Key Points
MicroRNA and plasma levels of the target gene CXCL13 differ between ITP and controls indicating that microRNA may be important in ITP.
Abstract
MicroRNA are small noncoding RNA molecules that regulate gene expression. To investigate the role of microRNA in immune thrombocytopenia (ITP), we performed genome-wide expression analyses of mRNA and microRNA in T cells from ITP patients and controls. We identified 1915 regulated genes and 22 regulated microRNA that differed between ITP patients and controls. Seventeen of the 22 regulated microRNA were linked to changes in target gene expression; 57 of these target genes were associated with the immune system, eg, T-cell activation and regulation of immunoglobulin production. CXCL13 and IL-21 were two microRNA target genes significantly increased in ITP. We could demonstrate increased plasma levels of CXCL13 and others have reported increased plasma levels of interleukin-21 in ITP. Thus, regulated microRNA were significantly associated with both gene and protein expression of molecules in immunological pathways. We suggest that microRNA may be important regulatory molecules involved in the loss of tolerance in ITP.
Introduction
Immune thrombocytopenia (ITP) is an autoimmune disease characterized by low platelet count and increased bleeding tendency.1 The pathophysiology of ITP is more complex than initially believed and includes both antibody-mediated and T cell–mediated platelet and/or megakaryocyte destruction.2-4 An insufficient thrombopoietin production in ITP also contributes to the thrombocytopenia.5
MicroRNA are short (19-25 nucleotides) evolutionary conserved single-stranded RNA molecules that regulate the expression of genes involved in diverse biological processes. The effect of microRNA on messenger (mRNA) is mediated through the binding of the microRNA to the ribonucleoprotein complex RNA-induced silencing complex that in addition also bind to the 3′ untranslated region of complementary mRNAs.6 The double-stranded complex between the microRNA and mRNA are then degraded, which leads to decreased protein translation.7 Approximately 30% of the human genome is estimated to be regulated by microRNA, and a single microRNA can potentially regulate hundreds of protein.8,9 More than 1000 microRNA have been identified in mammals and have been implicated in a wide range of biological functions10,11 that contribute to the pathophysiology of a number of important human diseases such as cancer,12-15 cardiac and neurodegenerative diseases, diabetes, inflammation, and diseases of the immune system.16 However, the in vivo function of most microRNA is mainly unknown. This is the first report on microRNA as potential regulators of T-cell gene expression in ITP patients.
Study design
Patient characteristics and detailed methods are given in “Supplemental Methods.” All individuals involved in this study gave informed consent in accordance with the Declaration of Helsinki. The study was approved by the regional ethics committee in Gothenburg, Sweden.
In brief, T-cell isolation and extraction of RNA was performed as previously described.2 Twenty nanograms of RNA was reverse transcribed, amplified, and labeled using the Ovation amplification system V2 (NuGEN Technologies Inc, San Carlos, CA); the corresponding cDNA was fragmented and biotinylated using the Encore biotin module (NuGEN) and hybridized to human genome U133 plus 2.0 arrays (Affymetrix, Santa Clara, CA) according to the manufacturer's instructions. For the microRNA analysis, 1000 ng T-cell RNA from each individual was biotin labeled using the FlashTag Biotin HSR kit (Genisphere, Hatfield, PA) according to the manufacturer's instructions and hybridized to microRNA 2.0 arrays (Affymetrix).
The DNA and microRNA microarrays (GSE43179 [for mRNA]; GSE43178 [for microRNA]) were normalized using RMA and PLIER algorithms; significantly regulated genes and microRNA were essentially detected using Student t test (“Supplemental Information”). To identify the global biological processes that differed between patients and controls a reporter algorithm was applied to the Gene Ontology (GO) network resulting in an enrichment score.17 GO terms that had enrichment P values < .001, using the R software, were considered and selected in the construction of a heat map (supplemental Figure S1).
The mirBase (http://www.mirbase.org) was used to identify microRNA functions and microRNA target mRNA using TargetScan and Miranda algorithms. To achieve high-confidence microRNA-mRNA associations and to evaluate the impact of each microRNA on the gene expression, the predicted target genes of each microRNA were identified and combined with the mRNA transcriptome from ITP patients and controls in an analysis using the Kolmogorov-Smirnov test (Table 1). The target genes from the microRNA identified as significant (P < .05) in the Kolmogorov-Smirnov analysis were cross-referenced against the list of significantly regulated mRNA between ITP patients and controls identified in the T-cell gene expression analysis. The resulting immune genes according to GO were classified further according to functional enrichment based on Immune System Gene Ontology18 by modular enrichment analysis (supplemental Table 3 and supplemental Figures 2 and 3).
Results and discussion
Autoimmune diseases consist of more than 80 variable and serious illnesses that collectively affect more than 5% of the population, often with debilitating effects.19 Loss of tolerance, where the immune system is misdirected and attacks organs or cells instead of protecting them, is the common denominator in these diseases. To better understand the genes and mechanisms involved in the organ-specific autoimmune disease ITP, we studied gene and microRNA expression in T cells from chronic ITP patients and healthy controls.
Initially, we identified 1915 significantly regulated genes in peripheral blood T cells between ITP patients and controls by DNA microarray analysis (P < .05). The GO project is a collaborative effort to address the need for consistent descriptions of gene products in different databases. One of these annotations is a biological process that is defined as series of events accomplished by 1 or more ordered assemblies of molecular functions. Therefore, the significantly regulated genes were classified according to biological process in GO, which demonstrated that ITP was associated with several significantly enriched biological processes involved in the immune system (supplemental Figure 1). In the next step, we compared differences in expression of microRNA in peripheral blood T cells and found that 22 microRNA differed significantly between ITP patients and controls (P < .05). In addition, 16 small nucleolar RNA such as small nucleolar RNA and small Cajal body–specific RNA also differed between ITP patients and controls (P < .05; supplemental Table 2).
