Key Points
Different pathogen-specific CD4 T cells manifest remarkable difference in susceptibility to HIV infection.
Distinct gene-expression profiles of pathogen-specific CD4 T cells are associated with their susceptibilities to HIV infection.
Abstract
In HIV infection, CD4 responses to opportunistic pathogens such as Candida albicans are lost early, but CMV-specific CD4 response persists. Little is currently known about HIV infection of CD4 T cells of different pathogen/antigen specificities. CFSE-labeled PBMCs were stimulated with CMV, tetanus toxoid (TT), and C albicans antigens and subsequently exposed to HIV. HIV infection was monitored by intracellular p24 in CFSElow population. We found that although TT- and C albicans–specific CD4 T cells were permissive, CMV-specific CD4 T cells were highly resistant to both R5 and X4 HIV. Quantification of HIV DNA in CFSElow cells showed a reduction of strong-stop and full-length DNA in CMV-specific cells compared with TT- and C albicans–specific cells. β-Chemokine neutralization enhanced HIV infection in TT- and C albicans–specific cells, whereas HIV infection in CMV-specific cells remained low despite increased entry by β-chemokine neutralization, suggesting postentry HIV restriction by CMV-specific cells. Microarray analysis (Gene Expression Omnibus accession number: GSE42853) revealed distinct transcriptional profiles that involved selective up-regulation of comprehensive innate antiviral genes in CMV-specific cells, whereas TT- and C albicans–specific cells mainly up-regulated Th17 inflammatory response. Our data suggest a mechanism for the persistence of CMV-specific CD4 response and earlier loss of mucosal Th17-associated TT- and C albicans–specific CD4 response in AIDS.
Introduction
HIV infection is characterized by a progressive depletion of CD4 T cells, ultimately leading to the onset of a variety of opportunistic infections and AIDS.1-3 Although the cause of in vivo CD4 depletion is highly complex, direct HIV infection and robust viral replication is considered to be one of the major factors that drives the depletion of memory CD4 T cells.4,5 Previous studies have reported that HIV infects various subsets of CD4 T cells, such as central memory (CM; CD45RA−CCR7+CD27+) and transitional memory (CD45RA−CCR7−CD27+) CD4 T cells.6-8 It was shown more recently that CXCR3+CCR6+ and CCR4+CCR6+ peripheral CD4 T cells were preferentially infected by HIV.9 Despite considerable advances into the impact of HIV infection on total memory CD4 cells and their subsets, little is known about how HIV infects CD4 T cells with different pathogen/antigen specificity.
HIV infection is associated with progressive and ordered susceptibility to opportunistic pathogens. For example, resistance to Candida species and to Mycobacterium tuberculosis (MTB) is typically lost early when total CD4 T-cell counts remain relatively high.10-13 In contrast, CMV disease only becomes evident in late-stage patients when total CD4 T-cell numbers are low.14-16 The underlying mechanism of this temporal hierarchy of immunodeficiency is less clear. Given the importance of the CD4 T-cell response in control of these infections, it has been proposed that memory CD4 populations specific to different opportunistic pathogens are depleted at different rates by HIV. It was shown recently that in chronic HIV infection, CMV-specific CD4 T cells remained readily detectable in the periphery, whereas concurrently the MTB-specific CD4 response was lost.13,16-18 However, it remains unknown whether differential depletion of pathogen-specific CD4 T cells in vivo is attributable to their differences in susceptibility to HIV infection and viral replication or to other bystander effects. There are no data comparing the susceptibility of different pathogen-specific CD4 populations to productive HIV infection. Furthermore, Hansen et al reported recently that a rhesus CMV-based simian immunodeficiency virus (SIV) vaccine could elicit a persistent effector memory T-cell response that was insensitive to CD4 depletion and stringently controlled SIV infection.19 These observations prompted an examination of the susceptibility of pathogen-specific CD4 T cells to HIV infection.
Using an in vitro system in which HIV infection of antigen-specific CD4 T cells in the same PBMC was determined by intracellular p24 expression in CFSElow, proliferating CD4 T cells, we have shown herein that although tetanus toxoid (TT)– and Candida albicans–specific CD4 T cells were permissive, CMV-specific CD4 T cells were highly resistant to both R5 and X4 HIV independent of co-receptor usage. Microarray analysis further identified distinct gene-expression profiles associated with differential susceptibility of pathogen-specific CD4 T cells to HIV infection.
Methods
Cells, viruses, and antigens
PBMCs from HIV− healthy subjects enrolled in an institutional review board–approved protocol (RV229) at Walter Reed Army Institute of Research were maintained in complete RPMI medium (Invitrogen) supplemented with 10% human serum, 100 U/mL of penicillin G, 100U/mL of streptomycin sulfate, and 1.7mM sodium glutamine. PBMC samples with positive responses to CMV, TT, and C albicans were chosen for this study (n = 6). R5 HIV (US1) and X4 HIV (92/[MU]G/029) were obtained from the US Military HIV Research Program. Antigens included CMV lysates (Advanced Biotechnologies), pp65 peptides (JPT Peptide Technology), TT (Statens Serum Institut) and C albicans sonicate (Greer Laboratories). This study was conducted in accordance with the Declaration of Helsinki.
CFSE staining, antigen stimulation, and HIV infection of PBMCs
PBMCs were CFSE labeled as described previously with slight modifications.20 A total of 30 × 106 PBMCs were washed and stained in 1 mL of 1% normal human serum–RPMI medium containing 1μM CFSE for 8 minutes at 25°C. Cells were then quenched with 2 mL of warm normal human serum for 5 minutes. CFSE-labeled PBMCs were pulsed with antigens (5 μg/mL of CMV, 25 μg/mL of TT, and 1:200 C albicans) at a concentration of 10 × 106 cells/mL for 4 hours at 37°C, followed by dilution to 2 × 106 cells/mL for culture for 3-4 days. Unstimulated PBMCs were included as a control. Cells were infected with pretitrated R5 HIV or X4 HIV and free viruses were washed 24 hours after HIV exposure. Three days after infection, cells were subjected to flow cytometric p24 analysis. In some experiments, anti–MIP-1α (5 μg/mL; clone 93321; R&D Systems), anti–MIP-1β (5μg/mL; clone 24006; R&D Systems), or anti-RANTES (5μg/mL; clone 21418; R&D Systems) was added into culture alone or in combination.
