Abstract
Long-living memory stem T cells (TSCM) with the ability to self-renew and the plasticity to differentiate into potent effectors could be valuable weapons in adoptive T-cell therapy against cancer. Nonetheless, procedures to specifically target this T-cell population remain elusive. Here, we show that it is possible to differentiate in vitro, expand, and gene modify in clinically compliant conditions CD8+ TSCM lymphocytes starting from naive precursors. Requirements for the generation of this T-cell subset, described as CD62L+CCR7+CD45RA+CD45R0+IL-7Rα+CD95+, are CD3/CD28 engagement and culture with IL-7 and IL-15. Accordingly, TSCM accumulates early after hematopoietic stem cell transplantation. The gene expression signature and functional phenotype define this population as a distinct memory T-lymphocyte subset, intermediate between naive and central memory cells. When transplanted in immunodeficient mice, gene-modified naive-derived TSCM prove superior to other memory lymphocytes for the ability to expand and differentiate into effectors able to mediate a potent xenogeneic GVHD. Furthermore, gene-modified TSCM are the only T-cell subset able to expand and mediate GVHD on serial transplantation, suggesting self-renewal capacity in a clinically relevant setting. These findings provide novel insights into the origin and requirements for TSCM generation and pave the way for their clinical rapid exploitation in adoptive cell therapy.
Key Points
Identification of the biologic requirements for memory stem T cell (TSCM) generation and expansion from naive precursors ex vivo.
Differentiation, expansion, and genetic manipulation of human TSCM for cancer adoptive cellular therapy.
Introduction
Adaptive immunity is a potent and flexible system able to combat microbes and cancer cells.1,2 In the presence of infections or cancer, antigen-specific lymphocytes expand and differentiate into effectors devoted to rapidly clearing the pathogen and memory cells able to persist long-term to patrol the entire organism for recurrence and minimal residual disease.3,4 However, the mechanism and hierarchical differentiation path underlying the generation of memory precursors and terminal effector cells remain to be fully elucidated.5 This process has been proposed to involve a self-renewing, stem cell–like memory T-cell subset capable of differentiating into effectors on antigen reencounter.6,7 This T-cell subset, referred to as memory stem T cells (TSCM), and initially described in mice,8,9 begins to be unveiled in humans.10 TSCM potential biodistribution and long-term persistence represent appealing features to overcome the current limitations of cancer adoptive immune-gene therapy.11-13 At present, clinical-grade protocols able to obtain or preserve TSCM functional and phenotypic characteristics remain to be defined. We previously showed that costimulation of unselected T cells and culture with γ-chain cytokines allow the preferential generation of gene-modified T cells with a functional central memory (TCM) phenotype, superior to effector/effector memory (TEM) counterparts for expansion potential and antitumor activity.14,15 Compared with TCM and TEM lymphocytes, naive T cells (TN) are endowed with the highest developmental plasticity and are unique in the ability to generate daughter cells with potential to enter the entire spectrum of immunologic memory, including TSCM. We thus hypothesized that, starting from naive precursors, we could differentiate and genetically engineer human TSCM. We report that IL-7 and IL-15 support the generation of postmitotic costimulated CD8+ T cells with molecular and functional features of TSCM cells. These cells—defined by the expression of CD45RA, CD45R0, CD62L, CCR7, IL-7Rα, and CD95—can be identified among healthy subjects, are selectively enriched in hematopoietic stem cell transplant (HSCT) recipients, and reveal a phenotypic and functional profile distinct from that of TCM and TEM cells for extensive expansion capacity and ability to differentiate into potent effectors, self-renew, and expand in serial transplant recipients.
Methods
T-cell isolation and sorting
Peripheral blood mononuclear cells (PBMCs) were harvested from healthy donors after informed consent in accordance with the Declaration of Helsinki. PBMCs were isolated by Ficoll-Hypaque gradient separation (Lymphoprep; Fresenius), and enriched for CD3+ or CD8+ cells with the Pan T Cell Isolation Kit II or the CD8 Isolation Kit, respectively (Miltenyi Biotec) following the manufacturer's instructions. Cells were labeled with anti-CD3, anti-CD45RA (BD Biosciences), and anti-CD62L (Exbio) fluorescent antibodies and FACS-purified into TN, TCM, and TEM on a MoFlo MLS cell sorter (DAKO). Postsorting analysis of purified subsets (comprising CD45RA, CD62L, and CD45R0 flow cytometric evaluation) revealed greater than 95% purity.
Cell stimulation and transduction
Sorted T lymphocytes were activated by 30 ng/mL plate-bound anti-CD3 antibody (OKT3; OrthoBiotech) or anti-CD3/CD28–conjugated magnetic beads (bCD3/CD28, ClinExVivo; CD3/CD28, Invitrogen) in a 3:1 bead/T-cell ratio and then cultured with: (1) recombinant human (rh) IL-2 at 300 IU/mL; (2) rhIL-7 at 5 ng/mL (PeproTech), or (3) rhIL-7 and rhIL-15 at 5 ng/mL each (PeproTech). Cytokines and medium were replaced every 3-4 days. Transduction was performed 48 hours after initial stimulation using either a HSV-Tk/ΔLNGFR-encoding retroviral vector (SFCMM-3 Mut2, MolMed)14 or a lentiviral vector (LV) encoding a transgenic WT1-specific TCR.16 Transduction efficiency was assessed at day 7 after stimulation by flow cytometry. Cells were counted every 3-4 days by Trypan blue dye exclusion.
Flow cytometry, antibodies, and intracellular staining
Cells were labeled with fluorescent antibodies against CD3, CD4, CD8, CD45RA (Biolegend); CD45R0, CD27, CD28, CD95, CD271, CCR5, CCR7, CXCR4, HLA-DR, IL-2Rβ, 7AAD (BD Biosciences); CD62L (Exbio); CXCR3 (R&D Systems); IL-7Rα, Vβ21 (Beckman Coulter). CMV pp65-specific T cells were detected with HLA-A*0201-NLVPMVATV PE-conjugated pentamers (Proimmune). For intracellular staining, cells were stimulated with 50 ng/mL PMA (Sigma-Aldrich) and 1 μg/mL ionomycin (Sigma-Aldrich), brefeldin 10 μg/mL (Sigma-Aldrich) was added for 2 hours, cells were stained with surface antibodies, washed and fixed with 1% paraformaldehyde. Intracellular staining was performed with anti–IL-2, γ-IFN, granzyme A, and perforin-specific antibodies (BD Biosciences) in PBS containing 0.05% saponin (Sigma-Aldrich). Data were acquired using a FACSCanto II (BD Biosciences) and analyzed with FlowJo Version 8.7 software (TreeStar).
Antigen-specific stimulation and rechallenge
Two weeks after bCD3/CD28 stimulation and transduction with an LV encoding for a WT1-specific TCR,17 lymphocyte subpopulations (0.5 × 106) were cocultured with WT1126-134-pulsed (100μM) irradiated (100 Gy) T2 cells (0.2 × 106) in the presence of irradiated (30 Gy) autologous PBMCs (2 × 106). Cells were cultured with IL-7 and IL-15 (5 ng/mL) for 16 days. Cytokines were replaced every 3-4 days. Lymphocytes were then tested simultaneously for cytokine production and memory subset composition, on short in vitro stimulation with cognate peptide. Briefly, 0.2 × 106 T cells were preincubated with 50μM TAPI-2 (Enzo Life Sciences) for 1 hour as previously described.18 Cells were then coincubated with WT1126-134–pulsed (100μM) irradiated T2 cells in 1:1 ratio for 4 hours. After adding brefeldin 10 μg/mL for an additional 2 hours, cells were stained with surface antibodies, fixed, and stained for intracellular IL-2 and γ-IFN.
Quantification of sjTRECs
Real-time quantitative PCR (qPCR) for single-joint TCR excision circles (sjTRECs) was performed as described previously,19 using GAPDH as a control to standardize DNA content. Briefly, amplification reactions were performed in a final volume of 25 μL containing 50 ng of genomic DNA isolated from the different T-cell subsets, TaqMan universal PCR master mix (PerkinElmer/Applied Biosystems), and the appropriate primers and probes. The number of sjTRECs per 100 ng of DNA was determined on the basis of a standard curve developed in-house by cloning the sequence of α1 circles from human cord blood genomic DNA into a plasmid vector and diluting the plasmid into human DNA from a cell line devoid of TRECs (K562).
