Key Points
Genome-wide binding profiles of FLI1, ERG, GATA2, RUNX1, SCL, LMO2, and LYL1 in human HSPCs reveals patterns of combinatorial TF binding.
Integrative analysis of transcription factor binding reveals a densely interconnected network of coding and noncoding genes in human HSPCs.
Abstract
Genome-wide combinatorial binding patterns for key transcription factors (TFs) have not been reported for primary human hematopoietic stem and progenitor cells (HSPCs), and have constrained analysis of the global architecture of molecular circuits controlling these cells. Here we provide high-resolution genome-wide binding maps for a heptad of key TFs (FLI1, ERG, GATA2, RUNX1, SCL, LYL1, and LMO2) in human CD34+ HSPCs, together with quantitative RNA and microRNA expression profiles. We catalog binding of TFs at coding genes and microRNA promoters, and report that combinatorial binding of all 7 TFs is favored and associated with differential expression of genes and microRNA in HSPCs. We also uncover a previously unrecognized association between FLI1 and RUNX1 pairing in HSPCs, we establish a correlation between the density of histone modifications that mark active enhancers and the number of overlapping TFs at a peak, we demonstrate bivalent histone marks at promoters of heptad target genes in CD34+ cells that are poised for later expression, and we identify complex relationships between specific microRNAs and coding genes regulated by the heptad. Taken together, these data reveal the power of integrating multifactor sequencing of chromatin immunoprecipitates with coding and noncoding gene expression to identify regulatory circuits controlling cell identity.
Introduction
The hierarchy and stages that hematopoietic stem cells (HSCs) traverse while they differentiate to various terminal lineages has been mapped in exquisite detail.1-3 Coupled with the advantage that large numbers of hematopoietic stem and progenitor cells (HSPCs) can be isolated using flow cytometry, the hematopoietic system is ideally suited for the analysis of the global architecture of cell differentiation. This knowledge has been exploited to construct draft maps of the regulatory circuitry of human hematopoiesis using gene expression profiles of purified populations of cells.4,5 These studies testify to the modular architecture of gene expression signatures in various cell populations and the roles that key transcription factors (TFs) play within these modules. However, without distinguishing direct from indirect interactions, it is not possible to address the redundancy and temporal dynamics of transcriptional networks that control cell identity.
Combinatorial interactions of TFs are key determinants of cell identity. The ability to reprogram terminally differentiated cells into either pluripotent or tissue specific stem cells, using defined sets of TFs, testifies to the power of combinatorial TF interactions.6,7 Surprisingly, given the long history of accumulated knowledge in hematopoiesis, functional HSCs have not as yet been generated from terminally differentiated cells by this technology. Indeed, although the roles of individual hematopoietic TFs both during developmental and adult hematopoiesis have been carefully cataloged,8 the essential combination of factors that contributes to HSC “stemness” is not known. A kernel of 3 TFs (FLI1, GATA2, and SCL/TAL1) were shown to form a densely interconnected transcriptional circuit in the aorta-gonad-mesonephros during HSC specification.9 However, TFs that are required during HSC specification are no longer essential by themselves for HSC maintenance.10,11 Recognizing that combinatorial interactions were probably required to maintain HSC “stemness,” genome-wide high-resolution binding profiles were recently generated in a mouse hematopoietic progenitor cell line for 10 TFs that revealed a previously unknown combinatorial interaction of 7 TFs (FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2).12 We have recently shown that this heptad of TFs contributes to stem cell signatures in acute myeloid leukemia and is associated with an adverse prognosis.13 Despite the availability of a number of ultra–high-resolution maps for histone modifications in human CD34+ HSPCs,14 corresponding high-quality genome-wide binding profiles for multiple hematopoietic TFs do not exist and have impaired efforts to model the transcriptional regulatory circuitry in these cells.
To evaluate the role of combinatorial TF binding in regulating transcription in human CD34+ HSPCs, we first generated high-quality genome-wide DNA binding profiles for FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2, using massive parallel sequencing of chromatin immunoprecipitates (ChIP-seq) and quantitative sequencing of long and short transcripts (RNA-seq and microRNA [miRNA]-seq, respectively) as a valuable data resource. These data were then integrated with data from the Human Epigenome Atlas15 and Encyclopedia of DNA Elements (ENCODE)16 to construct a network model that incorporates transcriptional and posttranscriptional regulation to expand our molecular understanding of transcriptional control of HSPCs.
Materials and methods
Sample collection
Granulocyte-colony stimulating factor mobilized apheresis samples from normal donors were collected with informed consent in accordance with the Declaration of Helsinki. The CD34+ fraction was obtained by magnetic bead separation using an automated CliniMACS cell separation system (Miltenyi Biotec, Bergist Gladbach, Germany). Cell purity was assessed by flow cytometry and was ≥98%. Collection of bone marrow from normal donors was approved by the Northern Sydney Area Human Research Ethics Committee based at the Prince of Wales Hospital, Sydney, and was endorsed by the Human Research Ethics Committee at the University of New South Wales, Sydney, Australia (see supplemental Data, available on the Blood Web site).
ChIP
Chromatin immunoprecipitation (ChIP) assays were performed as previously described13 with 20 × 106 per condition. Libraries were prepared and sequenced using the Illumina HiSeq2000 analyzer (BGI-Hong Kong) (see supplemental Data).
ChIP-seq analysis
Two publicly available peak-finding programs Model-Based Analysis for ChIP-seq (MACS [version 1.0.1])17 and FindPeaks (version 4.2),18 as well as the commercially available package Partek Genomics Suite (version 6.6), were used to call peaks. Peaks called by at least 2 algorithms were identified as high confidence binding sites for downstream analysis. De novo motif discovery was performed using Multiple EM for Motif Elicitation (MEME [version 4.9.0])19 and motifs were compared with the JASPAR-CORE database.20 Sequences were also interrogated for specific motifs using Find Individual Motif Occurrences (FIMO).21 High confidence binding regions were assigned as regulatory regions to at most two genes using annotations provided by the genomic regions enrichment of annotations tool (GREAT) analysis package.22 Gene set enrichment analysis was performed using the Gene Set Enrichment Analysis (GSEA) software23 (see supplemental Data). The data have been deposited in Gene Expression Omnibus as GSE45144.
Quantitative RNA and short RNA sequencing
Total RNA extraction and purification was performed using miRNeasy mini kits (QIAGEN). Total RNA was amplified using the Ovation RNA-seq system V2 (NuGEN) prior to sequencing. Reads were aligned to the human genome (hg19) using TopHat24 and transcripts were quantified using HOMER.18 Three publicly available algorithms (miRanda,25 TargetScan26 and PicTar [online version27 ]) were used for miRNA target prediction (see supplemental Data).