TZM-bl infection
TZM-bl cells were plated at the concentration of 5 × 104 cells/mL. Twelve hours later, supernatants containing HIV virus were added at 1:2 serial dilutions in the presence of 40 μg/mL of DEAE-dextran hydrochloride (Sigma-Aldrich). Virus infectivity was determined 48 hours after inoculation by measuring the level of firefly luciferase activity expressed in infected cells (Bright-Glo; Promega).
Flow cytometric intracellular p24 analysis
Antigen-stimulated and HIV-infected PBMCs were stained with aqua blue (Invitrogen) and antibody cocktails to surface antigens, including CD4 (Beckman Coulter), CD8, CD14, CD19, CD25, CD27, CD45RO, CD57, CCR5, or CXCR4 (all BD Biosciences), and PD-1 (eBiosciences). Cells were then fixed, permeabilized (BD Biosciences) and stained for CD3 (BD Biosciences) and p24 (Beckman Coulter). Antibody capture compensation beads (BD Biosciences) stained with individual antibodies were acquired for compensation. Data were analyzed using FlowJo Version 9.2 software (TreeStar).
Cell sorting
PBMCs were divided into 2 aliquots. One aliquot was infected with HIV. Twenty-four hours later, cells were fixed and stained with aqua blue and an antibody cocktail including CD4, CD3, CD8, and CD14/19. The other aliquot contained uninfected, nonfixed PBMCs. Live cells were stained with aqua blue and the same antibody cocktail as that for HIV-infected PBMCs. CFSElowCD3+CD8− T cells were sorted with a FACSAria cell sorter (BD Biosciences).
Quantification of cell-associated HIV DNA
Sorted, HIV-infected CD4 T cells were subject to DNA extraction using crude cell lysis buffer with freshly added proteinase K (1 mg/mL). DNA quantification was performed using 2× TaqMan Universal PCR Master Mix and the 7500 Real Time PCR System (Applied Biosystems). Cycling parameters were: 95°C for 10 minutes, 50 cycles of 95°C for 15 seconds, and 60°C for 1 minute. Amplifications were multiplexed with a GAPDH primer/probe set to both normalize sample input and serve as a DNA integrity control. Sequences for primers (Sigma-Aldrich) and probes (Sigma Aldrich and Applied Biosystems) are shown in Table 1. The final primer/probe concentrations were 100/200nM for HIV DNA and 75/100nM for GAPDH. Normalized relative target expression was calculated as the fold difference from cognate control values by 2(−ΔΔCt), where ΔΔCt = (ΔCt of the sample) − (ΔCt of the control); ΔCt = (average Ct of HIV target) − (average Ct of corresponding GAPDH).
Microarray data acquisition and analysis
RNA was extracted from sorted CFSElow CD4 cells using the RNeasy Plus Mini Kit (QIAGEN). RNA reverse transcription and synthesis of biotin-labeled amplified RNA were performed using the GeneChip IVT Express Kit (Affymetrix). aRNA was fragmented and hybridized to GeneChip Human Genome-U133 plus 2.0 array (Affymetrix; Gene Expression Omnibus accession number: GSE42853). The array was then washed, stained with a PE-streptavidin conjugate, and scanned. Data processing and analysis were performed using the R computing environment Version 2.12.2 with BioConductor packages. Gene-expression data were normalized into Robust Multichip Average expression measures21 and were compared between antigen specificities (CMV vs combined TT and C albicans). Statistical analysis was performed using Significance Analysis of Microarrays (SAM) Version 2.0.22 The false discovery rate (FDR) or the expected proportion of false positives among all significant genes was estimated based on SAM scores. Genes with an FDR < 0.05 were considered significant. Functional category and gene ontology enrichment analysis were performed using the online tool DAVID.
Statistical analysis for nonmicroarray data
Data were summarized as means and SD or using box and whisker plots. Statistical analysis was performed using Prism Version 5.0c software (GraphPad). Comparisons of 2 groups (ie, CMV vs TT and CMV vs C albicans) were performed using the Mann-Whitney test. P < .05 was considered significant.
