Abstract
A longstanding endeavor to define the genetic lesions that drive myeloid malignances has stimulated a period of remarkable discovery. Enabled by technological advances that have sharply decreased the cost of DNA sequencing, the full compendium of common, recurrent somatic mutations in the coding genome of myeloid malignancies is nearly complete. As the focus of genetic discovery shifts to the noncoding genome, renewed attention is being applied to the clinical and biological implications of recent genomic advances. Although the potential for this newfound knowledge to influence the care of patients has not yet been realized, broad genetic surveys of patient samples are now being used to improve the accuracy of disease diagnosis, define a molecular taxonomy of myeloid malignancies, refine prognostic and predictive models, and identify novel therapeutic strategies. Here, we will review recent advances in the genetics of myeloid malignancies and discuss their potential impact on clinical practice.
Introduction
The myeloid malignancies are a group of clonal bone marrow disorders with diverse clinical and pathological manifestations affecting hematopoietic stem cell (HSC) function and lineage-specific differentiation. The landscape of common, recurrent somatic alterations in the coding genome of many myeloid malignancies has been extensively characterized, revealing a central role for specific mutations in driving the distinctive features of disease biology.1-5 Other mechanisms that likely contribute to disease pathogenesis, including alterations of the noncoding genome, epigenome, and bone marrow microenvironment, will not be addressed here.
Integration of a highly complex and dynamic set of genetic findings into the diagnosis and nosology of myeloid malignancy diseases is now a central challenge. Following the observation in 1868 that the bone marrow is the source for normal and neoplastic hematopoiesis, and Ehrlich’s development of the triacid stain in 1877 enabling visualization of cellular morphology, hematologic malignancies have been classified primarily on the basis of cellular morphology and peripheral blood counts. Subsequent advances in chromosomal analysis permitted the identification of recurrent, somatic karyotype alterations with disease-defining phenotype associations. Despite the development of finely wrought diagnostic schema, however, substantial clinical heterogeneity remains within current classification schemes.6 The close association of specific somatic genetic lesions with specific clinical and hematologic phenotypes is thus likely to stimulate major advances in the diagnosis and categorization of myeloid malignancies (Figure 1).
Unbiased genome surveys of myeloid malignancies have revealed several key underlying principles. First, mutations affect a range of core, often interrelated, cellular pathways, including transcription, signaling, RNA splicing, and the DNA damage response. Second, myeloid malignancies are characterized by epigenome alterations due, at least in part, to genetic mutations in epigenetic modifiers. Third, some mutations cause genetic instability, promoting the acquisition of subsequent genetic lesions. Finally, the sequential acquisition of genetic alterations enables the development of multiple, genetically related clones during the course of disease progression.
Recurrent somatic mutations in specific biological pathways
Somatic mutations occur in the genomes of HSCs at a low but detectable frequency during normal DNA replication. Although most mutations are rapidly corrected by DNA repair mechanisms, those that persist are propagated during HSC self-renewal. Individual HSCs accumulate exonic mutations at a rate of ∼0.13 per year, meaning that by age 60, each HSC has accumulated 8 mutations with the potential to alter protein-coding sequence.7 In a cell that possesses the capacity for self-renewal, any genetic alteration that causes a selective advantage relative to other self-renewing cells will lead to clonal dominance. Among myeloid malignancies, such mutations are seen recurrently within a small group of genes and are termed “driver” mutations. Other mutations have no impact on clonal dominance, are not seen recurrently in specific genes, and are termed “passenger” mutations. The recurrence of somatic mutations within a particular gene more frequently than would be expected by background mutation rates provides powerful genetic evidence for its causative role in disease development.