Results
Number and distribution of genomic targets for FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 in human CD34+ HSPCs
Fli1, Erg, Gata2, Runx1, Scl/Tal1, Lyl1, and Lmo2 have been previously shown to form a heptad of TFs that combinatorially bind multiple targets in the genome of a mouse hematopoietic progenitor cell line, HPC-7.12 To directly investigate potential combinatorial interactions in primary human HSPCs, we purified CD34+ cells (≥98% pure) from normal donors, as detailed in our “Materials and methods” section. ChIP-seq technology was then used to generate genome-scale catalogs of sequences bound by FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 in primary human HSPCs. ChIP material was validated by quantitative polymerase chain reaction (supplemental Data) prior to sequencing. We generated more than 20 million mappable reads for each TF, and identified high confidence binding sites against an immunoglobulin G (IgG) ChIP-seq background. Taken together, we identified 28 793 binding sites, including 16 597 FLI1, 13 993 RUNX1, 9311 GATA2, 4803 ERG, 2588 LMO2, 1929 LYL1, and 1315 SCL/TAL1 peaks (see supplemental Table 1 for peak coordinates).
To characterize the distribution of binding events across genomic features, we partitioned peaks binding within 10 kb of transcription start sites (TSS) as promoter peaks (further segregated into 2 groups based on proximity to the TSS [see Figure 1]), with the remainder into those within and between genes (intragenic and intergenic, respectively [see Figure 1]). Taken as a whole, 60% to 75% of binding peaks for each TF were distributed within a region 10 kb upstream of TSS and the 3′untranslated regions of genes. Binding within 1 kb of TSS ranged from 5% to 15% for each TF with LYL1 and GATA2 at the lower end and SCL/TAL1 and RUNX1 at the higher end.
The number and distribution of genomic targets for FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 heptad in human CD34+HSPCs. The number of peaks for each transcription factor was determined as outlined in the supplemental Data. Each peak was then assigned to be either within a gene promoter (proximal [red] or distal [blue] based on distance from the TSS), intragenic (green), or intergenic (purple). For lists of peaks and associated genes see the supplemental Data.
The number and distribution of genomic targets for FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 heptad in human CD34+HSPCs. The number of peaks for each transcription factor was determined as outlined in the supplemental Data. Each peak was then assigned to be either within a gene promoter (proximal [red] or distal [blue] based on distance from the TSS), intragenic (green), or intergenic (purple). For lists of peaks and associated genes see the supplemental Data.
Analysis of combinatorial binding identifies the heptad as the prevalent pattern in human CD34+ HSPCs
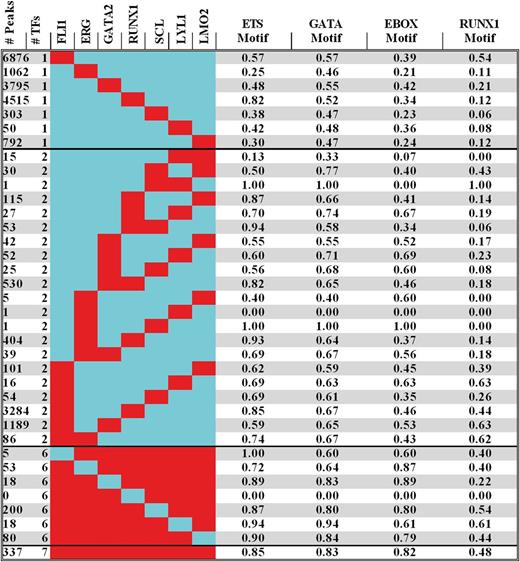
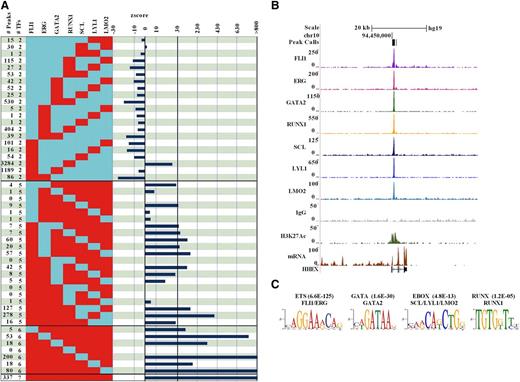
From the compiled peak lists, it was clear that many of the 28 793 binding sites were occupied by more than 1 TF. To formally evaluate combinatorial interactions in human HSPCs, we determined the number of overlapping peaks for all 119 possible combinations involving 2 or more TFs (Figure 2A left; supplemental Table 2) and found that 17 393, 6070, 2720, 1255, 644, 374, and 337 were bound by 1, 2, 3, 4, 5, 6, and 7 TFs, respectively. To address statistical significance, we used a lower end estimate of 80 000 regions potentially available for binding28 to determine the expected frequencies for all 119 binding patterns and calculated the significance of deviation between observed and expected values (Figure 2A right; supplemental Table 2). In general, binding of only 2 TFs was not favored, but binding of the closely related E-twenty six (ETS) family members FLII and ERG together, in the absence of any other member of the heptad, was least likely. The only pairwise combination that was favored in human HSPCs was that of FLI1 and RUNX1, which was found at 3284 out of 28 793 regions. In general, multi-TF combinations were favored, with overlapping binding of all 7 TFs being the most prevalent pattern in HSPCs, suggesting an important role for the heptad in transcriptional control of these cells. For example, the HHEX+1 HSPC enhancer, which was bound by the heptad in mouse HPC7 cells, was also bound by all 7 TFs in human HSPCs (Figure 2B). We next used regions bound by the heptad for de novo motif discovery, which recovered the expected ETS, GATA, E-BOX, and RUNX consensus motifs as not only the most significantly enriched, but also the only significantly enriched motifs, again demonstrating the high quality of the dataset and accuracy of peak identification (Figure 2C).
Analysis of combinatorial binding identifies the heptad as the prevalent pattern in human CD34+HSPCs. (A) The number of peaks for combinations of 2, 5, 6, and all 7 TFs is shown on the left of the figure (red = bound; blue = unbound). A complete set of all 119 combinations involving binding of 2 or more factors is shown in supplemental Table 2. Z scores on the right indicate significance of deviation between observed and expected events for the corresponding patterns. (B) Raw ChIP-seq reads for each transcription factor, control IgG, and H3K27ac are displayed as density plots in the University of California Santa Cruz (UCSC) genome browser above tracks for gene structure and expression in hCD34. The black vertical bar (in the Peak Calls row) above the density plots corresponds to peak coordinates at HHEX +1, a previously verified hematopoietic enhancer that was also bound by the heptad in mouse HPC7 cells. (C) De novo motif discovery performed on the set of regions bound by all 7 TFs identifies ETS, GATA, E-box, and RUNX motifs, corresponding to the 4 motifs that would be expected for these factors as those most significantly enriched.