Results
CMV-specific CD4 T cells are highly resistant to both R5 and X4 HIV infection, whereas TT- and C albicans–specific CD4 T cells are permissive
To compare the susceptibility of different pathogen-specific CD4 T cells to HIV infection, CFSE-labeled PBMCs from the same HIV− donors were stimulated with different pathogen-specific antigens for 3-4 days and then exposed to HIV. Antigens were chosen to cover viral, bacterial, and fungal pathogens. HIV infection of antigen-specific, proliferating CD4 T cells was determined by monitoring the intracellular p24 in CFSElow cells. The proliferative response of CFSElow cells was mainly driven by antigen-specific stimulation with small bystander proliferation, because no proliferation was observed when PBMCs from donors with no CD4 recall responses to that particular antigen were stimulated. Furthermore, after one round of antigen-specific stimulation, the CFSElow cells responded rapidly to a second stimulation with the same antigen by expressing the same cytokine (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
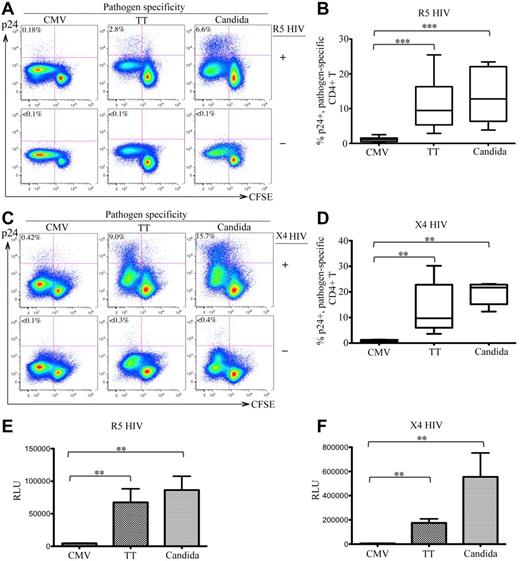
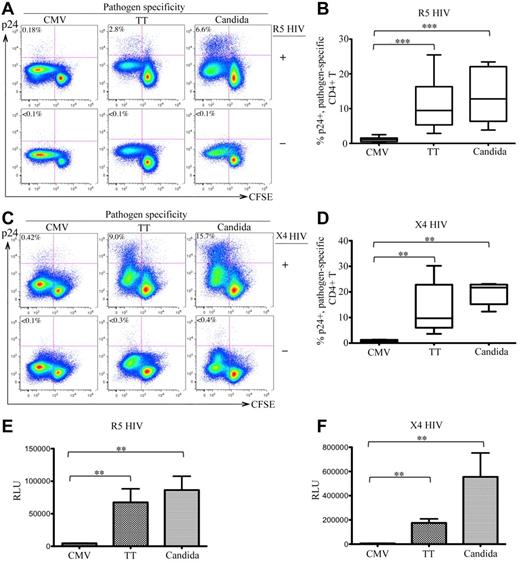
After infection with R5 HIV, 2.8% and 6.6% of TT- and C albicans–specific CD4 T cells were intracellular p24+, respectively, whereas only 0.18% of CMV-specific CD4 T cells expressed p24. No intracellular p24 was detected in mock-infected (HIV−) control PBMCs (Figure 1A bottom panels). Experiments were performed with 6 donors and demonstrated similar results. The difference in R5-HIV infection among pathogen-specific CD4 T cells was statistically significant (P < .005 for CMV vs TT and for CMV vs C albicans; Figure 1B). To determine whether this difference was independent of co-receptor usage, we also investigated X4 HIV infection and observed similar results (CMV: 0.42%; TT: 9.0%; C albicans: 15.7% for p24 rate in CFSElow cells; P < .01 for CMV vs TT and CMV vs C albicans; Figure 1C-D). Analysis of p24 in the supernatants showed results consistent with the intracellular p24 (data not shown). Comparing infections between R5 and X4 HIV, we consistently observed better viral replication of X4 viruses in antigen-stimulated CD4 T cells (Figure 1A and C). This result is compatible with the finding that X4 HIV is associated with more rapid CD4 loss and accelerated progression to AIDS.23
In vitro HIV infection of pathogen-specific CD4 T cells in PBMCs. (A,C) Representative flow cytometric plots are shown for intracellular p24 expression in CFSElow population of CD3+CD4 T cells in antigen-stimulated PBMCs with (top) or without (bottom) infection by R5-HIV (A) or X4-HIV (C). Only live CD14-19−CD3+CD8− T cells were gated for analysis. The data are expressed as a proportion of intracellular p24+ cells in the CFSElow population. (B,D) Comparison of the proportion of p24+CFSElow cells between CMV-, TT-, and C albicans–specific CD4 T cells from multiple subjects are shown as a box and Whisker plots in B (R5 HIV) and D (X4 HIV). (E-F) Relative quantification of infectious R5 HIV (E) or X4 HIV (F) viruses produced by pathogen-specific CD4 T cells in supernatants. Quantification was based on infection of TZM-bl cells and the data are expressed as relative light units (RLU). A total of 6 donors and at least 6 independent experiments were performed. Statistical analysis was performed using the Mann-Whitney test. *** P < .005; **P < .01; *P < .05.
In vitro HIV infection of pathogen-specific CD4 T cells in PBMCs. (A,C) Representative flow cytometric plots are shown for intracellular p24 expression in CFSElow population of CD3+CD4 T cells in antigen-stimulated PBMCs with (top) or without (bottom) infection by R5-HIV (A) or X4-HIV (C). Only live CD14-19−CD3+CD8− T cells were gated for analysis. The data are expressed as a proportion of intracellular p24+ cells in the CFSElow population. (B,D) Comparison of the proportion of p24+CFSElow cells between CMV-, TT-, and C albicans–specific CD4 T cells from multiple subjects are shown as a box and Whisker plots in B (R5 HIV) and D (X4 HIV). (E-F) Relative quantification of infectious R5 HIV (E) or X4 HIV (F) viruses produced by pathogen-specific CD4 T cells in supernatants. Quantification was based on infection of TZM-bl cells and the data are expressed as relative light units (RLU). A total of 6 donors and at least 6 independent experiments were performed. Statistical analysis was performed using the Mann-Whitney test. *** P < .005; **P < .01; *P < .05.
To quantify the infectious HIV particles produced by CD4 T cells, supernatants of antigen-stimulated, HIV-infected PBMCs were used to infect TZM-bl cells. Significantly more infectious HIV was produced in TT- and C albicans–stimulated PBMCs than CMV-stimulated PBMCs for both R5 (Figure 1E) and X4 (Figure 1F) HIV. These results demonstrate that TT- and C albicans–specific CD4 T cells were permissive and produced de novo functional HIV particles, whereas CMV-specific CD4 T cells were highly resistant to both R5 and X4 HIV, suggesting that there are marked differences in susceptibility to HIV infection between pathogen-specific CD4 T cells.
For CMV, viral lysates and pp65 peptides yielded similar results, indicating that the resistance of CMV-specific CD4 T cells to HIV infection is independent of antigen preparation. We also performed the experiments using CD8-depleted PBMCs and found that the relative susceptibility of CMV-, TT-, and C albicans–specific CD4 T cells to HIV infection was not significantly affected (data not shown).