In vivo model of GVHD in NOD/Scid mice
The protocol was approved by the Institutional Animal Care and Use Committee (IACUC). At day −1, 6- to 8-week-old female NOD/Scid mice (Charles River Italia) were given 1 mg of blocking anti-mouse IL-2Rβ monoclonal antibody intraperitoneally (IP) to neutralize residual natural killer activity. At day 0, mice received total body irradiation (250 cGy) and were infused IP with 5 × 106 T cells. Mice were clinically monitored and weighed 3 times per week. Human chimerism on peripheral blood was assessed weekly by flow cytometry. At time of severe GVHD, defined as the concomitant presence of human chimerism greater than 10% and weight loss greater than 5%, mice were killed, and chimerism was assessed on bone marrow (BM) and spleen. In serial transplantation experiments, BM- and spleen-derived cells were reinfused in secondary recipients after the same procedure.
Microarray hybridization and analysis
Transcriptional profiling was determined in 12 samples of T cells, using Affymetrix HG-U133 Plus 2.0 GeneChip arrays. RNA was isolated using the RNeasy Plus Mini kit (QIAGEN) and reverse transcribed by the GeneChip 3′IVT Express kit. Affymetrix HG-U133 Plus 2.0 GeneChip array hybridization, staining, and scanning were performed using Affymetrix standard protocols. Fluorescence signals were recorded by Affymetrix scanner 3000, and image analysis was performed with the GeneChip Operating Software (GCOS) software. All data analyses were performed in R using Bioconductor libraries and statistical packages. Probe-level signals have been converted to expression values using the robust multiarray average procedure (RMA).20 Specifically, intensity levels have been background adjusted, and normalized using quantile normalization, and log2 expression values were calculated using median polish summarization and the custom-definition files for Human Gene U133 Plus 2.0 arrays based on Entrez (Version 13; http://brainarray.mbni.med.umich.edu/Brainarray/default.asp).21 The final dataset was composed of 12 samples [3 replicates each for TN, TCM, T(TN), and T(TCM)] and 18 185 Entrez gene IDs. Differentially expressed genes have been identified using significance analysis of microarray (SAM) algorithm coded in the samr R package. In SAM, we estimated the percentage of false-positive predictions with 1000 permutations and selected those transcripts whose q value (ie, false discovery rate [FDR]) was equal to 0 (or ≤ 0.05) and absolute fold change was ≥ 2. Gene set enrichment analysis was performed with GSEA software (http://www.broadinstitute.org/gsea/index.jsp)22 on log2 expression data of TN, TCM, T(TN), and T(TCM) samples. Gene sets comprise BioCarta pathways derived from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp) and the genes up-regulated in TCM cells of the CD8+ T-cell memory signature (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Signatures have been considered significantly enriched at FDR ≤ 25% when using default parameters and 5000 permutations of gene sets. Gene Ontology classification has been assessed using DAVID functional annotation (http://david.abcc.ncifcrf.gov/), grouped under broad categories, and plot as pie chart. The list of human cell differentiation molecules has been obtained from UniProt (http://www.uniprot.org/docs/cdlist). The independent dataset from Gattinoni et al10 has been downloaded as.CEL files from GEO GSE23321 and the expression values quantified using RMA and the custom definition files for Human Gene 1.0 ST arrays based on Entrez (Version 13). Entrez-based probe sets for Human Gene U133 Plus 2.0 and 1.0 ST arrays have been matched before merging the 2 datasets obtaining a meta-dataset comprising 21 samples (12 samples from this study and 9 samples for TN, TSCM, TCM from GSE23321) and 17 186 Entrez gene IDs. The meta-dataset has been normalized using ComBat, an empirical Bayes framework for adjusting data for batch effects (http://jlab.byu.edu/ComBat).23 Microarray data generated in this study are available at Gene Expression Omnibus GSE41909.
CFSE-dilution assay
Sorted T cells were loaded with CFSE 5μM (Invitrogen) and then cultured for 5 days with titrated (1-0.001 μg/mL) plate-bound OKT3, with or without anti-CD28 antibody (2 μg/mL; BD Biosciences) in the presence of IL-2 (300 IU/mL), IL-7, and IL-15 (5 ng/mL). Cells were then harvested, stained with 7AAD to exclude nonviable cells, and analyzed by flow cytometry.
Patient characteristics
Seven patients with high-risk acute leukemias referred to our Hematology Unit and treated with myeloablative allo-HSCT were studied. Patients received a treosulfan- and total body irradiation–based myeloablative conditioning followed by an unmanipulated peripheral blood graft from a matched unrelated donor (3 of 7) or an HLA-haploidentical related donor (4 of 7). The conditioning regimen included in vivo T- and B-cell depletion by ATG-Fresenius (10 mg/kg for 3 times) and Rituximab (a single 500-mg dose); GVHD prophylaxis consisted of Rapamycin (target level: 8-15 ng/mL, until day +60) and Mycophenolate Mofetil (15 mg/kg 3 times a day, until day +30). Peripheral blood was harvested 1 month after transplantation (median day after transplantation 33, range 25-50), on written informed consent.
Statistical analysis
Statistical analysis was performed with Prism 5 (GraphPad Software). Data are shown as the mean ± SEM. One-way ANOVA with Bonferroni correction and the 2-tailed unpaired t test were used for comparison of 3 or more groups, or 2 groups, respectively. Correlation was estimated by 2-tailed Pearson coefficients and significance. GVHD-free survival of mice infused with human lymphocytes was analyzed with log-rank and Gehan-Breslow-Wilcoxon tests.
Results
IL-7 and IL-15 instruct the expansion of a novel CD62L+CD45RA+ memory T-cell population from naive precursors
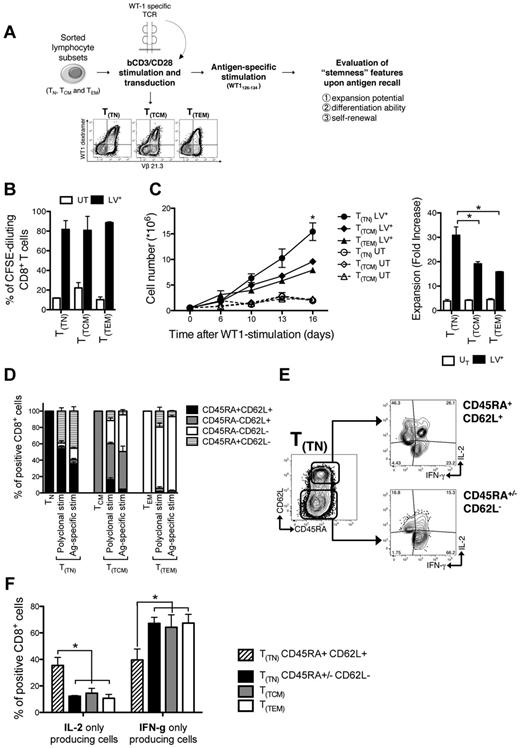
TN, TCM, and TEM were FACS-sorted to greater than 95% purity and activated with anti-CD3/anti-CD28–conjugated cell-size beads and low doses of IL-7 and IL-15 (Figure 1A). We then tested the expansion potential and sensitivity to genetic manipulation of individual T-cell subsets. All subsets revealed sensitive to both lentiviral and retroviral transduction (Figure 1B), with naive-derived transduced CD3+ T cells (T(TN)) expanding significantly more than TCM- and TEM-derived transduced cells (T(TCM) and T(TEM), respectively; P < .05; Figure 1C and supplemental Figure 1A for CD8+ cells). Of note, among T(TN) cells, a consistent fraction of CD8+ lymphocytes retained surface expression of CD45RA and CD62L (Figure 1D), a phenotype traditionally associated with naive T cells, for up to 30 days in culture (P < .05; Figure 1E). Most of these genetically modified CD62L+CD45RA+ cells also acquired the expression of CD45R0, the CD45 isoform associated with antigen-experienced T lymphocytes (Figure 1F-G), while preserving CCR7 expression (supplemental Figure 1B). This was in contrast to T(TCM) and T(TEM) cells, which mostly appeared CD45RA− and CCR7− (P < .001; supplemental Figure 1B). Expression of CD27 and CD28 costimulatory molecules was not significantly different among CD8+ T(TN) and T(TCM), whereas was lower in T(TEM) cells (P < .01; supplemental Figure 1C). While displaying a naive-like surface phenotype, functionally T(TN) revealed hallmarks of memory cells, such as undetectable sjTREC content and the ability to proliferate on TCR triggering in the absence of costimulation (supplemental Figure 2 panels A and B, respectively). To reveal the biologic determinants associated with this phenotype and dissect their relative contribution, we applied to FACS-purified CD45RA+CD62L+ cells different stimulation (costimulation vs TCR triggering alone) and culture conditions (IL-7 and IL-15 together, IL-7, IL-15, or IL-2 alone). Costimulation was critical for TN cell expansion because cells failed to robustly proliferate in response to OKT3 (P < .001; Figure 1H). Furthermore, while IL-7 was critical for CD8+ T cells to preserve a CD45RA+CD62L+ phenotype in response to bCD3/CD28 (P < .01; Figure 1I), IL-15 best supported their expansion (Figure 1H). Thus, TN appear unique for the ability to differentiate into T cells characterized by the stable expression of CD45RA, CD45R0, CD62L, CCR7 surface phenotype and high proliferative capacity. Mechanistically, costimulation, IL-7, and IL-15 appear critical and nonredundant for the instruction and expansion of this population.