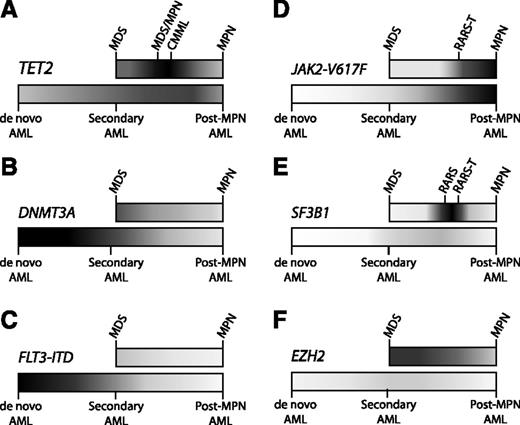
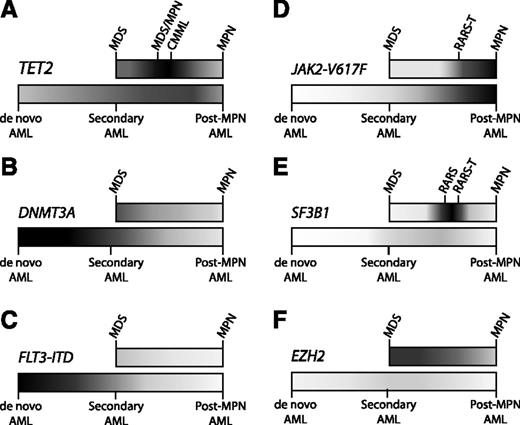
Distribution of mutations in myeloid malignancies. Some somatic mutations occur with relatively equal frequency across the spectrum of myeloid malignancies, while other mutations are enriched in particular pathological subtypes. Higher frequencies of mutation are portrayed with darker shading, and lower frequencies in lighter shading, along the spectrum of MDS to MPN (top bar) and AML (bottom bar). AML ontogeny is represented as a spectrum of de novo AML (no antecedent hematologic disorder), secondary AML (antecedent MDS), and post-MPN AML (antecedent MPN). (A) TET2 is an important regulator of stem cell self-renewal and myelomonocytic differentiation, consistent with the ubiquitous presence of TET2 mutations throughout myeloid malignancies, and with its striking enrichment in diseases with marked monocytosis, such as CMML. (B) DNMT3A mutations can be identified in a broad range of myeloid diseases, but are most frequent in de novo AML. (C) The FLT3 internal tandem duplication (FLT3-ITD) mutation drives myeloblast proliferation without inducing significant morphologic dysplasia, consistent with its strong association with de novo AML, but not with MDS, MPNs, or secondary AML. (D) The JAK2-V617F mutation causes a myeloproliferative phenotype, characteristic of MPNs, such as essential thrombocythemia (ET), polycythemia vera (PV), and primary myelofibrosis (PMF). (E) Mutations in SF3B1 are strongly associated with the presence of ring sideroblasts. Combinations of mutations can drive the composite features of overlap diseases, such as refractory anemia with ring sideroblasts (RARS) associated with marked thrombocytosis (RARS-T), where ∼50% possess the JAK2-V617F mutation and 70% harbor SF3B1 mutations.11 (F) EZH2 mutations are found most commonly across the MDS-MPN spectrum. Despite the adverse prognostic impact of EZH2 mutations in MDS, these mutations are exceptionally rare in AML, implicating mechanisms other than leukemic transformation for their negative clinical effect.
Distribution of mutations in myeloid malignancies. Some somatic mutations occur with relatively equal frequency across the spectrum of myeloid malignancies, while other mutations are enriched in particular pathological subtypes. Higher frequencies of mutation are portrayed with darker shading, and lower frequencies in lighter shading, along the spectrum of MDS to MPN (top bar) and AML (bottom bar). AML ontogeny is represented as a spectrum of de novo AML (no antecedent hematologic disorder), secondary AML (antecedent MDS), and post-MPN AML (antecedent MPN). (A) TET2 is an important regulator of stem cell self-renewal and myelomonocytic differentiation, consistent with the ubiquitous presence of TET2 mutations throughout myeloid malignancies, and with its striking enrichment in diseases with marked monocytosis, such as CMML. (B) DNMT3A mutations can be identified in a broad range of myeloid diseases, but are most frequent in de novo AML. (C) The FLT3 internal tandem duplication (FLT3-ITD) mutation drives myeloblast proliferation without inducing significant morphologic dysplasia, consistent with its strong association with de novo AML, but not with MDS, MPNs, or secondary AML. (D) The JAK2-V617F mutation causes a myeloproliferative phenotype, characteristic of MPNs, such as essential thrombocythemia (ET), polycythemia vera (PV), and primary myelofibrosis (PMF). (E) Mutations in SF3B1 are strongly associated with the presence of ring sideroblasts. Combinations of mutations can drive the composite features of overlap diseases, such as refractory anemia with ring sideroblasts (RARS) associated with marked thrombocytosis (RARS-T), where ∼50% possess the JAK2-V617F mutation and 70% harbor SF3B1 mutations.11 (F) EZH2 mutations are found most commonly across the MDS-MPN spectrum. Despite the adverse prognostic impact of EZH2 mutations in MDS, these mutations are exceptionally rare in AML, implicating mechanisms other than leukemic transformation for their negative clinical effect.