Phenotypic characterization of CMV-, TT-, and C albicans–specific CD4 T cells and the associated HIV infection
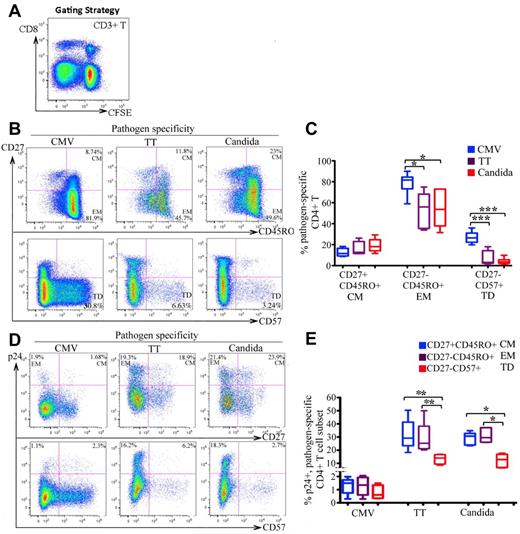
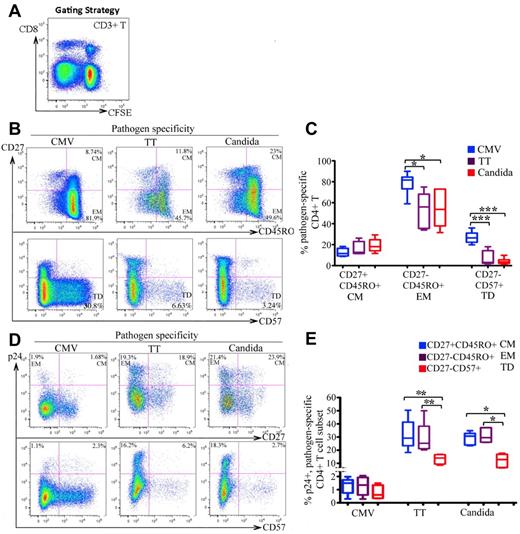
We next determined the cellular differentiation of CMV-, TT-, and C albicans–specific CD4 T cells and the associated HIV infection. CFSElowCD3+CD8− T cells were gated for analysis (Figure 2A).24 Compared with TT-and C albicans–specific T cells, CMV-specific CD4 T cells were more mature, as indicated by the more frequent expression of CD27−CD45RO+ (EM; P < .01) and CD27−CD57+ (terminally differentiated; P < .005) phenotypes (Figure 2B-C). For HIV infection, no significant difference was found between the CM and EM subsets in any of the 3 pathogen specificities (Figure 2D top and E). However, there was substantially less HIV infection in CD57+ cells than in CD57− memory cells, particularly in TT- and C albicans–specific CD4 cells. The difference was less pronounced in CMV-specific CD4 T cells (Figure 2D bottom and 2E).
Differentiation phenotype of pathogen-specific CD4 T cells and the associated HIV infection. (A) CFSElowCD3+CD8− T cells in antigen-stimulated PBMCs were gated for analysis. (B-C) Representative flow cytometric plots (B) and statistical comparison (C), shown as box and Whisker plots, for the frequencies of CM (CD27+CD45RO+), EM (CD27−CD45RO+), and terminally differentiated (TD: CD27−CD57+) subsets between pathogen-specific CD4 T-cell populations. (D-E) Representative flow cytometric plots (D) and statistical comparison (E), shown as box and Whisker plots, for HIV infection of different differentiation subsets between pathogen-specific CD4 T-cell populations. The HIV infection rate is expressed as the proportion of each subset expressing p24. For panels D and E, only the CFSElow CD3+CD8−CD45RO+ memory population was gated for analysis. A total of 6 donors and at least 6 independent experiments were performed. Statistical analysis was performed using the Mann-Whitney test. ***P < .005; **P < .01; *P < .05.
Differentiation phenotype of pathogen-specific CD4 T cells and the associated HIV infection. (A) CFSElowCD3+CD8− T cells in antigen-stimulated PBMCs were gated for analysis. (B-C) Representative flow cytometric plots (B) and statistical comparison (C), shown as box and Whisker plots, for the frequencies of CM (CD27+CD45RO+), EM (CD27−CD45RO+), and terminally differentiated (TD: CD27−CD57+) subsets between pathogen-specific CD4 T-cell populations. (D-E) Representative flow cytometric plots (D) and statistical comparison (E), shown as box and Whisker plots, for HIV infection of different differentiation subsets between pathogen-specific CD4 T-cell populations. The HIV infection rate is expressed as the proportion of each subset expressing p24. For panels D and E, only the CFSElow CD3+CD8−CD45RO+ memory population was gated for analysis. A total of 6 donors and at least 6 independent experiments were performed. Statistical analysis was performed using the Mann-Whitney test. ***P < .005; **P < .01; *P < .05.
It has been shown that CD25 expression is associated with HIV infection in CD4 T cells.18,26-28 We observed that, compared with TT- and C albicans–specific CD4 T cells, in which CD25 expression was evident, CMV-specific CD4 T cells expressed little CD2518,25 (supplemental Figure 2A). This low CD25 expression on CMV-specific cells may be because CMV-specific CD4 T cells produced very little endogenous IL-2 in response to recall antigen stimulation18 (data not shown). For HIV infection, more HIV was observed in CD25+ than in CD25− TT-, and C albicans–specific CD4 T cells; the difference was less pronounced in CMV-specific CD4 T cells (supplemental Figure 2). Exogenous IL-2 increased CD25 expression on all 3 groups of pathogen-specific CD4 T cells. However, significant HIV infection was found in the CD25+ subsets in the presence of exogenous IL-2 for TT- and C albicans–specific CD4 T cells, but scant HIV infection was detected in CMV-specific CD25+CD4 T cells (supplemental Figure 2).
PD-1high CD4 T cells have been recently recognized as an important HIV reservoir in vivo and are enriched for HIV proviral DNA compared with corresponding PD-1low cells.6 We found that CMV-specific CD4 T cells expressed markedly lower PD-1 than TT- and C albicans–specific CD4 T cells (supplemental Figure 3A). HIV infection was substantially higher in PD-1high cells than in PD-1low cells for CMV-, TT-, and C albicans–specific CD4 T cells. In TT- and C albicans–specific cells, approximately 70% of PD-1+ cells were p24+ (supplemental Figure 3B).