IL-7 and IL-15 instruct the expansion of a novel CD62L+CD45RA+ memory T-cell population from naive precursors. (A) Diagram summarizing the procedure to generate T(TN), T(TCM), and T(TEM) subsets. FACS-sorted TN (CD62L+CD45RA+), TCM (CD62L+CD45RA−), and TEM (CD62L−CD45RA−) lymphocytes were activated with anti-CD3/CD28 beads (bCD3/CD28) and cultured with IL-7 (5 ng/mL) and IL-15 (5 ng/mL). Forty-eight hours after activation, cells were transduced with retroviral (RV) or lentiviral (LV) vectors. (B) RV and LV transduction efficiency measured in each T-cell subset by flow cytometry. (C) T-cell expansion (measured as fold increase) analyzed for each cell subset. (D) Representative FACS plots of CD45RA and CD62L expression. (E) Quantification of CD62L+CD45RA+ double-positive cells 16 days after initial activation (left) and longitudinal analysis of CD62L/CD45RA coexpression over 30-day culture (right) in each T-cell subset. Graphs depict the percentages of CD62L+CD45RA+ cells gated on CD8+ T cells. (F) Representative FACS plots of CD45RA and CD45R0 coexpression gated on CD62L+CD8+ T(TN), T(TCM), and T(TEM) subsets. (G) Quantification of CD62L+CD45RA+CD45R0+ triple-positive CD8+ cells 16 days after activation. (H) Absolute numbers of CD62L+CD45RA+CD8+ T cells generated from purified TN treated with different stimulation (bCD3/CD28 in black or OKT3 in gray) and culture conditions (IL-7 and IL-15 together, IL-7, IL-15, or IL-2 alone). (I) Percentages of CD62L+CD45RA+ cells (gated on CD8+ T cells) on bCD3/CD28-stimulated TN cells cultured with different cytokines. Mean data from at least 3 healthy donors are presented, and error bars represent SEM.
IL-7 and IL-15 instruct the expansion of a novel CD62L+CD45RA+ memory T-cell population from naive precursors. (A) Diagram summarizing the procedure to generate T(TN), T(TCM), and T(TEM) subsets. FACS-sorted TN (CD62L+CD45RA+), TCM (CD62L+CD45RA−), and TEM (CD62L−CD45RA−) lymphocytes were activated with anti-CD3/CD28 beads (bCD3/CD28) and cultured with IL-7 (5 ng/mL) and IL-15 (5 ng/mL). Forty-eight hours after activation, cells were transduced with retroviral (RV) or lentiviral (LV) vectors. (B) RV and LV transduction efficiency measured in each T-cell subset by flow cytometry. (C) T-cell expansion (measured as fold increase) analyzed for each cell subset. (D) Representative FACS plots of CD45RA and CD62L expression. (E) Quantification of CD62L+CD45RA+ double-positive cells 16 days after initial activation (left) and longitudinal analysis of CD62L/CD45RA coexpression over 30-day culture (right) in each T-cell subset. Graphs depict the percentages of CD62L+CD45RA+ cells gated on CD8+ T cells. (F) Representative FACS plots of CD45RA and CD45R0 coexpression gated on CD62L+CD8+ T(TN), T(TCM), and T(TEM) subsets. (G) Quantification of CD62L+CD45RA+CD45R0+ triple-positive CD8+ cells 16 days after activation. (H) Absolute numbers of CD62L+CD45RA+CD8+ T cells generated from purified TN treated with different stimulation (bCD3/CD28 in black or OKT3 in gray) and culture conditions (IL-7 and IL-15 together, IL-7, IL-15, or IL-2 alone). (I) Percentages of CD62L+CD45RA+ cells (gated on CD8+ T cells) on bCD3/CD28-stimulated TN cells cultured with different cytokines. Mean data from at least 3 healthy donors are presented, and error bars represent SEM.
T(TN) lymphocytes reveal distinct homing, survival, and functional properties compared with T(TCM) and T(TEM) cells
To further define the phenotype and function of T(TN) cells, we analyzed the expression of IL-7Rα, a marker for T-cell fitness and persistence,24,25 and of chemokine receptors implied in homeostatic (CXCR4) or inflammation-driven (CCR5) migration on T(TN), T(TCM), and T(TEM) CD8+ subsets. For T(TN), we analyzed the entire cellular population as well as the specific CD45RA+CD62L+ cellular fraction. IL-7Rα was expressed by a higher proportion of CD8+ T(TN) than T(TCM) and T(TEM) (P < .01; Figure 2A) for up to 30 days in culture (P < .001; Figure 2B). Likewise, CD8+ T(TN) expressed higher levels of CXCR4 than CD8+ T(TCM) and T(TEM), while CCR5 expression was lower in T(TN) than in CD8+ T(TCM) and T(TEM) cells (Figure 2C). CD8+ T(TN) displayed also a significantly lower expression of HLA-DR compared with other memory counterparts (P < .01; Figure 2C). In contrast, CXCR3 and IL-2Rβ were similarly expressed in all T-cell subsets (supplemental Figure 1 panels D and E, respectively).
T(TN) lymphocytes reveal distinct homing, survival, and functional properties compared with T(TCM) and T(TEM) cells. (A) Expression of IL-7Rα on T-cell subsets gated on CD8+ lymphocytes: representative FACS plot (left) and average histograms (right) including also the analysis performed on CD45RA+CD62L+ T(TN). (B) Longitudinal analysis of cell-surface IL-7Rα expression on CD8+ T cells. (C) Panel of cell-surface markers analyzed on CD8+ T(TN), T(TCM), and T(TEM) subsets. From top to bottom: CXCR4, CCR5, and HLA-DR. For each marker representative FACS plots (left) and average histograms [right, including also the analysis performed on CD45RA+CD62L+ T(TN)] are depicted. (D) Representative FACS plots of IL-2, IFN-γ, granzyme-A, and perforin production are shown. Plots are gated on CD8+ cells and quadrant frequencies are indicated. (E) Quantitation of IL-2, IFN-γ, and granzyme-A production by T(TN), T(TCM), and T(TEM) cells. Average percentages of CD8+ cytokine-producing cells are indicated. (F) Negative correlation of IFN-γ production with CD62L+CD45RA+ coexpression (top) and IL-7Rα cell-surface expression (bottom). All FACS analyses were performed 16 days after stimulation. Gray histograms represent fluorochrome-matched isotype controls. Mean data from at least 3 healthy donors are presented, and error bars represent SEM.