Analysis of combinatorial binding identifies the heptad as the prevalent pattern in human CD34+HSPCs. (A) The number of peaks for combinations of 2, 5, 6, and all 7 TFs is shown on the left of the figure (red = bound; blue = unbound). A complete set of all 119 combinations involving binding of 2 or more factors is shown in supplemental Table 2. Z scores on the right indicate significance of deviation between observed and expected events for the corresponding patterns. (B) Raw ChIP-seq reads for each transcription factor, control IgG, and H3K27ac are displayed as density plots in the University of California Santa Cruz (UCSC) genome browser above tracks for gene structure and expression in hCD34. The black vertical bar (in the Peak Calls row) above the density plots corresponds to peak coordinates at HHEX +1, a previously verified hematopoietic enhancer that was also bound by the heptad in mouse HPC7 cells. (C) De novo motif discovery performed on the set of regions bound by all 7 TFs identifies ETS, GATA, E-box, and RUNX motifs, corresponding to the 4 motifs that would be expected for these factors as those most significantly enriched.
Motif analysis of peak regions reveals clustering of consensus sites and anticipation of ETS, GATA, E-BOX, and RUNX TF binding
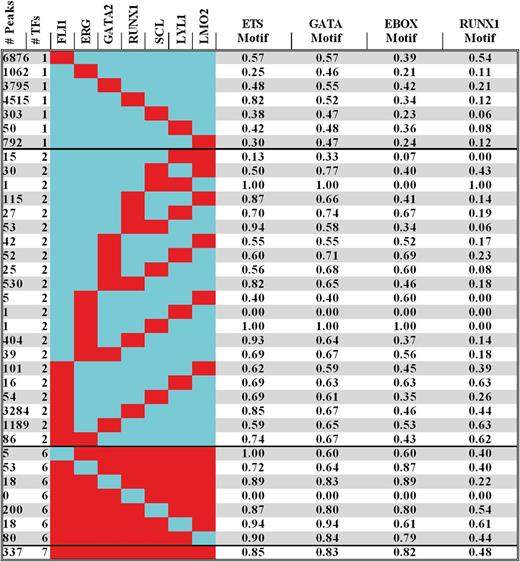
Given the concentration of ETS, GATA, E-BOX, and RUNX motifs at regions bound by the heptad and prior knowledge that members of the heptad form multiple protein-protein interactions, we systematically interrogated all regions bound by 1 or more factors for the presence of these motifs using the Find Individual Motif Occurrences (FIMO) algorithm,21 as detailed in the supplemental Data (supplemental Table 3; Figure 3). Discounting combinations for which there were less than 20 genome-wide binding events (eg, SCL/LYL1 or ERG/SCL or ERG/LYL1 for which there was only a single recorded instance), genomic regions corresponding to combinatorial TF binding were enriched with ETS, GATA, and E-Box motifs, ranging from 0.5 to 0.95, from 0.55 to 0.9, and from 0.35 to 0.9, respectively. At first, this variance (P ≤ .001) suggested a poor correlation between the presence of a consensus motif within a peak region and actual binding of at least 1 TF representative of a particular class. However, when viewed globally, FLI1, ERG, GATA2, SCL, LYL1, and LMO2 binding occurred significantly (P < .0001) more often at sites with a cognate binding motif than without (see supplemental Methods). Nevertheless, these data are consistent with previous reports that there is not an absolute requirement for a motif to be present for an individual TF to bind, as multiprotein complexes engage regions through cognate motifs for 1 or more proteins that constitute a complex.29 The clustering of ETS/GATA/E-BOX motifs at cis-regulatory regions also underscores the role they play as components of an evolutionarily conserved genomic code that instructs gene expression during hematopoiesis.30
Motif analysis of peak regions reveals clustering of consensus sites and anticipation of ETS, GATA, E-BOX, and RUNX TF binding. The number of peaks for combinations involving binding of 1, 2, 6, or all 7 factors are shown on the left of the figure (red = bound; blue = unbound). A complete set of all combinations involving binding of 1 or more factors is shown in supplemental Table 3. On the right are the fractions of regions with Ets, Gata, E-Box, and Runx motifs corresponding to the binding pattern on the left. Regions bound by 1 class of TF in human HSPCs have motifs for other members of the heptad, irrespective of whether these other factors are bound at that region or not.
Motif analysis of peak regions reveals clustering of consensus sites and anticipation of ETS, GATA, E-BOX, and RUNX TF binding. The number of peaks for combinations involving binding of 1, 2, 6, or all 7 factors are shown on the left of the figure (red = bound; blue = unbound). A complete set of all combinations involving binding of 1 or more factors is shown in supplemental Table 3. On the right are the fractions of regions with Ets, Gata, E-Box, and Runx motifs corresponding to the binding pattern on the left. Regions bound by 1 class of TF in human HSPCs have motifs for other members of the heptad, irrespective of whether these other factors are bound at that region or not.
RUNX consensus motifs ranged from 0.05 to 0.65 (P < .0001) with the striking observation that FLI1-bound regions showed a higher likelihood for the presence of RUNX motifs than RUNX1 binding per se, a feature not replicated by ERG, an ETS factor closer related to FLI1. Indeed, in contrast with the other 6 members of the heptad, RUNX1 was bound more often at sites without a RUNX binding motif (P < .001). To explore whether FLI1 binding was permissive for subsequent RUNX1 binding, we evaluated recently published genome-wide binding profiles of FLI1 and RUNX1 in primary megakaryocytes cultured from human CD34+ HSPCs.31 Megakaryocyte-specific RUNX1 peaks were significantly more likely to be acquired at sites with prior FLI1 binding in human HSPCs, than at random promoter/enhancers (P < .00001) or at prior GATA2 (P < .0001) or SCL (P < .0045) bound regions (see supplemental Data). Indeed, 18% of FLI1 peaks in megakaryocytes are FLI1/RUNX1 pairs as opposed to 1.8% of GATA2 and 2.7% of SCL peaks.