Lower amounts of cell-associated HIV DNA were detected in CMV-specific CD4 T cells compared with TT- and C albicans–specific CD4 T cells after in vitro HIV infection
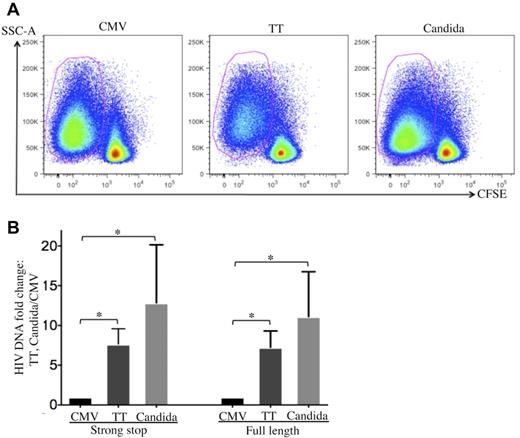
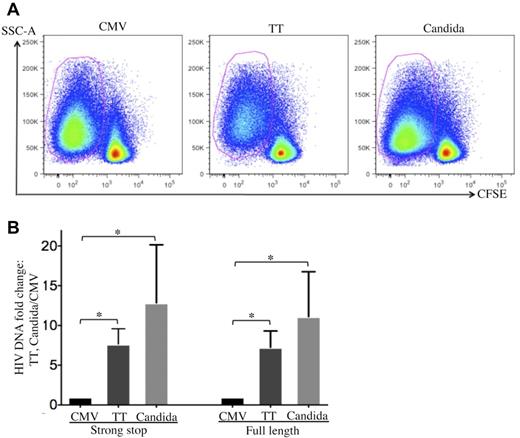
To investigate how HIV is restricted in CMV-specific CD4 T cells, cell-associated HIV DNA in FACS-sorted, pathogen-specific CD4 T cells was quantified by real-time PCR 24 hours after R5 tropic HIV infection (Figure 3A). CMV-specific CD4 T cells demonstrated a significant reduction in both strong-stop and full-length HIV DNA compared with TT- or C albicans–specific CD4 T cells (Figure 3B), suggesting that there is a more profound block at entry or early stages before reverse transcription in CMV-specific CD4 T cells than in TT- and C albicans–specific CD4 T cells. The strong-stop/full-length HIV DNA ratio was comparable between pathogen specificities (Figure 3B), suggesting that reverse transcription is not preferentially impaired in CMV-specific CD4 T cells.
Quantification of cell-associated HIV DNA in pathogen-specific CD4 T cells after in vitro HIV infection. (A) Pathogen-specific CD4 T cells were sorted from PBMCs with a FACSAria cell sorter based on CFSElow. (B) Quantification of cell-associated strong-stop and full-length HIV DNA in sorted pathogen-specific CD4 T cells. The results are expressed as the fold increase in HIV DNA copies for TT- and C albicans–specific CD4 T cells relative to CMV-specific CD4 T cells (mean and SD). Three independent experiments for each donor with a total of 3 donors were performed. Statistical analysis was performed using the Mann-Whitney test. *P < .05.
Quantification of cell-associated HIV DNA in pathogen-specific CD4 T cells after in vitro HIV infection. (A) Pathogen-specific CD4 T cells were sorted from PBMCs with a FACSAria cell sorter based on CFSElow. (B) Quantification of cell-associated strong-stop and full-length HIV DNA in sorted pathogen-specific CD4 T cells. The results are expressed as the fold increase in HIV DNA copies for TT- and C albicans–specific CD4 T cells relative to CMV-specific CD4 T cells (mean and SD). Three independent experiments for each donor with a total of 3 donors were performed. Statistical analysis was performed using the Mann-Whitney test. *P < .05.
CMV-specific CD4 T cells demonstrate postentry restriction of HIV replication in addition to entry block, whereas TT- and C albicans–specific CD4 T cells restrict HIV largely at entry
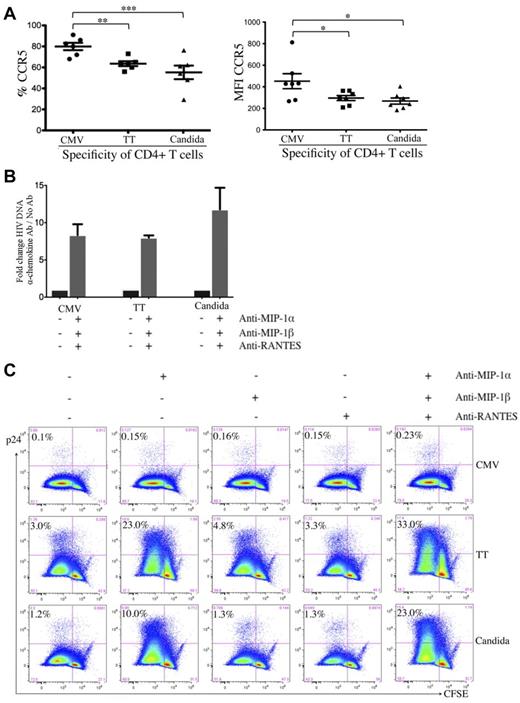
To investigate the stages of HIV restriction and the associated cellular factors, we assessed the surface expression of CD4 and HIV co-receptors. CD4 expression was comparable between pathogen-specific CD4 T cells (data not shown). For co-receptors, CMV-specific CD4 T cells expressed even higher surface CCR5 (Figure 4A) and CXCR4 (data not shown) than TT- and C albicans–specific CD4 T cells.
