathway in Ph+ ALL. Effects of imatinib-treatment (IM) on phosphorylation status of survival signaling molecules (Stat5, AKT, SRC, RPS6) were compared with effects of Sox4 deletion (A). In the absence of Sox4, activation of AKT (p-AKTS473), S6 (p-S6S235/6), and SRC (p-SRCY416) were decreased. Effects of Cre-mediated deletion on protein levels of Arf (in the presence and absence of imatinib-treatment) and p53 were studied by Western blot in panels B and C. In panel D, Sox4fl/fl ALL cells were transduced with an expression vector for Bcl-xL on induction of Cre-mediated deletion of Sox4. The effects of Bcl-xL leukemia cell survival (D) and quantitative analysis (E; P = .004) are shown.
Sox4 mediates leukemic transformation in vivo and is a predictor of poor clinical outcome
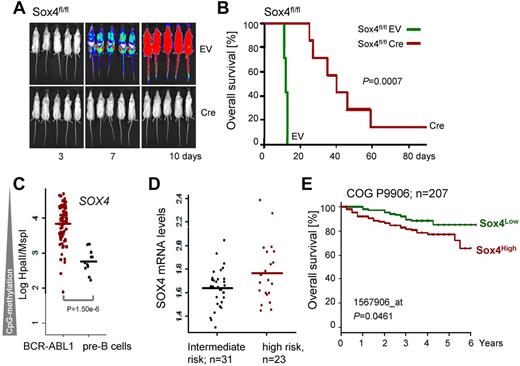
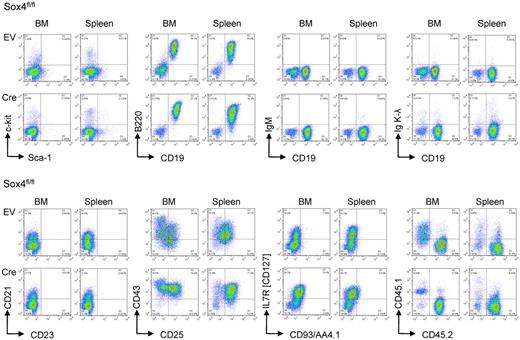
To test whether Cre-mediated deletion of Sox4 affects the course of leukemia development in vivo, 1 million Sox4fl/fl leukemia cells carrying 4-OHT-inducible Cre or an empty vector control were injected into sublethally irradiated NOD/SCID mice. Leukemia cells were labeled with firefly luciferase and leukemic expansion was monitored by luciferase bioimaging. Sox4fl/fl leukemia cells rapidly expanded (Figure 6A) and caused lethal disease in all recipient mice within 15 days (Figure 6B). Inducible deletion of Sox4 did not cause significant phenotype changes of leukemia (Figure 7) but delayed the onset of disease and substantially prolonged overall survival of recipient mice (Figure 6A-B). These findings are also relevant to human disease because promoter CpG methylation analysis (HpaII tiny fragment enrichment by ligation-mediated PCR [HELP] assay)20 revealed lower levels SOX4 promoter methylation in human Ph+ ALL cells compared with their normal pre-B cell counterparts (Figure 6C). In addition, samples from patients with high risk ALL (n = 31) show a trend toward higher SOX4 mRNA levels compared with patients with intermediate risk ALL (n = 23, P = .07, Figure 6D; ALL REZ BFM 2002,13 GSE4698). Analyzing data from a larger study based on 207 patients with ALL (COG P9906)12 showed a statistically significant association between high mRNA levels of SOX4 and poor clinical outcome (shorter overall survival; Figure 6E and supplemental Figure 8).
Sox4 promotes leukemic transformation in vivo and is correlated with poor clinical outcome. Sox4fl/fl ALL cells were transduced with 4-OHT–inducible Cre or an empty vector control (EV) and labeled with firefly luciferase. One × 106 cells were injected into sublethally irradiated (2.5 Gy) NOD/SCID mice and leukemic expansion was tracked by luciferase bioimaging (A). In panel B overall survival of transplant recipient mice is shown in a Kaplan-Meier analysis (7 mice per group; P = .0007). The methylation status of the SOX4 promoter was studied (HELP assay20 ; C) in normal human bone marrow pre-B cells (n = 12) and bone marrow biopsies of human Ph+ ALL (n = 83; P = 1.5 ×10−6; data from ECOG E2993 and ECOG leukemia tissue bank). In panel D, samples from patients with high risk ALL (n = 31) are compared with patients with intermediate risk ALL in relation to their SOX4 mRNA levels (n = 23, P = .07; ALL REZ BFM 2002,12 GSE4698). In panel E, we analyzed clinical and gene expression data from the pediatric ALL trial COG P990614 (GSE36543). Samples from 207 patients were divided into 2 groups according to higher (red curve; n = 103) or lower (green; n = 104) than mean expression of Sox4 among all patients. The Kaplan-Meier analysis of overall survival shows a significant difference between the 2 groups (P = .046; H).