The density of H3K27 acetylation and size of the histone-free region surrounding a peak increases with the number of overlapping TFs at that peak
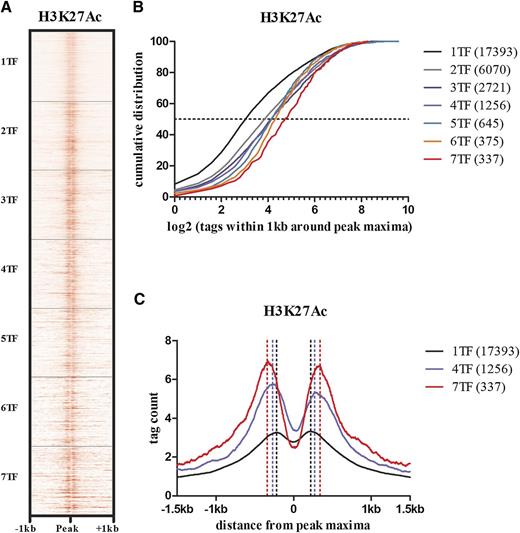
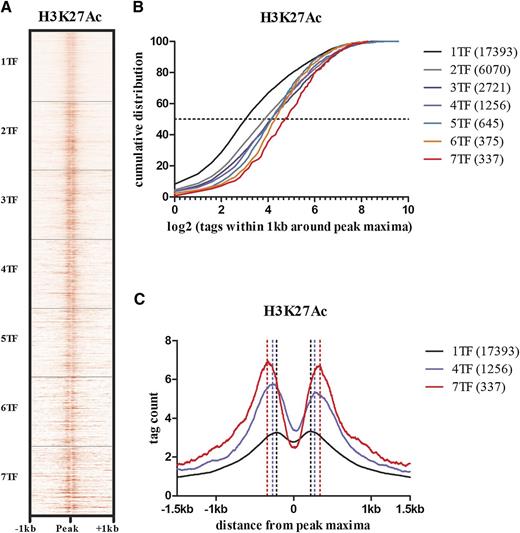
The dynamic relationship between TF binding, nucleosome depletion, and histone modifications at enhancers is complex and is not entirely resolved as reviewed elsewhere.32 However, pioneer TFs are recognized as a subgroup that can bind nucleosomal DNA and cooperatively relieve chromatin condensation by recruiting other TFs. Although active histone marks are not a prerequisite for pioneer TF binding, they facilitate binding of both pioneer and other factors that can further enrich or deplete these histone marks.33 ETS, GATA, and RUNX factors have the ability to bind condensed chromatin, expand the linker region between nucleosomes, and promote local histone modifications.34 Of these various histone modifications, H3K27ac has been reported to distinguish active enhancers from inactive/poised enhancer elements containing H3K4me1 alone.35 To evaluate the relationship between combinatorial TF binding and histone modifications that mark enhancers, we overlapped our TF ChIP-seq data with H3K4me1 and H3K27ac ChIP-seq data made available by the Human Epigenome Atlas. There was progressive increase in H3K27ac and H3K4me1 densities surrounding the TF peak maxima with the number of overlapping TFs within a peak (Figure 4A-B; supplemental Figure 1). Furthermore, as the number of TFs within a peak increased, there was progressive expansion of the linker region between nucleosomes (Figure 4C).
The density of H3K27 acetylation and size of the histone-free region surrounding a peak increases with the number of overlapping TFs at that peak. (A) A read density heat map of H3K27ac at 28 793 TF-bound regions. Although the vertical axis indicates the number of bound sites by different numbers of TFs, it is scaled to equal size. H3K27ac densities adjacent to a peak increase with the number of overlapping TFs at that peak. (B) Quantitative distribution of H3K27ac densities for regions bound by 1 or more TFs. In regions bound by only 1 TF, approximately 10% show no detectable H3K27ac and only ∼8 (log23) reads in up to 50%. In regions bound by all 7 TFs >90% of regions have more than ∼8 reads. (C) The average distances between H3K27ac peak densities flanking the TF peak maxima increases with the number of overlapping TFs within a peak. The corresponding increase in depth of the H3K27ac troughs reflects the increased H3K27ac read densities associated with a higher number of TFs.
The density of H3K27 acetylation and size of the histone-free region surrounding a peak increases with the number of overlapping TFs at that peak. (A) A read density heat map of H3K27ac at 28 793 TF-bound regions. Although the vertical axis indicates the number of bound sites by different numbers of TFs, it is scaled to equal size. H3K27ac densities adjacent to a peak increase with the number of overlapping TFs at that peak. (B) Quantitative distribution of H3K27ac densities for regions bound by 1 or more TFs. In regions bound by only 1 TF, approximately 10% show no detectable H3K27ac and only ∼8 (log23) reads in up to 50%. In regions bound by all 7 TFs >90% of regions have more than ∼8 reads. (C) The average distances between H3K27ac peak densities flanking the TF peak maxima increases with the number of overlapping TFs within a peak. The corresponding increase in depth of the H3K27ac troughs reflects the increased H3K27ac read densities associated with a higher number of TFs.
The heptad factors form a densely interconnected network of factors
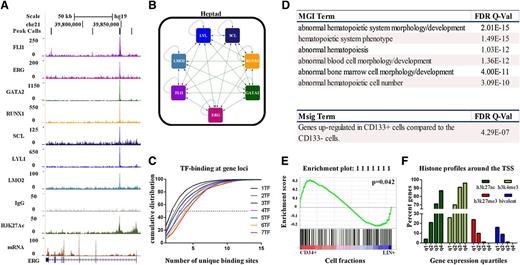
Having recognized combinatorial FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 binding at active enhancer elements in human HSPCs, next, we investigated the connectivity within the core network formed by these 7 factors. The heptad is enriched at known HSC enhancers, such as ERG +85 (Figure 5A).13 Indeed, most loci were bound by all 7 TFs (Figure 5B; supplemental Figure 2) with overlapping binding at ERG +8513 (7 TFs), RUNX1 +2336 (7 TFs), GATA2 +3.537 (7 TFs), FLI1-15 (7 TFs), TAL1 +4038 (5 TFs), LMO2 −2539 (5 TFs), and the LYL1 promoter40 (6 TFs). All of these regions, with the exception of FLI1-15, were known hematopoietic regulatory elements. To test whether binding patterns were predictive of hematopoietic enhancer activity, corresponding mouse sequences from FLI1-15 were tested in transgenic assays. The Fli1-15/SV/lacZ transgene was selectively active in endothelial cells along the ventral surface of the dorsal aorta, from which blood stem/progenitors emerge, and in a small proportion of blood cells in the fetal liver (supplemental Figure 3). These enhancers have been tested in hematopoietic cell lines in transfection assays (ERG +85,13,41 RUNX1 +23,36 TAL1 +40,38 LYL1 promoter40 ), where the mutation of ETS, GATA, or E-Box elements resulted in the loss of enhancer activity, implying positive regulation by their transactivating factors. Heptad binding at these regions in hCD34+ cells coincides with active chromatin marks. Taken together, these data support the presence of a densely interconnected circuit of these TFs in hCD34+ cells (Figure 5B). A particular gene may appear in multiple lists as loci have more than 1 peak of TF binding. Indeed, gene loci that are bound by peaks with increasing numbers of TFs show higher numbers of associated peaks (Figure 5C). This increased complexity of TF binding in the gene set targeted by the heptad probably also signifies its importance in regulating cellular processes in HSPCs.