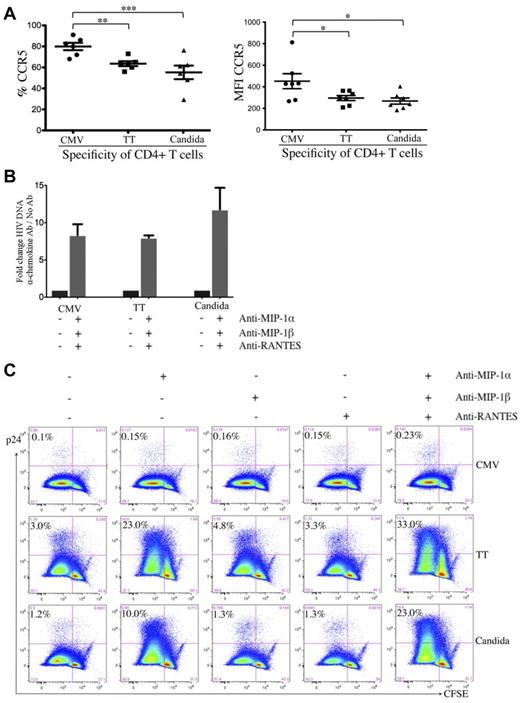
Effect of β-chemokine neutralization on cell-associated HIV DNA and p24 contents in pathogen-specific CD4 T cells. (A) Expression of CCR5 on pathogen-specific CD4 T cells expressed as a proportion (left) or as intensity (right). (B) Effect of β-chemokine neutralization on cell-associated HIV DNA content in pathogen-specific CD4 T cells. Pathogen-specific CD4 T cells with or without treatment by neutralizing antibodies (anti–MIP-1α, anti–MIP-1β, and anti-RANTES) were subjected to real-time PCR for quantification of cell-associated full-length HIV DNA. The results are expressed as the fold change in HIV DNA copies for cells with neutralization relative to no neutralization treatment within each pathogen-specificity. (C) Effect of β-chemokine neutralization on intracellular p24 content. CFSE-loaded PBMCs were antigen stimulated and HIV infected in the absence or presence of individual anti–β-chemokine antibodies alone or in combination and subjected to p24 flow cytometric analysis. The results are expressed as the proportion of p24+CFSElow cells in each group of pathogen-specific CD4 T cells and the representative flow data are shown. A total of 6 donors and at least 3 independent experiments were performed. Statistical analysis was performed using the Mann-Whitney test. ***P < .005; **P < .01; *P < .05.
Effect of β-chemokine neutralization on cell-associated HIV DNA and p24 contents in pathogen-specific CD4 T cells. (A) Expression of CCR5 on pathogen-specific CD4 T cells expressed as a proportion (left) or as intensity (right). (B) Effect of β-chemokine neutralization on cell-associated HIV DNA content in pathogen-specific CD4 T cells. Pathogen-specific CD4 T cells with or without treatment by neutralizing antibodies (anti–MIP-1α, anti–MIP-1β, and anti-RANTES) were subjected to real-time PCR for quantification of cell-associated full-length HIV DNA. The results are expressed as the fold change in HIV DNA copies for cells with neutralization relative to no neutralization treatment within each pathogen-specificity. (C) Effect of β-chemokine neutralization on intracellular p24 content. CFSE-loaded PBMCs were antigen stimulated and HIV infected in the absence or presence of individual anti–β-chemokine antibodies alone or in combination and subjected to p24 flow cytometric analysis. The results are expressed as the proportion of p24+CFSElow cells in each group of pathogen-specific CD4 T cells and the representative flow data are shown. A total of 6 donors and at least 3 independent experiments were performed. Statistical analysis was performed using the Mann-Whitney test. ***P < .005; **P < .01; *P < .05.
It is possible that the CCR5 on CMV-specific CD4 T cells is less accessible to HIV for entry because of cellular factors such as β-chemokines.24,29 We investigated the effect of β-chemokine neutralization on HIV entry in pathogen-specific CD4 T cells by quantifying cell-associated HIV DNA. Neutralization of MIP-1α, MIP-1β, and RANTES led to a substantial increase in HIV DNA in CD4 T cells specific for all 3 pathogens compared with no β-chemokine neutralization (fold increase: 8.3 for CMV, 8.0 for TT, and 11.8 for C albicans; Figure 4B), indicating that the receptors for HIV entry on CMV-, TT-, and C albicans–specific CD4 T cells are functional and that all 3 groups of pathogen-specific CD4 T cells are able to restrict HIV infection at entry through β-chemokine blockade of the CCR5 co-receptor.
To determine whether there is a difference in the postentry restriction of HIV replication, we assessed intracellular p24 in the presence of β-chemokine neutralization. Neutralization of MIP-1α, but not MIP-1β or RANTES, led to a substantial increase in p24 in TT- and C albicans–specific CD4 T cells (TT: 3.0%-23.0%; C albicans: 1.2%-10.0%) and neutralizing all 3 β-chemokines had a synergistic effect leading to the highest p24 (TT: 33.0%; C albicans: 23.0%; Figure 4C). Together with the observation shown in Figure 4B, these results suggest that HIV infection of TT- and C albicans–specific CD4 T cells in PBMCs is largely restricted at entry, after which time these 2 groups of pathogen-specific CD4 T cells are permissive to HIV replication. In contrast, despite increased HIV entry in β-chemokine–neutralized, CMV-specific CD4 T cells (Figure 4B), p24 expression in these cells remained low (0.1% vs 0.23%; Figure 4C), indicating that, unlike TT- and C albicans–specific CD4 T cells, CMV-specific CD4 T cells demonstrate potent postentry restriction of HIV replication.