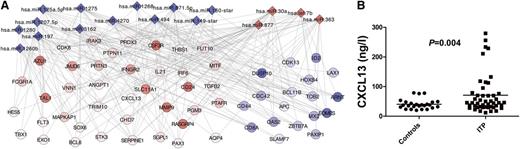
First, to better understand the role of the regulated microRNA in ITP patients, we identified the predicted target genes of the significantly regulated microRNA using TargetScan and Miranda algorithms. Second, to investigate the impact of the 22 significantly changed microRNA on gene expression, we performed Kolmogorov-Smirnov tests using the target genes identified in TargetScan and Miranda on all transcripts in the gene expression dataset. This resulted in 17 microRNA that were significantly associated with the expression of target genes (Table 1). Third, the identified target genes of the 17 significantly changed microRNA were cross-referenced against the significantly regulated mRNA that differed between ITP patients and controls, resulting in 991 genes. Fourth, the cross-referenced genes were classified according to function by GO, and the 57 genes classified as being involved in the immune system (Figure 1A) were analyzed using functional module enrichment based on Immune System Gene Ontology.18 This resulted in 7 modules that were enriched with the following functions: T-cell activation involved in immune response, natural killer cell differentiation, regulation of immunoglobulin production, positive regulation of leukocyte activation, lymphocyte activation involved in immune response, lymphocyte differentiation, and lymphocyte costimulation (supplemental Figure 2A-B). Regulation of immunoglobulin production is in agreement with one of the known mechanisms behind ITP, namely production of platelet autoantibodies seen in approximately 50% to 60% of all chronic ITP patients.20 The other enriched processes such as T-cell activation involved in immune response, positive regulation of leukocyte activation, and lymphocyte co-stimulation, further highlight the importance of T cells in this disease. This supports previous findings such as proliferation of T cells and production of cytokines in response to stimulation with whole platelets or fragments of GPIIb/IIIa and GPIIIa in ITP,21-25 and that cytotoxic T cells can lyse platelets in patients with ITP.2,26-28 That both B-cell and T-cell mechanisms are important pathophysiologic mechanisms in ITP has also been shown in an elegant animal model of ITP.26
MicroRNA regulate mRNA and protein expression in T cells. (A) A computational method to identify microRNA function and mRNA targets in T cells. Target genes of significantly regulated microRNA between ITP patients and controls, using TargetScan and Miranda algorithms, were compared with significantly regulated mRNA from peripheral blood T cells between patients and controls. Diamonds are microRNA and circles are mRNA. Red indicates up-regulated and blue indicates down-regulated genes. (B) Plasma levels of CXCL13, which was a significantly regulated target gene of a microRNA that differed in expression between ITP patients and controls, were significantly increased in plasma from patients with ITP compared with controls.
MicroRNA regulate mRNA and protein expression in T cells. (A) A computational method to identify microRNA function and mRNA targets in T cells. Target genes of significantly regulated microRNA between ITP patients and controls, using TargetScan and Miranda algorithms, were compared with significantly regulated mRNA from peripheral blood T cells between patients and controls. Diamonds are microRNA and circles are mRNA. Red indicates up-regulated and blue indicates down-regulated genes. (B) Plasma levels of CXCL13, which was a significantly regulated target gene of a microRNA that differed in expression between ITP patients and controls, were significantly increased in plasma from patients with ITP compared with controls.
To test whether these changes in microRNA expression were accompanied by changes of corresponding proteins, we determined plasma levels of CXCL13 in ITP patients and controls. The plasma level of this protein was found to be significantly increased in ITP patients compared with controls (Figure 1B). Our data also suggested that interleukin-21 (IL-21) was a target of the regulated microRNA and that IL-21 expression would be increased in patients with ITP. Indeed, increased plasma levels of IL-21 in ITP patients compared with controls has previously been demonstrated by Zhu et al, which supports our present data.29
In conclusion, regulated microRNA in ITP significantly affect both gene and protein expression in T cells, indicating that they may be important regulatory molecules involved in the loss of immune tolerance in ITP.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work is supported by grants from the Swedish Research Council (K2009-65X-15424-05-3, K2011-X-20401-05-6), the Swedish federal government under the LUA/ALF agreement, the Foundations of the National Board of Health and Welfare, Torsten and Ragnar Söderberg Foundation, Clas Groschinsky Foundation, the Arosenius Foundation, Åke Wiberg Foundation, Jeansson Foundation, Tore Nilsson Foundation for Medical Research, Magnus Bergvall Foundation, Wilhelm and Martina Lundgren Science Foundation, the Knut and Alice Wallenberg Foundation, and the Chalmers Foundation.
Authorship
Contributions: M.J. designed and coordinated the study, performed all laboratory work, analyzed data, and wrote the paper; I.N. analyzed and interpreted data and wrote the paper; H.W. collected the patient material, interpreted the data, and wrote the paper; and B.O. designed the study, analyzed and interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interest.
Correspondence: Margareta Jernås, Department of Internal Medicine, Institute of Medicine, Sahlgrenska Academy at University of Gothenburg, Vita Stråket 12, SE-413 45 Gothenburg, Sweden; e-mail: margareta.jernas@medic.gu.se.