T(TN) lymphocytes reveal distinct homing, survival, and functional properties compared with T(TCM) and T(TEM) cells. (A) Expression of IL-7Rα on T-cell subsets gated on CD8+ lymphocytes: representative FACS plot (left) and average histograms (right) including also the analysis performed on CD45RA+CD62L+ T(TN). (B) Longitudinal analysis of cell-surface IL-7Rα expression on CD8+ T cells. (C) Panel of cell-surface markers analyzed on CD8+ T(TN), T(TCM), and T(TEM) subsets. From top to bottom: CXCR4, CCR5, and HLA-DR. For each marker representative FACS plots (left) and average histograms [right, including also the analysis performed on CD45RA+CD62L+ T(TN)] are depicted. (D) Representative FACS plots of IL-2, IFN-γ, granzyme-A, and perforin production are shown. Plots are gated on CD8+ cells and quadrant frequencies are indicated. (E) Quantitation of IL-2, IFN-γ, and granzyme-A production by T(TN), T(TCM), and T(TEM) cells. Average percentages of CD8+ cytokine-producing cells are indicated. (F) Negative correlation of IFN-γ production with CD62L+CD45RA+ coexpression (top) and IL-7Rα cell-surface expression (bottom). All FACS analyses were performed 16 days after stimulation. Gray histograms represent fluorochrome-matched isotype controls. Mean data from at least 3 healthy donors are presented, and error bars represent SEM.
In functional analyses, we found that a substantial proportion of CD8+ T(TN) secreted IL-2 in the absence of IFN-γ, and stained mostly negative for granzyme-A and perforin (Figure 2D-E). By sharp contrast, a larger proportion of CD8+ T(TCM) and T(TEM) cells produced IFN-γ, granzyme-A, and perforin (Figure 2D-E). Thus, T(TN) revealed a less differentiated phenotype compared with T(TCM) and T(TEM) cells and lacked immediate effector functions. Accordingly, IFN-γ production negatively correlated with CD45RA/CD62L or IL7-Rα surface expression (r2 = 0.8077 and P = .0002; r2 = 0.7954 and P = .0002, respectively; Figure 2F). Thus, genetically modified T(TN) cells display phenotypic and functional characteristics of early differentiated cells.
T(TN) cells proliferate, differentiate, and self-renew on antigen encounter
The phenotypic and functional profile suggests that T(TN) cells represent a population of antigen-experienced cells at an early stage of differentiation. Therefore, we evaluated their expansion potential, differentiation, and self-renewal abilities in an antigen-specific model. Purified T-cell subsets were activated with bCD3/CD28 and engineered to express a transgenic TCR specific for a HLA-A2–restricted peptide from the WT1 tumor antigen.17 TCR-transduced lymphocytes were then stimulated with T2 cells pulsed with cognate peptide (Figure 3A). Although all gene-modified cells proliferated at high and similar extent on antigen encounter (Figure 3B), T(TN) cells expanded significantly more than T(TCM) and T(TEM) (P < .05; Figure 3C). At day 16 after antigen-specific stimulation, TCR-transduced T cells were challenged with the cognate peptide and simultaneously assessed for cytokine production and memory subset composition. Of note, while the majority of T(TN) differentiated into TCM- and TEM-like cells (Figure 3D), a substantial fraction of T(TN) cells preserved the CD45RA+CD62L+ phenotype, which was absent in T(TCM) and T(TEM) counterparts. According to surface phenotype, within T(TN) cells, differentiated CD45RA+/−CD62L− T cells produced high amounts of IFN-γ (Figure 3E), while the CD45RA+CD62L+ T cells were characterized by enhanced IL-2 secretion and low IFN-γ production (P < .05; Figure 3F). Altogether, these data suggest that T(TN) are able to proliferate, differentiate, and self-renew on antigen encounter.
T(TN) cells proliferate, differentiate, and self-renew on antigen encounter. (A) Diagram summarizing the TCR transfer model: purified T-cell subsets were bCD3/CD28-stimulated and LV-transduced in the presence of IL-7 and IL-15, to express a TCR recognizing the HLA-A2–restricted WT1126-134 peptide. Transduced cells were stimulated with irradiated T2 cells pulsed with WT1126-134 peptide and tested for: (B) proliferation expressed as CFSE dilution, evaluated 5 days after stimulation; (C) expansion, expressed as absolute counts (left panel) and fold increase at day 16 (right panel); (D) memory subset compositions (based on CD45RA and CD62L expression) before and after transduction, and after antigen-specific stimulation. Stacked histograms represent mean values ± SEM of memory subset composition from 3 independent donors. (E) Representative FACS plots of the cytokine secretion profile of T(TN) cells gated on CD45RA+CD62L+ and CD45RA+/−CD62L− CD8+ lymphocytes, and (F) mean histograms from 3 independent donors. UT indicates untransduced cells; and LV+, TCR-transduced cells.
T(TN) cells proliferate, differentiate, and self-renew on antigen encounter. (A) Diagram summarizing the TCR transfer model: purified T-cell subsets were bCD3/CD28-stimulated and LV-transduced in the presence of IL-7 and IL-15, to express a TCR recognizing the HLA-A2–restricted WT1126-134 peptide. Transduced cells were stimulated with irradiated T2 cells pulsed with WT1126-134 peptide and tested for: (B) proliferation expressed as CFSE dilution, evaluated 5 days after stimulation; (C) expansion, expressed as absolute counts (left panel) and fold increase at day 16 (right panel); (D) memory subset compositions (based on CD45RA and CD62L expression) before and after transduction, and after antigen-specific stimulation. Stacked histograms represent mean values ± SEM of memory subset composition from 3 independent donors. (E) Representative FACS plots of the cytokine secretion profile of T(TN) cells gated on CD45RA+CD62L+ and CD45RA+/−CD62L− CD8+ lymphocytes, and (F) mean histograms from 3 independent donors. UT indicates untransduced cells; and LV+, TCR-transduced cells.
T(TN) engraft, expand, and exhibit xenoreactivity and self-renewing ability on transfer into immunodeficient mice
To further investigate the self-renewal potential of T(TN) cells, we moved to an in vivo setting and evaluated T(TN) ability to mediate xenogeneic GVHD26 in serial transplantation experiments. We adoptively transferred equal numbers (5 × 106 CD3+/mouse) of T(TN) and T(TCM) into irradiated (250 cGy) and NK-ablated immunodeficient mice. As controls, cohorts of mice were infused with unmanipulated FACS-sorted TN, TCM or with unselected lymphocytes in vitro activated with bCD3/CD28 plus IL-7 and IL-15 [T(PBLs)]. The CD4:CD8 ratios of infused cells were: T(TN), 0.71 ± 0.42; T(TCM), 1.65 ± 0.65; TN, 0.63 ± 0.24; TCM, 3.57 ± 1.73; T(PBLs), 2.12 ± 0.91.
One week after infusion, comparable frequencies of circulating human T cells were observed in all mice (supplemental Figure 3). After an additional week, T(TN), but not T(TCM) or T(PBLs) cells persisted in vivo to levels comparable with those of unmanipulated human T cells (Figure 4A). Human chimerism correlated with GVHD. Indeed, T(TN), but not T(TCM), induced severe GVHD in the majority of mice, with kinetics and intensity comparable with those observed in mice infused with unmanipulated cells (Figure 4B). At time of GVHD, the majority of circulating CD8+ T(TN) cells revealed a central memory (CD45RA−CD62L+) or effector memory (CD45RA−CD62L−) phenotype, indicating their ability to differentiate in vivo (Figure 4C). Of note, in T(TN)-infused mice, a fraction of lymphocytes retained CD45RA and CD62L coexpression up to the day of sacrifice, whereas CD45RA+CD62L+ cells could not be detected in mice that received other T-cell subsets. Circulating T(TN) cells showed higher frequencies of IL-7Rα, compared with T(TCM) (P < .001; Figure 4D). This correlated with a higher human CD3+ cell chimerism in the bone marrow (Figure 4E) and in the spleens of T(TN)-infused mice, which was approximately 5-fold higher than that found in mice infused with T(TCM) cells (Figure 4F). Thus, T(TN) cells engraft and expand in vivo and mediate a potent xeno-GVHD activity.