Distinct gene-expression profiles of CMV-, TT-, and C albicans–specific CD4 T cells
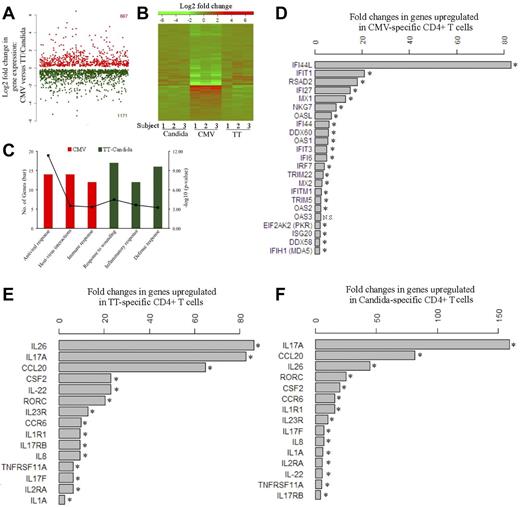
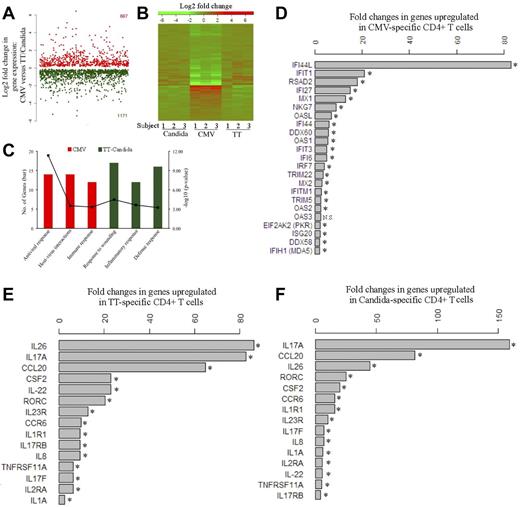
Global gene-expression profiling was performed to identify the cellular factors that differentially regulate HIV infection of pathogen-specific CD4 T cells. Five to 6 days after antigen stimulation, CFSElow, CD4 T cells were sorted from PBMCs and subjected to microarray analysis. We found that CMV-specific CD4 T cells demonstrated a distinct gene-expression profile from TT- and C albicans–specific CD4 T cells (Figure 5A-B). Functional category and gene ontology enrichment analysis revealed that the profile of CMV-specific CD4 T cells was dominated by responses linked to antiviral response and host-virus interactions, whereas the TT- and C albicans–specific profiles were mainly characterized by inflammatory and defense responses (Figure 5C). For CMV-specific CD4 T cells, a broad array of innate antiviral responses were activated, such as type-I IFN responses (eg, IFI44L, IFIT1, IFI27, IFI44, IFIT3, and IFI6), antiviral RNA responses (eg, OASL, OAS1, OAS2, OAS3, PKR, and MDA5), antiviral defenses (eg, RSAD2 and MX1), and HIV/SIV restriction factors (eg, TRIM22 and TRIM5; Figure 5D). IFI44L showed the greatest induction, with a more than 80-fold increase in expression in CMV-specific CD4 T cells, followed by IFIT1 (> 20-fold), RSAD2, IFI27, and MX1 (Figure 5D). Several of these genes have some activity against HIV/SIV,30-36 providing a plausible explanation for restriction of HIV replication by CMV-specific CD4 T cells. In contrast, the genes up-regulated in TT- and C albicans–specific CD4 T cells were mainly associated with the Th17 inflammatory response, such as IL-17A, IL-17F, and the Th17-specific transcription factor RORC37 (Figure 5E-F). IL-17A expression was up-regulated by more than 80- and 150-fold in TT- and C albicans–specific cells, respectively. RORC was up-regulated by more than 20-fold in both TT- and C albicans–specific CD4 T cells. Other Th17-associated cytokines, including IL-22 and IL-26, and the genes induced by Th17 signaling, CCL20 and CCR6, were also significantly up-regulated in TT- and C albicans–specific cells (Figure 5E-F). The Th17 phenotype of TT- and C albicans–specific CD4 T cells was confirmed by intracellular IL-17 cytokine expression in CFSElow cells after antigen restimulation (supplemental Figure 4).
Transcriptomic analysis of pathogen-specific CD4 T cells. (A) Global view of the fold changes in gene expression for genes that were significantly (FDR q-value < 0.05) up-regulated in CMV-specific CD4 T cells (667, red) or in TT- and C albicans–specific CD4 T cells (1171, green). (B) Heat map for global comparison of gene-expression changes between CMV-specific (middle) and TT-specific (right) or C albicans–specific (left) CD4 T cells from 3 subjects. Relative up-regulation (red) and down-regulation (green) of mRNA level are shown. (C) Functional category and gene ontology enrichment analysis using DAVID based on significant genes identified by SAM. The number of significant genes and P value for each category are shown. (D) List of genes that are up-regulated in CMV-specific CD4 T cells and associated with antiviral responses. The fold increase of each gene is expressed as CMV relative to the combined TT and C albicans. (E-F) List of genes that are up-regulated in TT-specific (E) or C albicans–specific (F) CD4 T cells and associated with Th17 inflammatory responses. The fold increase is expressed as TT relative to CMV or C albicans relative to CMV. Data are shown as the geometric means of the fold increases for 3 subjects. Three donors and 3 independent microarray analyses were performed. NS indicates no significance. *FDR q-value < 0.05.
Transcriptomic analysis of pathogen-specific CD4 T cells. (A) Global view of the fold changes in gene expression for genes that were significantly (FDR q-value < 0.05) up-regulated in CMV-specific CD4 T cells (667, red) or in TT- and C albicans–specific CD4 T cells (1171, green). (B) Heat map for global comparison of gene-expression changes between CMV-specific (middle) and TT-specific (right) or C albicans–specific (left) CD4 T cells from 3 subjects. Relative up-regulation (red) and down-regulation (green) of mRNA level are shown. (C) Functional category and gene ontology enrichment analysis using DAVID based on significant genes identified by SAM. The number of significant genes and P value for each category are shown. (D) List of genes that are up-regulated in CMV-specific CD4 T cells and associated with antiviral responses. The fold increase of each gene is expressed as CMV relative to the combined TT and C albicans. (E-F) List of genes that are up-regulated in TT-specific (E) or C albicans–specific (F) CD4 T cells and associated with Th17 inflammatory responses. The fold increase is expressed as TT relative to CMV or C albicans relative to CMV. Data are shown as the geometric means of the fold increases for 3 subjects. Three donors and 3 independent microarray analyses were performed. NS indicates no significance. *FDR q-value < 0.05.