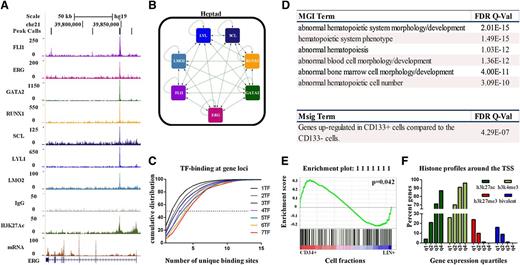
FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 form a densely interconnected network and are associated with differential gene expression. (A) Density plots for ChIP-seq reads in hCD34+ HSPCs displayed in the UCSC genome browser with corresponding RNA-seq expression shows overlapping binding of the heptad at the ERG +85 hematopoietic stem cell enhancer (also see supplemental Figure 2). (B) The heptad TFs show binding at each constituent locus (also see Figure 5A; supplemental Figure 2). The edges show binding of TF protein (node) at the target gene. (C) Cumulative density distribution illustrating the number of unique sites associated with a gene bound by least 1 TF and having at least 1 site bound by 1 to 7 TFs. For example, the ERG locus shown in panel A is associated with the binding of 7 TFs at the +85 enhancer (red curve) and has an additional 4 binding sites at approximately +70, +84, +120, and + 175. Thecumulative density distribution shows that heptad-associated genes are bound at a median of 4 additional sites, whereas genes associated with only 1 TF have a median of 1 additional bound region. (D) Correlation of the GREAT output with Mouse Genome Informatics and Molecular Signatures databases for phenotype-/disease-type associations (see supplemental Table 4 for the full set of enriched terms). (E) GSEA shows significant enrichment for the expression of genes that are targets of the heptad. (F) Correlation of gene expression levels quantified by RNA-seq and divided into expression quartiles with histone marks around the transcription start sites (q1 = lowest expressed; q4 = highest expressed) (also see supplemental Figures 4-6).
FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 form a densely interconnected network and are associated with differential gene expression. (A) Density plots for ChIP-seq reads in hCD34+ HSPCs displayed in the UCSC genome browser with corresponding RNA-seq expression shows overlapping binding of the heptad at the ERG +85 hematopoietic stem cell enhancer (also see supplemental Figure 2). (B) The heptad TFs show binding at each constituent locus (also see Figure 5A; supplemental Figure 2). The edges show binding of TF protein (node) at the target gene. (C) Cumulative density distribution illustrating the number of unique sites associated with a gene bound by least 1 TF and having at least 1 site bound by 1 to 7 TFs. For example, the ERG locus shown in panel A is associated with the binding of 7 TFs at the +85 enhancer (red curve) and has an additional 4 binding sites at approximately +70, +84, +120, and + 175. Thecumulative density distribution shows that heptad-associated genes are bound at a median of 4 additional sites, whereas genes associated with only 1 TF have a median of 1 additional bound region. (D) Correlation of the GREAT output with Mouse Genome Informatics and Molecular Signatures databases for phenotype-/disease-type associations (see supplemental Table 4 for the full set of enriched terms). (E) GSEA shows significant enrichment for the expression of genes that are targets of the heptad. (F) Correlation of gene expression levels quantified by RNA-seq and divided into expression quartiles with histone marks around the transcription start sites (q1 = lowest expressed; q4 = highest expressed) (also see supplemental Figures 4-6).
To evaluate the nature of candidate target genes regulated by the heptad and other occupancy patterns in human HSPCs shown in Figure 2A, TF binding peaks were mapped to nearby genes using the genomic regions enrichment of annotations tool (GREAT),22 which defines genomic neighborhoods for TF-bound peaks by assigning weights to flanking genes based on their distance to the peak. The tool then directly uses the identified gene lists to calculate enrichment against various databases. The highest enriched terms from the Mouse Genome Informatics and Molecular Signatures databases were reported. The 337 regions bound by the heptad showed strong enrichment for a number of hematopoietic phenotypes (Figure 5D; supplemental Table 4). With heptad binding being the most significant occupancy pattern and the most highly enriched for CD34+ genes, concurrent binding by these factors is likely to represent an important control mechanism in these cells.
To identify those sets of target genes most likely to be important for maintaining the HSPC phenotype, next, we performed GSEA.23 To this end, genome-wide expression profiles for 38 distinct purified human hematopoietic cell populations were obtained from the Gene Expression Omnibus (GSE24759).5 The microarray data were categorized into Lin− CD34+ and Lin+ CD34−, and this classification was used to generate a ranked gene list (Lin− CD34+ vs Lin+). Peak-to-gene annotation from GREAT were used to construct 119 different gene sets for each distinct pattern of combinatorial TF binding (eg, the heptad gene set contains all genes associated with heptad-binding peaks). The GSEA package was used to test enrichment of these gene sets against the ranked gene list (see supplemental Data). The only occupancy pattern with a gene set that was enriched in CD34+ HSCPs was the heptad (Figure 5E, full interactive results at: http://149.171.101.136/python/arrangeout/submission/).
A number of genes differentially expressed in Lin+ cells were also targets for the heptad (Figure 5E). To evaluate this further, we ordered heptad target genes into quartiles based on gene expression (q1-q4 = low-high), and we assessed histone marks at their promoters. The fraction of genes with active histone marks (H3K4me3 and H3K27ac) at their promoters increased with increasing gene expression (Figure 5F; supplemental Figure 4). The fraction of genes with the inactive H3K27me3 mark was the highest in the lowest expressed quartile, and it diminished as expression increased. Significantly, genes that are targets of the heptad, with bivalent histone marks at their promoters (supplemental Figure 5), are lowly expressed in CD34+ cells and are upregulated in 1 or another Lin+ cell type (supplemental Figure 6). Taken together, the heptad is not only associated with genes that are highly expressed in HSPCs, but it also appears to prime gene loci for subsequent expression in Lin+ cells.
Expression of noncoding RNAs in human CD34+ HSPCs is associated with heptad binding
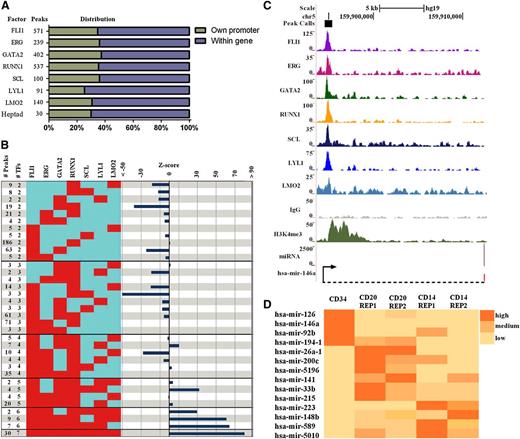
To directly investigate the hypothesis that combinatorial TF binding promotes the miRNA expression program in HSCs, we mapped binding peaks of each TF to putative TSS of primary miRNA transcripts (pri-miRNAs) as detailed in the “Materials and methods” section (Figure 6A). FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 peaks were mapped to 571, 239, 402, 537, 100, 91, and 140 miRNAs, respectively. More than half of these miRNAs were within a coding gene (Figure 6A).