T(TN) cells engraft, expand, and exhibit xenoreactivity and self-renewing ability on transfer into immunodeficient mice. NOD/Scid mice were conditioned, infused IP with 5 × 106 human T lymphocytes, and followed for human chimerism and incidence of severe GVHD. (A) Flow cytometric analysis of human chimerism on peripheral blood (PB) at week 2 after infusion. (B) Mice were followed for severe GVHD-free survival over time (defined as weight loss greater than 5% concomitant to human chimerism greater than 10%). (C) Expression of CD45RA and CD62L on circulating human CD8+ T cells 2 weeks after infusion. FACS plots from representative T(TN)– and T(TCM)–infused mice (left) and average histograms (right) are shown. (D) Expression of IL-7Rα on circulating human CD8+ T cells from T(TN)– and T(TCM)–infused mice: representative plot (left) and average histograms (right). Gray histograms represent fluorochrome-matched isotype controls. (E) Percentages of human T cells retrieved from spleen and BM of T(TN)– and T(TCM)–infused mice at sacrifice. (F) Absolute numbers of human T cells retrieved per spleen of T(TN)– and T(TCM)–infused mice at sacrifice. The dashed line indicates the number of cells infused. (G) Results of serial transplantation experiments; 5 × 106 BM- and spleen-retrieved human T cells from first transplantation were infused into secondary recipients and human chimerism on PB was assessed 1 week after infusion. (H) GVHD-free survival in secondary recipients. (I) Representative plots of CD45RA/CD62L phenotype (left) and IL-7Rα (right) on circulating CD8+ T cells from T(TN) secondary recipients. (J) Human chimerism on BM and spleen at sacrifice.
T(TN) cells engraft, expand, and exhibit xenoreactivity and self-renewing ability on transfer into immunodeficient mice. NOD/Scid mice were conditioned, infused IP with 5 × 106 human T lymphocytes, and followed for human chimerism and incidence of severe GVHD. (A) Flow cytometric analysis of human chimerism on peripheral blood (PB) at week 2 after infusion. (B) Mice were followed for severe GVHD-free survival over time (defined as weight loss greater than 5% concomitant to human chimerism greater than 10%). (C) Expression of CD45RA and CD62L on circulating human CD8+ T cells 2 weeks after infusion. FACS plots from representative T(TN)– and T(TCM)–infused mice (left) and average histograms (right) are shown. (D) Expression of IL-7Rα on circulating human CD8+ T cells from T(TN)– and T(TCM)–infused mice: representative plot (left) and average histograms (right). Gray histograms represent fluorochrome-matched isotype controls. (E) Percentages of human T cells retrieved from spleen and BM of T(TN)– and T(TCM)–infused mice at sacrifice. (F) Absolute numbers of human T cells retrieved per spleen of T(TN)– and T(TCM)–infused mice at sacrifice. The dashed line indicates the number of cells infused. (G) Results of serial transplantation experiments; 5 × 106 BM- and spleen-retrieved human T cells from first transplantation were infused into secondary recipients and human chimerism on PB was assessed 1 week after infusion. (H) GVHD-free survival in secondary recipients. (I) Representative plots of CD45RA/CD62L phenotype (left) and IL-7Rα (right) on circulating CD8+ T cells from T(TN) secondary recipients. (J) Human chimerism on BM and spleen at sacrifice.
Given the rapid onset of lethal GVHD, we could not evaluate long-term persistence of T(TN) lymphocytes. We thus performed serial transplantations. BM- and spleen-retrieved human CD3+ cells (5 × 106 CD3+/mouse) from engrafted primary recipients were injected into irradiated and NK-ablated secondary mice. Circulating T lymphocytes were detected 1 week after infusion in secondary recipients of T(TN) cells. T(PBLs) also engrafted but to a lesser extent, while T(TCM) failed to be detected (P < .05; Figure 4G). Accordingly, only T(TN)-infused mice experienced lethal GVHD (P < .01; Figure 4H), characterized by a faster kinetics compared with first recipients. Most of circulating CD8+ lymphocytes in T(TN)-infused mice acquired a central memory phenotype (mean percentage of CD45RA−CD62L+ CD8+ T cells: 77.2%, SEM 0.96), with a small fraction of the cells maintaining IL-7Rα expression (mean percentage of IL-7Rα+ CD8+ T cells: 4.1%, SEM 0.53; Figure 4I). T(TN) proved able to home to the spleen and BM also in secondary recipients (Figure 4J). Thus, T(TN) cells engraft, expand, and exhibit xenoreactivity in serial transplantations, suggesting self-renewal abilities.
Molecular profiling defines a signature of memory differentiation shared by natural TCM, T(TN), and T(TCM) lymphocytes
To further characterize T(TN) cells, we performed a genome-wide microarray analysis of CD8+ T(TN) and T(TCM) subsets and compared them to the recently reported TSCM10 and to unmanipulated CD8+ TN and TCM as controls. We first compared the expression pattern of natural TN and TCM to overcome the noise secondary to transduction and in vitro culture of T(TN) and T(TCM). SAM algorithm identified a human memory signature defined by a set of 65 genes up- and down-regulated during the differentiation of naive CD8+ cells into memory lymphocytes (supplemental Table 1). The memory signature grouped TN, TCM, T(TN), and T(TCM) samples as depicted in Figure 5A. In particular, hierarchical clustering showed that T(TN) lymphocytes were partitioned in the memory branch of the dendrogram, indicating that T(TN) are part of the memory compartment. However, within the memory arm, while TCM and T(TCM) clustered together, T(TN) constituted a separate branch which still conserved some homology with unmanipulated TN cells. To precisely place T(TN) in the T-cell hierarchy, we analyzed their relationships with the other subsets. First, to further reinforce that T(TN) cells represent a memory subpopulation different from naive precursors, we used gene set enrichment analysis (GSEA). Specifically, we interrogated the genes of the CD8+ T-cell memory signature up-regulated in the TCM subset (TCM gene set; supplemental Table 1) and gene sets extracted from the Molecular Signature Database for statistical association between gene signatures and our T-cell subsets. If T(TN) cells represent a memory subset with a naive surface phenotype, then the TCM gene set of the CD8+ T-cell memory signature should be more enriched in T(TN) than in TN cells. Indeed, GSEA evidenced that the TCM gene set is significantly overrepresented in T(TN) (FDR q value = 0), confirming the clustering indication that T(TN) cells are relatively more differentiated than their natural counterpart (Figure 5B). Next, we sought to validate also at the molecular level that T(TN) are hierarchically superior to TCM and T(TCM). Indeed, a closer comparison of gene expression profiles between T(TN) and the more differentiated TCM and T(TCM) revealed that genes less represented in T(TN) have known roles in effector T-cell functions, (eg, EOMES, T-bet, NK receptors and granzyme family members; supplemental Table 2). All genes differentially expressed (2-fold cutoff and q value ≤ 0.05) between T(TN) and memory cells [TCM and T(TCM)] were functionally classified under broad categories based on information of the Gene Ontology and of the Molecular Signature Database (enrichment analysis using GSEA). These analyses showed that T-cell differentiation and inflammation-related pathways are inversely correlated to the T(TN) phenotype (Figure 5C). Thus, by gene profiling analysis, TN-derived CD62L+CD45RA+CD45R0+ T cells prove to represent a novel memory subset distinct and hierarchically superior to TCM and T(TCM) cells and similar to naturally occurring putative TSCM (supplemental Figure 4). Finally, we validated selected genes from a list of cluster of differentiation (CD) molecules differentially expressed in TN compared with unmanipulated TCM (supplemental Table 3) by flow cytometry. We confirmed the differential expression of CCR7 and CCR5 in T(TN) and T(TCM). Among the markers screened, we identified CD95, a molecule associated with memory T cells,24 as one of the top-ranked antigens expressed by TCM and not by TN (supplemental Table 3). Flow cytometric analysis confirmed optimal expression of CD95 on T(TN), T(TCM), and TCM memory cells (Figure 5D-E). In contrast, only a minority of natural CD8+CD45RA+CD62L+ TN cells expressed CD95 (Figure 5D-E).