Sox4 promotes leukemic transformation in vivo and is correlated with poor clinical outcome. Sox4fl/fl ALL cells were transduced with 4-OHT–inducible Cre or an empty vector control (EV) and labeled with firefly luciferase. One × 106 cells were injected into sublethally irradiated (2.5 Gy) NOD/SCID mice and leukemic expansion was tracked by luciferase bioimaging (A). In panel B overall survival of transplant recipient mice is shown in a Kaplan-Meier analysis (7 mice per group; P = .0007). The methylation status of the SOX4 promoter was studied (HELP assay20 ; C) in normal human bone marrow pre-B cells (n = 12) and bone marrow biopsies of human Ph+ ALL (n = 83; P = 1.5 ×10−6; data from ECOG E2993 and ECOG leukemia tissue bank). In panel D, samples from patients with high risk ALL (n = 31) are compared with patients with intermediate risk ALL in relation to their SOX4 mRNA levels (n = 23, P = .07; ALL REZ BFM 2002,12 GSE4698). In panel E, we analyzed clinical and gene expression data from the pediatric ALL trial COG P990614 (GSE36543). Samples from 207 patients were divided into 2 groups according to higher (red curve; n = 103) or lower (green; n = 104) than mean expression of Sox4 among all patients. The Kaplan-Meier analysis of overall survival shows a significant difference between the 2 groups (P = .046; H).
Phenotype of pre-B ALL on Sox4 deletion in vivo. BCR-ABL1–transformed mouse pre-B ALL from bone marrow of Sox4fl/fl mice were transduced with 4-hydroxy tamoxifen (4-OHT)–inducible Cre (Cre) or an empty vector control (EV). Two × 106 leukemia cells were injected into NOD/SCID recipient mice in each group. Phenotypic changes on deletion of Sox4 were studied by flow cytometry after isolation of leukemia cells from killed animals and staining for c-kit, Sca-1, CD19, B220, IgM, Igκ, and Igλ light chains, CD21, CD23, CD43, IL2RA (CD25), IL7R (CD127), and AA4.1 (CD93). BCR-ABL1 transduced ALL cells from donor bone marrow of Sox4fl/fl mice express CD45.2 but not CD45.1. Donor-derived leukemia cells can be identified as CD45.2+ cells after injection into CD45.1+ NOD/SCID recipient mice.
Phenotype of pre-B ALL on Sox4 deletion in vivo. BCR-ABL1–transformed mouse pre-B ALL from bone marrow of Sox4fl/fl mice were transduced with 4-hydroxy tamoxifen (4-OHT)–inducible Cre (Cre) or an empty vector control (EV). Two × 106 leukemia cells were injected into NOD/SCID recipient mice in each group. Phenotypic changes on deletion of Sox4 were studied by flow cytometry after isolation of leukemia cells from killed animals and staining for c-kit, Sca-1, CD19, B220, IgM, Igκ, and Igλ light chains, CD21, CD23, CD43, IL2RA (CD25), IL7R (CD127), and AA4.1 (CD93). BCR-ABL1 transduced ALL cells from donor bone marrow of Sox4fl/fl mice express CD45.2 but not CD45.1. Donor-derived leukemia cells can be identified as CD45.2+ cells after injection into CD45.1+ NOD/SCID recipient mice.
To determine whether SOX4 expression represents an independent predictor of poor clinical outcome, we have performed a multivariate analysis of SOX4 expression comparing overall survival (OS) time in relation to age, white blood cell count (WBC), IKZF1, and CDKN2A expression in a multivariate Cox proportional hazards regression model in R package (R Development Core Team, 2009). These analyses indeed identified SOX4 expression levels as an independent predictor of outcome (OS time) with respect to age, WBC, IKZF1 expression, and CDKN2A expression (P = .0434, HR = 1.6481, 95% confidence interval 1.0150: 2.676; supplemental Figure 8).
Collectively, these findings identify SOX4 as a critical upstream regulator of survival signaling in Ph+ ALL. Pathways affected by SOX4 include PI3K/AKT signaling downstream of BCR-ABL1, activation of BCL-xL (BCL2L1) and negative regulation of Arf and p53.