Expression of noncoding RNAs in human CD34+HSPCs is associated with heptad binding. (A) Binding peaks for each TF were evaluated for their proximity to miRNA as outlined in the supplemental Data. The number of binding peaks associated with a miRNA is shown for each TF, and proportions of those with their own promoter (green) and those located within a coding gene (purple) are shown (also see supplemental Figure 7). (B) The numbers of peaks associated with miRNA for combinations involving binding of 2 or more TFs are shown on the left of the figure. Only combinations with at least 2 targets are shown, whereas all combinations are reported in supplemental Table 1. Z scores on the right indicate significant deviation between observed and expected events. The likelihood of miRNA associated regions with 6 or 7 overlapping TFs were overrepresented, whereas regions with 2 and 3 overlapping TFs were underrepresented. (C) Density plots of ChIP-seq reads at the promoter of pri-miR-146a in hCD34+ HSPCs displayed in the UCSC genome browser with corresponding miRNA expression. (D) A heat map showing differential expression across cell types for heptad target miRNAs that are expressed in at least 1 of the 3 cell types.
Expression of noncoding RNAs in human CD34+HSPCs is associated with heptad binding. (A) Binding peaks for each TF were evaluated for their proximity to miRNA as outlined in the supplemental Data. The number of binding peaks associated with a miRNA is shown for each TF, and proportions of those with their own promoter (green) and those located within a coding gene (purple) are shown (also see supplemental Figure 7). (B) The numbers of peaks associated with miRNA for combinations involving binding of 2 or more TFs are shown on the left of the figure. Only combinations with at least 2 targets are shown, whereas all combinations are reported in supplemental Table 1. Z scores on the right indicate significant deviation between observed and expected events. The likelihood of miRNA associated regions with 6 or 7 overlapping TFs were overrepresented, whereas regions with 2 and 3 overlapping TFs were underrepresented. (C) Density plots of ChIP-seq reads at the promoter of pri-miR-146a in hCD34+ HSPCs displayed in the UCSC genome browser with corresponding miRNA expression. (D) A heat map showing differential expression across cell types for heptad target miRNAs that are expressed in at least 1 of the 3 cell types.
From the compiled peak and associated miRNA lists for individual TFs (see supplemental Table 1), it was clear, as with coding genes, that many regions associated with miRNAs overlapped with one another. To formally evaluate combinatorial interactions in human HSPCs, we determined the number of overlapping peaks for all 119 combinations for 2 or more TFs associated with miRNAs (Figure 6B left). To address statistical significance, we again used a lower-end estimate of 80 000 regions potentially available for binding to determine the expected frequencies for all 119 binding patterns and calculated the significance of deviation between observed and expected values for combinations with at least 2 target miRNAs (Figure 6B right). Combinations of less than 4 TFs overlapping at miRNA TSS was not favored in human HSPCs, whereas the combination of all 7 TFs binding was the most favored. Of the 30 miRNAs regulated by the heptad, 9 had their own TSS (as opposed to being within another gene), and all of these had active H3K4me3 histone marks overlapping heptad binding (supplemental Figure 7). These include miR-146a, which has been shown to functionally impact on HSC survival and differentiation (Figure 6C).42 To evaluate differential expression of target miRNAs in CD34+ and lineage committed cells, we interrogated the ENCODE data for which CD34+, CD20+ (B-cell lineage), and CD14+ (monocyte lineage) miRNA-seq data were available. Expression levels were normalized across experiments as detailed in the supplemental Data, and differential expression across cell types is shown for the 14 miRNAs with at least 10 normalized replicate reads in any 1 of these cell types (Figure 6D; supplemental Figure 8).43 Although not expressed at this threshold in CD34+, CD20+, and CD14+ cells, it remains possible that the other 16 miRNAs are upregulated in alternate Lin+ cell types. Therefore, heptad binding at miRNA promoters is associated with both their expression in CD34+ cells and priming of expression at a later stage.
Heptad bound miRNA modulate components of the heptad and their target genes
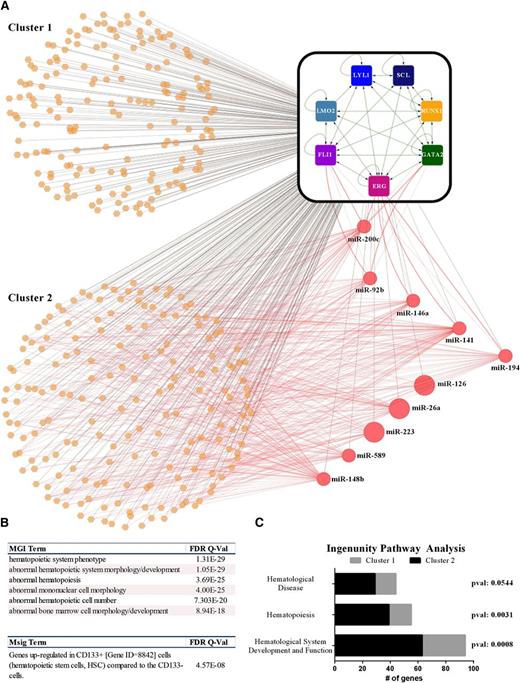
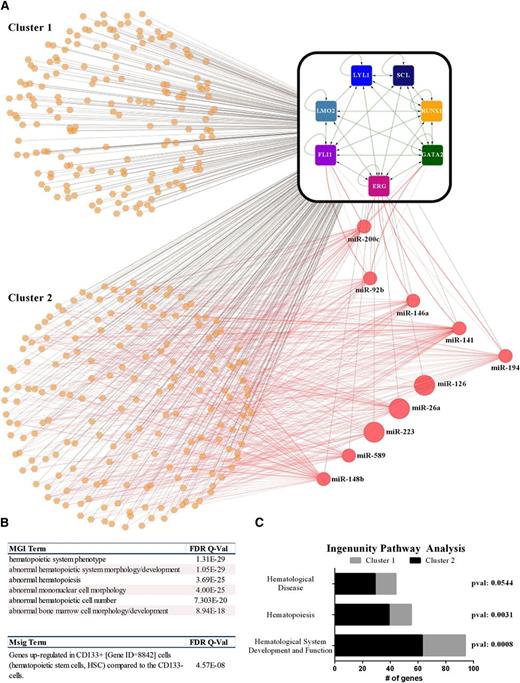
To investigate connectivity between heptad regulated miRNAs and coding genes, we analyzed miRNAs with at least 10 normalized reads (24-216 936) in CD34+ cells. Three target prediction algorithms were used as outlined in the supplemental Data and only those targets predicted by 2 or more databases were used to construct a regulatory network model illustrating the connectivity of all genes and miRNAs controlled by the heptad (Figure 7A). This analysis revealed a core set of genes that are regulated both by the heptad and miRNAs, which in turn are also regulated by the heptad. This network motif, where a regulator exerts both positive and negative effects on its target, is termed “incoherent feed-forward” regulation and provides a mechanism to fine-tune steady state levels and activation kinetics of target proteins.44 It is noteworthy that 5 of the 10 miRNAs form negative feedback loops with their transcriptional drivers. To evaluate possible biological differences between genes, nominally under tighter control by incoherent feed-forward regulation (cluster 2) and those that are not (cluster 1), we re-analyzed corresponding regions using GREAT. Cluster 2 returned the same biological processes and signatures as the entire cohort (ie, related to blood development), but with even higher significance (comparing Figure 7B with Figure 5D). Relevant enrichments were also noted for genes in this cluster by ingenuity pathway analysis (supplemental Figure 9). By contrast, cluster 1 as a group showed no enrichment of any cellular phenotype or signature in either the GREAT or ingenuity pathway analysis. Cluster 2 is also more likely to have genes associated with normal/abnormal hematopoiesis than cluster 1 (Figure 7C; supplemental Table 5). Taken together, these data show that a densely connected kernel of 7 TFs controls hundreds of effectors and that those associated with hematopoietic development are under tight control by miRNAs that are also regulated by the heptad.