Molecular profiling defines a signature of memory differentiation shared by natural TCM, T(TN), and T(TCM) lymphocytes. (A) Comparison of the expression pattern of natural TN and TCM identifies a human memory phenotype signature defined by a set of 65 genes up- and down-regulated during the differentiation of naive CD8+ T cells into memory lymphocytes. Hierarchical clustering of T(TN) and T(TCM) within the conserved CD8+ T-cell memory signature classifies T(TN) lymphocytes as a separate subgroup of the memory branch. (B) Enrichment profile of genes of the CD8+ T-cell memory signature (up-regulated in the TCM gene set) in TN versus T(TN) cells. GSEA evidences that the TCM gene set is significantly overrepresented in T(TN) cells (FDR q value = 0) compared with TN. (C) All genes differentially expressed (2-fold cutoff and q value ≤ 0.05) between T(TN) and memory lymphocytes (TCM and T(TCM); supplemental Table 2) are functionally classified under broad categories based on information of the Gene Ontology (pie chart) and of the Molecular Signature Database (enrichment analysis using GSEA). The normalized enrichment score (NES) indicates that all 11 pathways statistically enriched (at FDR q value ≤ 25%) are inversely correlated to the T(TN) phenotype. (D) Representative FACS plots of CD95 expression on TN, T(TN), TCM, and T(TCM). Cells are gated on CD8. Gray histograms represent fluorochrome-matched isotype controls. (E) Quantification of CD95+ cells among the different subsets.
Molecular profiling defines a signature of memory differentiation shared by natural TCM, T(TN), and T(TCM) lymphocytes. (A) Comparison of the expression pattern of natural TN and TCM identifies a human memory phenotype signature defined by a set of 65 genes up- and down-regulated during the differentiation of naive CD8+ T cells into memory lymphocytes. Hierarchical clustering of T(TN) and T(TCM) within the conserved CD8+ T-cell memory signature classifies T(TN) lymphocytes as a separate subgroup of the memory branch. (B) Enrichment profile of genes of the CD8+ T-cell memory signature (up-regulated in the TCM gene set) in TN versus T(TN) cells. GSEA evidences that the TCM gene set is significantly overrepresented in T(TN) cells (FDR q value = 0) compared with TN. (C) All genes differentially expressed (2-fold cutoff and q value ≤ 0.05) between T(TN) and memory lymphocytes (TCM and T(TCM); supplemental Table 2) are functionally classified under broad categories based on information of the Gene Ontology (pie chart) and of the Molecular Signature Database (enrichment analysis using GSEA). The normalized enrichment score (NES) indicates that all 11 pathways statistically enriched (at FDR q value ≤ 25%) are inversely correlated to the T(TN) phenotype. (D) Representative FACS plots of CD95 expression on TN, T(TN), TCM, and T(TCM). Cells are gated on CD8. Gray histograms represent fluorochrome-matched isotype controls. (E) Quantification of CD95+ cells among the different subsets.
CD95 allows the identification of the T(TN) natural counterpart in healthy donors and patients who received transplants
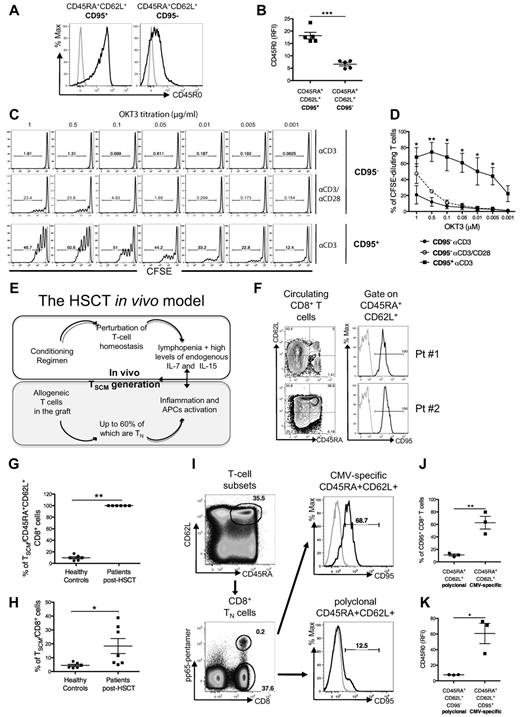
Among circulating CD45RA+CD62L+CD8+cells harvested from healthy donors, CD95+ cells expressed higher levels of CD45R0 than CD95− cells (measured as relative fluorescence intensity [RFI], P < .001; Figure 6A-B), thus resembling in vitro–generated T(TN) cells. Furthermore, FACS-sorted CD45RA+CD62L+CD95+ CD8+ lymphocytes, but not their CD95− counterpart, proliferated at very low concentrations of plate-bound anti-CD3 antibody in the absence of costimulation, recapitulating a memory hallmark (Figure 6C-D). Thus, a distinct subset of CD45RA+CD62L+CD45R0dim CD95+ cells displaying a naive surface phenotype associated with memory functional features can be identified in vivo.
CD95 helps the identification of T(TN) natural counterpart in vivo, in healthy donors and in recipients of allogeneic HSCT. Circulating CD95+CD45RA+CD62L+ cells express higher levels of CD45R0 compared with their CD95− counterpart: (A) representative FACS plots and (B) its quantification. Functional validation of CD95+CD45RA+CD62L+ cells: purified CFSE-labeled CD95+ and CD95− CD45RA+CD62L+ were challenged with decreasing doses of anti-CD3 antibody with or without costimulation. (C) Representative FACS plots of CFSE dilution and (D) quantification of CFSE dilution. (E) Proposed model for TSCM in vivo generation after HSCT: the conditioning regimen causes severe host lymphopenia, creating a milieu in which infused allogeneic lymphocytes are exposed to elevated levels of IL-7 and IL-15. Conditioning regimen favors systemic inflammation, leading to the activation of antigen-presenting cells (APCs), which become costimulation competent. (F) Representative plots of circulating CD8+ lymphocytes subpopulations (left) and CD95 expression on CD45RA+CD62L+ putative naive CD8+ cells (right) from 2 representative HSCT recipients. Gray histograms represent fluorochrome-matched isotype controls. (G) Proportion of CD45RA+CD62L+ CD8+ T lymphocytes expressing CD95 (TSCM) in HSCT recipients and healthy controls. (H) Proportion of CD45RA+CD62L+CD95+ lymphocytes gated on CD8+ T cells in HSCT recipients and healthy controls. (I) CD95 expression on antigen-specific T cells. Circulating T lymphocytes are gated on the CD45RA+CD62L+ population (top left plot) and then CMV pp65-specific and polyclonal CD8+ T cells are identified (bottom left plot). CD95 expression is enriched on CMV pp65-specific (top right plot) compared with polyclonal (bottom right plot) CD45RA+CD62L+ lymphocytes. (J) Scatter plot showing CD95 differential expression on CMV pp65-specific and polyclonal CD45RA+CD62L+ lymphocytes, gated on CD8+. (K) Scatter plot showing expression levels of CD45R0 on CMV pp65-specific CD45RA+CD62L+CD95+ lymphocytes and naive CD45RA+CD62L+CD95− lymphocytes, gated on CD8+.