Discussion
In this study, we identified Sox4 as a critical factor in survival signaling of Ph+ ALL. Sox4 regulates survival via Bcl-xL, Mapk8 (JNK1) and genes of the PI3K/AKT pathway. A recent study based on pharmacologic targeting of the PI3K/AKT downstream signaling highlighted the relevance of this pathway in human Ph+ ALL.21 The BCR-ABL1 oncogene has been shown to activate the JNK pathway22 and disruption of Mapk8 (JNK1) results in defective transformation of BCR-ABL1 driven pre-B ALL.23 In Ph+ leukemia, Bcl-xL represents an important survival molecule and Stat5-dependent transcriptional activation of Bcl-xL promotes inhibition of apoptosis.24 Therefore, the discovery of SOX4 as a critical upstream regulator to enable Bcl-xL, JNK1, and PI3K/AKT signaling in Ph+ ALL represents an important finding. The specific rescue by Bcl-xL and active mutants of AKT or PI3K of loss of viability on Sox4 deletion provides mechanistic evidence that Sox4 regulates leukemia cell survival through these components (Figure 5D-E, supplemental Figure 7). In addition, Sox4 negatively regulates p53/Arf (Figure 5B-C). Because our ChIP analyses did not reveal direct binding of Sox4 at CDKN2A (arf) and TP53 (p53) promoters, these effects may be indirect, for example, mediated via PI3K/AKT. Given that Arf and p53 represent critical negative regulators of self-renewal,25 Sox4-mediated negative regulation of Arf/p53 probably contributes to the important function of Sox4 in mediating leukemia colony formation in serial replating experiments (Figure 2C). In agreement with reduced expression of Bcl-xL (Bcl2l1) and increased levels of Arf/p53, the overall outcome of Sox4 deletion is rapid loss of viability and apoptosis in BCR-ABL1–transformed ALL but not normal pre-B cells (Figure 2A).
Previous studies demonstrated that Bcl-xL regulates survival during early B-cell development,26,27 with high Bcl-xL expression levels in pro-B cells.26 Likewise, Bcl-xL transgenic mice develop large B-cell expansions as a result of increased survival during immunoglobulin VH-DJH gene rearrangement at the pro to pre-B cell transition.27 The stage-specific function and gradient of BCL-xL expression and Sox4-dependent regulation of BCL-xL is consistent with the pro-B cell differentiation block in Sox4-deficient mice. In the absence of Sox4-induced BCL-xL, pro-B cells are destined to die when they undergo VH-DJH gene rearrangement at the pre-B cell checkpoint.
Sox4 amplifications have been associated with human cancers including lung, breast, prostate cancer.11,28-30 The correlation of high SOX4 expression with poor clinical outcome (Figure 6D-E) is novel and suggests that SOX4 and SOX4-mediated signaling represent a potential therapeutic target for patients with ALL. Although transcription factors are typically considered intractable for pharmacologic inhibition, work by us and others15,31 suggests that BCL6 may represent a notable exception, because targeting is based on protein-protein interactions with critical BCL6 cofactors. In the case of Sox4, we hope that identification of critical cofactors that mediate Sox4-mediated transcriptional activation of BCL2L1 (BCL-xL) and PI3K/AKT will make this oncogenic pathway accessible to small molecule targeting as previously exemplified for BCL6.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The murine Sox4 was amplified from MSCV-Sox4-puro vector, which was a kind gift from Drs Yang Du and Cynthia Dunbar (Hematology Branch, National Heart, Lung, and Blood Institute, Bethesda, MD). The authors thank Behzad Kharabi Masouleh for critical discussions. P.R.-R. and C.H. are PhD students and members of the Graduate Program, University of Freiburg Faculty of Biology, Freiburg, Germany.
This work is supported by the ECOG leukemia tissue bank (E.P.) and through National Institutes of Health(NIH)/National Cancer Institute (NCI) grants R01CA137060, R01CA139032, R01CA157644, R01CA169458, and R21CA152497 (to M.M.); NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grant R01AR54153 (to V.L.); Translational Research Program grants and a SCOR grant from the Leukemia & Lymphoma Society (grants 6132-09, 6097-10, 6221-12, 7005-11; to M.M.); the William Lawrence and Blanche Hughes Foundation and a Stand Up To Cancer-American Association for Cancer Research Innovative Research Grant (IRG00909, to M.M.); and the California Institute for Regenerative Medicine (CIRM; TR2-01 816 to MM). M.M. is a Scholar of the Leukemia & Lymphoma Society.
National Institutes of Health
Authorship
Contribution: P.R.-R., H.G., C.H., L.N.C., and Z.C. performed experiments and analyzed data; H.J. and V.L. provided critical reagents; A.M., E.P., W.L.C., and C.L.W. analyzed data and provided clinical information; and M.M. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Markus Müschen, Deptartment of Laboratory Medicine, University of California, San Francisco, 521 Parnassus Ave, San Francisco, CA 94143; e-mail: markus.muschen@ucsf.edu.