Heptad-bound miRNA modulate components of the heptad and their target genes. (A) FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 form a densely interconnected core circuit with multiple, positive-feedback loops. The heptad TFs are illustrated by square nodes and interactions (green edges) between heptad TFs, and heptad coding genes are illustrated (as detailed in Figure 5B). Genes simultaneously bound by the heptad TFs are illustrated as orange hexagons and miRNAs as red circles, whereas these interactions are shown as black edges. Potential posttranscriptional regulatory interactions between miRNAs and gene transcripts (messenger RNAs) are shown as red edges. The size of the circles representing each miRNA in the network is proportionate to their normalized expression level in CD34+ cells. A number of miRNAs form feedback loops with ERG, FLI1, and GATA2. (B) Hematopoietic phenotypes and signatures were enriched (using GREAT) in cluster 2 but not in cluster 1. (C) The numbers of genes corresponding to normal/abnormal hematopoiesis in the ingenuity pathway analysis are enriched in cluster 2 (also see supplemental Figure 9).
Heptad-bound miRNA modulate components of the heptad and their target genes. (A) FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 form a densely interconnected core circuit with multiple, positive-feedback loops. The heptad TFs are illustrated by square nodes and interactions (green edges) between heptad TFs, and heptad coding genes are illustrated (as detailed in Figure 5B). Genes simultaneously bound by the heptad TFs are illustrated as orange hexagons and miRNAs as red circles, whereas these interactions are shown as black edges. Potential posttranscriptional regulatory interactions between miRNAs and gene transcripts (messenger RNAs) are shown as red edges. The size of the circles representing each miRNA in the network is proportionate to their normalized expression level in CD34+ cells. A number of miRNAs form feedback loops with ERG, FLI1, and GATA2. (B) Hematopoietic phenotypes and signatures were enriched (using GREAT) in cluster 2 but not in cluster 1. (C) The numbers of genes corresponding to normal/abnormal hematopoiesis in the ingenuity pathway analysis are enriched in cluster 2 (also see supplemental Figure 9).
Discussion
Here we provide new high-resolution genome-wide binding maps of 7 key TFs in human CD34+ HSPCs and report that combinatorial binding of all 7 TFs is favored and correlates with genes that are differentially expressed in HSPCs. We also report that there is anticipatory binding of the heptad at distal enhancers of genes that show bivalent histone marks at promoters and are primed for expression. The heptad forms a dense autoregulatory core in human HSPCs and collectively regulates miRNAs that in turn target components of the heptad and genes regulated by the heptad. Taken together, these data reveal a network of tightly regulated coding and noncoding genes in human HSPCs, which are centered on a group of key transcriptional regulators.
One of the limitations of our dataset and those in the ENCODE and Human Epigenome Atlas, is that hCD34+ cells are not a homogenous cell population and represent a collection of stem and progenitor cells. The distribution of HSC and progenitor proportions within the hCD34 bone marrow fraction (1:2:5:5:5:1 for HSC:MPP:CMP:GMP:MEP:CLPs, respectively) is variable depending on age and site of sampling.45 To our knowledge, the proportions in mobilized peripheral blood have not been reported, but are also likely to vary from individual to individual. It is possible that we are capturing activity of a regulatory element that is active in only a subpopulation of hCD34+ cells. It is also possible, that the same element is bound by different sets of these 7 TFs in distinct subpopulations. The expression levels of the heptad do not vary significantly between HSCs (Lin−CD34+CD38−CD90+CD45RA−) and MPPs (Lin−CD34+CD38−CD90−CD45RA−), but they do vary from the Lin−CD34+C38+ progenitor fractions46 (supplemental Figure 11). However, it is reassuring that CD34+ cells cluster together as a group when expression levels of the heptad genes are used to sort a compendium of blood cell types by hierarchical clustering5 (supplemental Figure 12). Therefore, the draft map that is presented here is a reflection of the CD34+ collective and subtle variations that apply only to a stem cell or particular progenitor fraction that would be missed. Indeed, if cell numbers were not a limitation, it would be useful to know the degree of variation of chromatin marks and heptad binding between HSCs and committed progenitors. Additional TFs expressed in specific subpopulations or cell specific posttranslational modifications of some of the heptad TFs may modify the composition of the multiprotein complexes binding to given genomic loci.