CD95 helps the identification of T(TN) natural counterpart in vivo, in healthy donors and in recipients of allogeneic HSCT. Circulating CD95+CD45RA+CD62L+ cells express higher levels of CD45R0 compared with their CD95− counterpart: (A) representative FACS plots and (B) its quantification. Functional validation of CD95+CD45RA+CD62L+ cells: purified CFSE-labeled CD95+ and CD95− CD45RA+CD62L+ were challenged with decreasing doses of anti-CD3 antibody with or without costimulation. (C) Representative FACS plots of CFSE dilution and (D) quantification of CFSE dilution. (E) Proposed model for TSCM in vivo generation after HSCT: the conditioning regimen causes severe host lymphopenia, creating a milieu in which infused allogeneic lymphocytes are exposed to elevated levels of IL-7 and IL-15. Conditioning regimen favors systemic inflammation, leading to the activation of antigen-presenting cells (APCs), which become costimulation competent. (F) Representative plots of circulating CD8+ lymphocytes subpopulations (left) and CD95 expression on CD45RA+CD62L+ putative naive CD8+ cells (right) from 2 representative HSCT recipients. Gray histograms represent fluorochrome-matched isotype controls. (G) Proportion of CD45RA+CD62L+ CD8+ T lymphocytes expressing CD95 (TSCM) in HSCT recipients and healthy controls. (H) Proportion of CD45RA+CD62L+CD95+ lymphocytes gated on CD8+ T cells in HSCT recipients and healthy controls. (I) CD95 expression on antigen-specific T cells. Circulating T lymphocytes are gated on the CD45RA+CD62L+ population (top left plot) and then CMV pp65-specific and polyclonal CD8+ T cells are identified (bottom left plot). CD95 expression is enriched on CMV pp65-specific (top right plot) compared with polyclonal (bottom right plot) CD45RA+CD62L+ lymphocytes. (J) Scatter plot showing CD95 differential expression on CMV pp65-specific and polyclonal CD45RA+CD62L+ lymphocytes, gated on CD8+. (K) Scatter plot showing expression levels of CD45R0 on CMV pp65-specific CD45RA+CD62L+CD95+ lymphocytes and naive CD45RA+CD62L+CD95− lymphocytes, gated on CD8+.
We also investigated the representation of CD45RA+CD62L+CD45R0dimCD95+ cells in patients with hematologic malignancies treated with allogeneic HSCT. This in vivo clinical setting is characterized by systemic inflammation and severe host lymphopenia.27 Alloreactive TN lymphocytes present in the graft are exposed to costimulation-competent antigen-presenting cells and elevated levels of IL-728 and IL-15,29,30 an environment that closely mimic the culture conditions adopted in our study (Figure 6E). Results indicate that 1 month after HSCT, the majority of circulating CD45RA+CD62L+CD8+ T lymphocytes express CD95 (P < .01; Figure 6F-G), and represent a high fraction of CD8+ T cells (P < .05; Figure 5H), if compared with healthy controls. Finally, we investigated whether CD45RA+CD62L+CD45R0dimCD95+ could also be defined within antigen-specific T cells. To this, we studied the phenotype of cytomegalovirus (CMV) pp65(NLVPMVATV)–specific CD8+ T cells in seropositive HLA-A02+ donors (Figure 6I), and found that within CD45RA+CD62L+CD8+ cells, CMV-specific T cells were significantly enriched for CD95+ lymphocytes compared with polyclonal cells (P < .01; Figure 6J). Furthermore, CD95+CD45RA+CD62L+CD8+ CMV-specific lymphocytes expressed significantly higher levels of CD45R0 compared with CD95− polyclonal counterparts. Thus, these data support the existence of CD45RA+CD62L+CD45R0+ CD95+ lymphocytes in vivo, and their accumulation in an IL-7/IL-15–enriched milieu. Finally, having identified CD95 as the discriminative marker for naive and memory CD45RA+CD62L+ lymphocytes, we formally proved that T(TN)/TSCM could be differentiated from natural CD95− TN cells. CD95+CD62L+CD45RA+CD45R0+ T cells were originated by bCD3/CD28 stimulation in the presence of IL-7 and IL-15 starting from purified naive CD95− precursors. Expansion, sensitivity to genetic manipulation, and phenotypic and functional features of CD62L+CD45RA+CD45R0+ T cells differentiated from CD95− naive precursors were similar to those of T(TN) lymphocytes generated from unfractionated CD62L+CD45RA+ cells (supplemental Figure 5, Figures 1-2). Likewise, the critical role of costimulation, IL-7, and IL-15 for the instruction of postmitotic CD62L+CD45RA+CD45R0+CD95+ T cells was confirmed starting from purified naive CD95− precursors (supplemental Figure 6). Costimulation was necessary for CD95− TN cell expansion regardless of the cytokine used (supplemental Figure 6A-C). IL-7 was instrumental for the generation of T(TN)/TSCM cells (supplemental Figure 6D), but their expansion required either IL-15 or IL-2 (supplemental Figure 6A). Importantly, IL-15 proved superior to IL-2 in supporting expansion coupled to preservation of the TSCM phenotype (P < .05; supplemental Figure 6B). Notably, interference with the glycogen synthase kinase-3β/Wnt pathway was significantly less effective than the combination of IL-7/IL-15 in expanding and sustaining TSCM cells generated from naive precursors (supplemental Figure 7). Altogether, our data support the existence of CD45RA+CD62L+CD45R0+CD95+ lymphocytes with stem-cell memory-like functions that differentiate from naive precursors and accumulate preferentially in the presence of IL-7 and IL-15. TSCM cells can be easily differentiated, expanded, and genetically modified thus representing optimal candidates for adoptive T-cell therapy.
Discussion
We identified a postmitotic CD8+ lymphocyte population, generated in vitro from TN precursors but yet traceable in nature, characterized by lack of immediate effector functions, high expansion potential, and self-renewal abilities. T(TN) are best defined as CD8+ lymphocytes expressing the CD45RA+CD45R0+CD62L+CCR7+CD95+ phenotype, and can be either induced starting from naive precursors in response to CD3 and CD28 engagement in the presence of IL-7 and IL-15 or isolated from the peripheral blood of healthy subjects. In clinical conditions characterized by high availability of IL-7 and IL-15, such as in HSCT recipients, TSCM cells are enriched.
The existence and origin of memory T cells with stem cell–like qualities have been debated for some time.6,7 In mice, memory stem T cells with the capacity of self-renewal and differentiation into TCM and TEM subsets have been identified among antigen-experienced CD44low CD62Lhigh CD8+ T cells.8,9 In humans, memory T cells with stem cell–like qualities have been recently reported.10 Recently described CCR7+CD62L+CD45RA+CD45R0−CD27+CD28+IL-7Rα+ putative TSCM cells mediated superior antitumor responses than other memory T cells in adoptive T-cell therapy attempts, thus representing valuable candidates for adoptive T-cell therapies. The naive-derived CD45RA+CD45R0+CD62L+CCR7+IL-7Rα+CD95+ lymphocytes described here (T(TN)) share many of the features reported for TSCM lymphocytes, as they preserved the CD45RA+CD62L+ phenotype and refrained to acquire effector functions. They solely differ from putative TSCM for CD45R0 expression. This discrepancy could be plausibly ascribed to the differences in the activation status at the time of CD45R0 assessment. While T(TN) are analyzed at the peak of their activation (ie, 16 days after stimulation), putative TSCM are circulating lymphocytes and therefore reasonably resting T cells. Indeed, we observed that putative TSCM, once activated, up-regulate CD45R0 (supplemental Figure 7C), supporting the hypothesis that CD45R0 expression in TSCM fluctuates as a function of T-cell activation status.
Here, we show that it is possible to generate, expand, and gene-modify TSCM lymphocytes, while preserving their critical stem cell–like features. Most importantly, we demonstrate for the first time, in serial transplantation studies, the multipotency, the differentiation ability, and possibly the self-renewal capacity of T(TN)/TSCM lymphocytes, declined in a clinically relevant setting, such as xeno-GVHD. Notably, while gene expression profiling clusters our expanded T(TN) cells with recently reported natural TSCM cells10 (supplemental Figure 4), in a direct comparison with the alternative proposed method for ex vivo TSCM generation,10 we show that costimulation, coupled to IL-7 and IL-15, is superior to glycogen synthase kinase-3β/Wnt inhibitors in promoting the expansion and transduction of T(TN)/TSCM cells (supplemental Figure 7). In addition, gene-modified T(TN) cells described here express markers predictive of clinical outcome in adoptive immunotherapy trials31,32 such as IL-7Rα, CD27, and CD28.