Discussion
The present study was a direct comparison of productive HIV infection among CD4 T cells specific to different antigens. This is also the first effort to characterize the distinct transcriptional profiles of sorted, antigen-specific CD4 T cells. Our results have demonstrated that TT- and C albicans–specific CD4 T cells were permissive, but CMV-specific CD4 T cells appeared highly resistant to X4 and R5 HIV. This restriction of infection in CMV-specific CD4 T cells occurred at the entry and postentry stages. Transcriptional profiling revealed that different pathogen-specific CD4 T cells manifest distinct gene-expression profiles: CMV-specific CD4 T cells selectively activate a broad array of innate antiviral responses, whereas TT- and C albicans–specific CD4 T cells predominantly up-regulate the components of the Th17 response.
The mechanisms of different sensitivities to HIV infection in pathogen-specific CD4 T cells are unknown. Despite comparable levels of HIV receptor/co-receptors, CMV-specific CD4 T cells harbored less HIV proviral DNA after infection with equivalent HIV inocula (Figure 3B). This is consistent with prior observations that CMV-specific CD4 T cells may be protected from HIV in vivo by autocrine production of β-chemokines.18,24 We have also shown that β-chemokines,24,29,38-40 produced in PBMCs after CMV stimulation, do lower HIV infection of CMV-specific CD4 T cells at entry (Figure 4B). β-Chemokine neutralization also led to increased HIV entry in TT- and C albicans–specific CD4 T cells, which may be because, despite TT- and C albicans–specific CD4 T cells being less capable of producing β-chemokines compared with CMV-specific CD4 T cells, other cell types such as monocytes and dendritic cells produce β-chemokines in TT- and C albicans–stimulated PBMCs (data not shown). We also found that CMV-specific CD4 T cells demonstrate postentry restriction of HIV replication, whereas TT- and C albicans–specific CD4 T cells were permissive to viral replication, suggesting that there may be anti-HIV factors in CMV-specific CD4 T cells that limit viral replication.
Gene-expression profiling has been useful in identifying host factors that modulate infections. Unlike previous microarray studies that were conducted with either whole PBMCs41,42 or a specific cell type (CD4 or CD8+ T cells),43-46 microarray analysis in our study was performed with FACS-sorted, antigen-specific CD4 populations. Microarray analysis revealed that CMV-specific CD4 T cells activated a broad array of innate antiviral genes or pathways that have known anti-HIV or SIV activities.30-36 For example, IFIT1 is a novel antiviral protein that recognizes 5′-triphosphate RNA and controls viral replication.30 Type-I IFNs were not detected in CMV-stimulated PBMCs (data not shown), favoring the hypothesis that the innate antiviral profile of CMV-specific CD4 T cells is not induced by exposure to type-I IFNs. Moreover, we tested the HIV infectivity of CD4 T cells specific to another virus that is closely related to CMV, HSV. The preliminary data show that, unlike CMV, the HSV-specific CD4 T cells were susceptible to HIV with infectivity closer to TT- and C albicans–specific CD4 T cells (data not shown), suggesting that the HIV-resistant phenotype and the broad antiviral profile of antigen-specific CD4 T cells may be unique to CMV and may not be generalizable to other viral pathogens.
TT- and C albicans–specific CD4 T cells derived from the same PBMCs as CMV-specific CD4 T cells displayed a predominant Th17-associated profile, which is typical of CD4 responses elicited by mucosal pathogens.8,37,47 Given previous observations that Th17 CD4 T cells are massively depleted in vivo at mucosal sites by HIV7 and SIV,48 it is perhaps reasonable to hypothesize that TT- and C albicans–specific Th17 CD4 T cells at mucosal sites may be preferentially depleted by HIV because of higher susceptibility. In addition, microarray analysis identified high expression of CCR6 and its ligand CCL20 in the permissive TT- and C albicans–specific CD4 T cells, supporting the results of a previous study showing preferential infection of peripheral CCR6+ CD4 T cells by HIV.9
The results of the present study have implications for HIV vaccine research. It was shown recently that an RhCMV-vectored SIV vaccine induces SIV-specific TEM responses that are persistent and stringently control SIV infection in monkeys. Interestingly, the RhCMV-associated SIV control was insensitive to CD4 depletion.19 Given the importance of CD4 help for sustaining long-term T-cell immunity, RhCMV-vectored SIV vaccine may have induced a functional population of antigen-specific CD4 T cells highly resistant to SIV depletion. Because early HIV infection is associated with infection and destruction of HIV-specific CD4 T cells,49 induction of CD4 T cells that are relatively resistant to HIV infection would have distinct advantages. The ability of certain viruses to induce CD4 T cells with the CMV-specific T-cell “HIV resistance” phenotype has implications for the selection of viral vectors for future HIV vaccines.
In conclusion, we describe herein a novel in vitro system for investigating the relative susceptibility of antigen-specific CD4 T cells to HIV infection and have identified a population of CMV-specific CD4 T cells highly resistant to R5 and X4 HIV. Our results suggest a potential mechanism for the earlier loss of Th17-associated mucosal TT- and C albicans–specific CD4 responses and the preservation of the CMV-specific CD4 response seen during the pathogenesis of AIDS. These findings raise the hypothesis that the selection of the viral vector for future HIV vaccines may subsequently influence the susceptibility of vaccine-induced CD4 T cells to HIV infection.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr J. Currier, Dr M. Eller, Dr V. Polonis, and Ms M. Wesberry for their kind assistance and advice and the PBMC donors for their contributions to this study.
The work was supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation and the US Department of Defense.
The views expressed herein are those of the authors and should not be construed to represent the positions of the US Army or Department of Defense.
Authorship
Contribution: H.H conceived of the study, designed and performed the experiments, analyzed the data, and wrote the manuscript; M.N., P.E, A.-L.C., and Y.Z. performed the microarray, HIV DNA quantification, TZM-bl infection, and quantification of type-I IFN and β-chemokines, respectively; C.M. performed p24 quantification in supernatants; Z.J.D. and Z.W. analyzed the microarray data; M.V., N.L.M., J.H.K., and M.M. discussed the data and wrote the manuscript; and S.R.-K. conceived of the study, discussed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Haitao Hu, MD, PhD, US Military HIV Research Program, Walter Reed Army Institute of Research, 503 Robert Grant Ave, Silver Spring, MD 20910; e-mail: hhu@hivresearch.org.