We were also constrained by the availability of cell numbers from performing replicate experiments. The TF ChIP experiments were performed using 20 million primary human CD34+ cells per antibody and approximately 160 million pooled hCD34+ cells in total (for 7 TFs and IgG). The high-resolution sequencing maps generally exceeded the guidelines set by the ENCODE and modENCODE consortia for ChIP-seq data47 (supplemental Experimental Procedures; supplemental Figure 13). It is also salient to note that when we report combinatorial binding, we are integrating in silico, the results of 7 independent TF ChIP-seq experiments. As such, any region bound by more than 1 TF, essentially, has a biological replicate, and a heptad bound region has been identified in 7 separate experiments. The overlap between our TF ChIP-seq binding peaks and ENCODE, and the Human Epigenome Atlas chromatin marks, in an entirely independent cohort of donor hCD34+ cells was also reassuring. To generate high-resolution binding maps for multiple TFs in highly purified CD34+ subpopulations will require significant technical advances as the number of cells required with current methodology is prohibitive. Even then, a potential caveat would be that highly purified primary cells that are phenotypically homogeneous are functionally heterogenous.48 However, one of the motivations for this study was to produce high-resolution genome-wide binding maps for a heptad of TFs, which individually and collectively have been associated with generating stem signatures in leukemic cells.13 Of note, these stem cell signatures are recognized in leukemic cells, despite their heterogeneity.49
Multifactor ChIP-seq data has been produced in a mouse progenitor cell line.12 Despite good concordance with signatures associated with particular cellular functions (supplemental Figure 9; supplemental Table 6), there was surprisingly modest overlap between regulatory elements. This underscores differences in how transcriptional programs are wired in mouse and human cells to generate the final transcriptome. The other surprise was the enrichment of binding motifs for companion factors of the heptad that were present at peaks, despite the actual factor not being bound in human HSPCs. A disproportionate number of these regions are subsequently bound by their corresponding heptad member during lineage specific differentiation. Indeed, as shown in developmental gene regulatory networks of lower organisms in particular and, to a lesser extent, in mammals, evolutionary conservation of regulatory elements with combinations of binding motifs, which are used and reused, occur as a design principle and not just by chance.50 Anticipatory binding of the heptad at genes with bivalent histone marks at their promoters is in line with a hierarchical model of TF organization, in which the basal TF network primes target genes for subsequent induction.51
Experimental validation of large scale miRNA–messenger RNA interactions have been performed using Argonaute high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitations.52 A similar approach in hCD34+ cells would facilitate the validation of the interactions that we describe in this manuscript. Nevertheless, the predicted autoregulatory loops in HSPCs are reminiscent of an autoregulatory loop consisting of OCT4, SOX2, NANOG, and TCF3, which was reported to regulate embryonic stem cell identity by controlling both coding genes and miRNAs that impact self-renewal and differentiation.53 The 3 miRNAs that are highly expressed in human HSPCs (miR-126, miR-146a, and miR-223), and form incoherent feed-forward loops with genes also regulated by the heptad, have all been functionally validated in HSCs as having a potent impact on HSC self-renewal and/or differentiation. Interestingly, ERG, an oncogenic transcription factor and a member of a poor prognostic stem-cell signature in leukemic cells13,49 is targeted by multiple miRNAs in the network. FLI1 and GATA2, the other members of the heptad, which are subject to feedback control, act at the top of the transcriptional network driving blood and endothelial development in the embryo.54,55 It is possible that a similar transcriptional hierarchy exists within the heptad and governs HSC maintenance. These data highlight the exquisite control of gene expression required to maintain identity in a stem/progenitor population, which is called on to replenish cells throughout life.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Emma Song and staff at the bone marrow transplant laboratory, Prince of Wales Hospital, Sydney for assistance with CD34+ cell collection.
This work was supported by research in the Pimanda Laboratory by the National Health and Medical Research Council of Australia, the Leukaemia Foundation, the Translational Cancer Research Network, and the Dr Tom Bee Stem Cell Research Fund. Dr Wong is supported by the Cancer Institute of New South Wales. Research in the Gottgens Laboratory is supported by the Biotechnology and Biological Sciences Research Council, Leukaemia and Lymphoma Research, The Leukaemia and Lymphoma Society, Cancer Research UK, and core support grants by the Wellcome Trust to the Cambridge Institute for Medical Research and Wellcome Trust - MRC Cambridge Stem Cell Institute.
Authorship
Contribution: D.B., J.A.I.T., D.P., J.S., K.K., S.J.K., N.K.W., A.U., and J.W.H.W. performed research and analyzed data; D.B., J.W.H.W., and B.G. contributed to study design, writing, and data interpretation; T.A.O. provided vital reagents; and J.E.P. designed the study, interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John E. Pimanda, Lowy Cancer Research Centre, University of New South Wales, Sydney, NSW 2052, Australia; e-mail: jpimanda@unsw.edu.au; and Jason W. H. Wong, Lowy Cancer Research Centre, University of New South Wales, Sydney, NSW 2052, Australia; e-mail: jason.wong@unsw.edu.au.
References
Author notes
J.A.I.T. and D.P. contributed equally to this study.

![Figure 1. The number and distribution of genomic targets for FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 heptad in human CD34+ HSPCs. The number of peaks for each transcription factor was determined as outlined in the supplemental Data. Each peak was then assigned to be either within a gene promoter (proximal [red] or distal [blue] based on distance from the TSS), intragenic (green), or intergenic (purple). For lists of peaks and associated genes see the supplemental Data.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/14/10.1182_blood-2013-03-490425/4/m_e12f1.jpeg?Expires=1766008521&Signature=lLPDuWvazQsMsO6Kd2MDUpNQ-KCGUmxQ6Wcvtw3VUfRChepo5GpjEfZbsDPRVpEqXhw1TsEP2V30TfXt9Xi6feg7azJMFgDuEkQz4zZSzi2tEGXyOUE7StFvQM8KcfM~npi09abAe7XjXViqggq7qmWJZhDv~tIsY4T68f9sKlgbSZ2DTTU23h~hrNePR~lB9nrj~tA5TnGWMppUtq95LLT6AXlutNJVDbxdCCluapVMtSpElJAwfzwXtd3GBhRd2bnmwRO7Ya-AKBiZ8X2pr6FzeMRgeenn9sftyPR3SnYzH-Y276znPxK171NHE6VNOcZ0-QJ-5j5aildx-Wy4cg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 1. The number and distribution of genomic targets for FLI1, ERG, GATA2, RUNX1, SCL/TAL1, LYL1, and LMO2 heptad in human CD34+ HSPCs. The number of peaks for each transcription factor was determined as outlined in the supplemental Data. Each peak was then assigned to be either within a gene promoter (proximal [red] or distal [blue] based on distance from the TSS), intragenic (green), or intergenic (purple). For lists of peaks and associated genes see the supplemental Data.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/14/10.1182_blood-2013-03-490425/4/m_e12f1.jpeg?Expires=1766008522&Signature=JkrFDTnJTvCzcrwvxcTiK9uuYYfGwyB12aWF4N7s4XCgYuZGkocO-c-fxODFSodXRKf9KI9RtfskJ-Xan4o8ehU4ct3-NLREhAPfdcCqmfvRqV-c~3Iu~ykmyzzeSRJZnVzxshgJmky7oLHcfh0LG4xOFLzIfmXctUw6Rer2qpVubJkcZgKf4XBCaiUwBVtKmHv1cwv4z~TFGDFbXIcttEspQslWVBJWkN6YLLTJJDaDiRnQM917w9Hcdt~RhyvYoemkON3GtN0li73TV9z3107LOXV52T7P6dzvPkBMeSpUy4yQ2eNCMXPcbarWS6ciw5p26~4N6XCbg8koLpaDvQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)