We identified a natural counterpart of in vitro–expanded cells, as the analysis of healthy subjects and of CMV+ donors revealed that CD45RA+CD45R0dimCD62L+CCR7+CD95+ lymphocytes represent a natural T-cell subset, distinct from naive and memory cells for phenotypic appearance, and clearly behaving as memory lymphocytes, for activation requirements.33 In contrast to TN, TSCM cells can proliferate in the absence of costimulation. We compared different activation stimuli and cytokines for the ability to foster TSCM generation and expansion in vitro. While CD28 costimulation and IL-15 were critical for optimal expansion of these cells, in line with their role in homeostasis and proliferation of CD8+ T cells,34-37 IL-7 appeared uniquely capable of instructing lymphocytes toward the stem-like phenotype. Accordingly, the finding that IL-7Rα was preserved by self-renewing T(TN) in serial transplantation experiments in vivo further supports IL-7 as a critical determinant for TSCM generation and maintenance. Among other possibly relevant signaling events, the glycogen synthase kinase-3β/Wnt pathway has been proposed as critical for the generation and maintenance of memory and stem cell–like properties. However, we failed to record up-regulation of Wnt-dependent genes in the T(TN) gene-profiling analysis. This might indicate the existence of posttranscriptional mechanisms of regulation or of alternative signaling pathways, possibly emanating from the IL-7R. Future challenges will be needed indeed to define the molecular pathways controlling TSCM generation. IL-7 has a pivotal role in controlling the size of the lymphoid population, and T cells compete for a limited amount of constitutively produced IL-7.38,39 Hence, it is plausible that, during a physiologic primary immune response, in which proinflammatory factors prevail, only the few TN cells that do not down-regulate IL-7Rα and gain access to IL-7, might give rise to TSCM cells. By contrast, our clinical-grade culture system, in which access to IL-7 is not limited, may enable a higher fraction of cells to differentiate into functional TSCM. In the HSCT clinical setting, IL-7 bioavailability is increased because of lymphodepletion28 in a context of systemic inflammation and immune activation27 that closely recapitulates our in vitro model. In this scenario, we documented a selective expansion of the TSCM compartment, which represented the totality of CD45RA+CD62L+ putative naive CD8+ lymphocytes. This clinical observation endorses the central role of IL-7 in TSCM generation.
Our study raises important questions on the mechanism of memory generation and maintenance. Our results suggest a precursor-product relationship between the CD45RA+CD62L+CD95+ postmitotic T(TN) cells and the other memory subsets, consistent with a linear differentiation model in which TN cells may differentiate first to TSCM, then to TCM and TEM cells. However, we cannot exclude that in vivo priming of TN cells could result in the generation of a mixed population of effector and memory cells, perhaps by asymmetric division, as suggested by other groups.40,41
An important aim of our study was to identify the optimal source of cells to be used for the genetic manipulation of lymphocytes suitable for adoptive T-cell therapy (ACT).12,42,43 When we directly compared FACS-sorted TN, TCM, and TEM, only TN cells gave rise to significant numbers of TSCM-like cells. Accordingly, T(TN) revealed higher in vivo engraftment, persistence, and xenoreactivity when directly compared with T(TCM) cells in primary and secondary transplant recipients. Thus, targeting naive T cells might have a direct impact on ACT.
In recent years, genetic manipulation of T cells produced important advances in the treatment of cancer patients. Expression of suicide genes, tumor-specific TCR, or chimeric antigen receptors, have significantly increased the safety and efficacy profile of T-cell products, and already produced objective clinical responses.44-46 Recent technological advances, including the use of zinc finger nucleases, allow today a sophisticated genetic surgery of T lymphocytes, aimed at substituting, instead of only adding, specific biologic functions, such as antigen specificity.16 In this context, unique in the ability to couple antitumor efficacy with enhanced safety, preservation of a TSCM functional phenotype will be critical for the generation of cellular products not only able to efficiently target cancer cells, but also capable of persisting and mediating a dynamic cancer immunosurveillance in treated patients. A rapid implementation of our protocol will be ensured by the availability of clinical-grade reagents required for T(TN) generation and genetic manipulation. Thus, TSCM cells could be readily edited with tumor antigen specificities and expanded according to our clinical-grade protocol, while preserving their expansion, multipotency, and differentiation abilities.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof T. Tanaka (Osaka University, Japan) for kindly providing the TMβ-1 hybridoma.
This work was supported by the Italian Ministry of Health (GR07-5 BO and RO10/07-B-1), the Italian Ministry of Research and University (FIRB-IDEAS, linked to ERC starting grants), Fondazione Cariplo, the Italian Association for Cancer Research (AIRC, AIRC 5x1000), and the EU-FP7 program (ATTACK, PERSIST).
N.C. conducted this study as partial fulfillment of the PhD in Cellular and Molecular Biology, San Raffaele University (Milan, Italy). G.O. conducted this study as partial fulfillment of the PhD in Molecular Medicine, San Raffaele University (Milan, Italy).
Authorship
Contribution: N.C. and C. Bonini designed the study and wrote the manuscript; N.C. conducted laboratory experiments; B.C., E.P., and A.B. helped to perform in vivo experiments; F. Cocchiarella, F.M., and A.R. performed microarrays; M.F. and S.B. performed bioinformatics analysis; G.O. performed and analyzed sjTREC quantification; C. Bordignon and A.M. contributed to manuscript drafting; and J.P., F. Ciceri, and M.T.L.-S. provided clinical data and samples.
Conflict-of-interest disclosure: C. Bordignon is an employee of MolMed SpA. C. Bonini has a research contract with MolMed SpA. The remaining authors declare no competing financial interests.
Correspondence: Dr Chiara Bonini, Experimental Hematology Unit, San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy; e-mail: bonini.chiara@hsr.it.


![Figure 2. T(TN) lymphocytes reveal distinct homing, survival, and functional properties compared with T(TCM) and T(TEM) cells. (A) Expression of IL-7Rα on T-cell subsets gated on CD8+ lymphocytes: representative FACS plot (left) and average histograms (right) including also the analysis performed on CD45RA+CD62L+ T(TN). (B) Longitudinal analysis of cell-surface IL-7Rα expression on CD8+ T cells. (C) Panel of cell-surface markers analyzed on CD8+ T(TN), T(TCM), and T(TEM) subsets. From top to bottom: CXCR4, CCR5, and HLA-DR. For each marker representative FACS plots (left) and average histograms [right, including also the analysis performed on CD45RA+CD62L+ T(TN)] are depicted. (D) Representative FACS plots of IL-2, IFN-γ, granzyme-A, and perforin production are shown. Plots are gated on CD8+ cells and quadrant frequencies are indicated. (E) Quantitation of IL-2, IFN-γ, and granzyme-A production by T(TN), T(TCM), and T(TEM) cells. Average percentages of CD8+ cytokine-producing cells are indicated. (F) Negative correlation of IFN-γ production with CD62L+CD45RA+ coexpression (top) and IL-7Rα cell-surface expression (bottom). All FACS analyses were performed 16 days after stimulation. Gray histograms represent fluorochrome-matched isotype controls. Mean data from at least 3 healthy donors are presented, and error bars represent SEM.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/4/10.1182_blood-2012-05-431718/4/m_zh89991200920002.jpeg?Expires=1767886098&Signature=L7hRGHiJycV3hkxWE6TgGPR~gMKaeR~y3RJ-OxWelCRB~rOwQrFRFUVNMg8j5ckPCivmhDcGhnykBy0qUlt2uo45o6c1gbAc7sogjC4ouTDA5ahEWrs0tdb3m9kmRtzICyH8lWHDbEzVxLGjw-DDW~GTw9MvPf5F0IZMDFyunMWeFbZRj7r06~dMlV7V~IFKwMAY7UmU03bSYwJXBY0xSBtbcVj4oxHLfgILvZwSz1~dnfWK8nuG4rT6KPoZN8Zy8pI3L6X7RuSwW3R-Nltq6rYK8af-gkbJmMVma~QiYJJnDSKVeFithMib-bG7sCUi0Xn1LOT~1MFWrW23CXS48Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)