Complete genomic characterization of a spectrum of myeloid malignancies has revealed a relatively restricted number of genes that are recurrently mutated; these genes encode proteins that are part of a small number of biological pathways that play a central role in myeloid malignancies. In de novo acute myeloid leukemia (AML), for example, only 10 genes are mutated at higher than 5% frequency.1 Well-known classes of genes that are altered in myeloid malignancies have been reviewed elsewhere, such as effectors of kinase signaling downstream of cytokine receptors and core transcriptional regulators of HSC and progenitor cell function.8,9 Here, we focus on several classes of genes identified by unbiased discovery efforts, including those implicated in RNA splicing, the cohesin complex, and epigenome modification.
RNA splicing
Unbiased genome and exome sequencing of samples from patients with myelodysplastic syndromes (MDSs) revealed mutations in genes encoding core components of the RNA spliceosome, including SF3B1, SRSF2, U2AF1, and ZRSR2.10-12 Mutations within this pathway are remarkably common in myeloid malignancies and generally enriched in diseases characterized by morphologic dysplasia, such as MDS, chronic myelomonocytic leukemia (CMML), therapy-related AML, and AML with myelodysplasia-related changes. By contrast, splicing mutations occur at low frequency in myeloid diseases without dysplasia, including de novo AML and the myeloproliferative neoplasms.
The presence of highly recurrent somatic mutations in multiple spliceosomal genes suggests that misregulated RNA splicing may drive clonal dominance in myeloid malignancies. Spliceosome mutations are largely restricted to proteins involved in 3′ splice-site recognition and U2 small nuclear ribonucleoprotein function, indicating that disruption of 3′ splice-site recognition could alter splicing in a systematic manner by affecting exon utilization or activating alternative splice sites. Spliceosome dysfunction could create novel protein isoforms with altered function, induce ectopic expression of tissue-inappropriate protein isoforms, or cause quantitative changes in gene expression by activation of nonsense-mediated decay.
Initial experiments, however, have not revealed clear, systematic patterns of mutation-associated splicing abnormalities, raising the possibility that these mutations may exert their function by disrupting noncanonical spliceosome functions. In addition to mediating precursor messenger RNA (mRNA) processing, spliceosomal proteins have been shown to influence histone modifications via physical interactions with Polycomb group proteins,13 activate transcription through direct and indirect regulation of RNA polymerase elongation,14,15 and coordinate aspects of cellular response to DNA damage.16,17 The mutual exclusivity of splicing pathway mutations indicates that these alterations are either functionally redundant or that more significant disruption of canonical or noncanonical spliceosome activity is not tolerated by the cell.
Cohesin complex
Recurrent mutations in the multiprotein cohesin complex were identified using unbiased, large-scale sequencing surveys of MDS and AML.7,18-20 Cohesin mutations are present in 10% to 15% of AML and can affect several components of the complex with mutual exclusivity, including RAD21, SMC1A, SMC3, and STAG2.1 The canonical function of cohesin in establishing and maintaining sister chromatid cohesion during cell division is required for proper chromosome segregation. As such, it has been suggested that cohesin dysfunction may induce a chromosomal instability that leads to cellular transformation.21 However, despite causing aneuploidy in other human cancer models, cohesin mutations are not associated with karyotype complexity in myeloid malignancies, suggesting that mechanisms other than sister chromatid cohesion may be important in these diseases.1,7,22
The role of cohesin in diverse cellular functions is unified by its ability to regulate chromosome topography. In consort with the sequence-specific DNA-binding protein CTCF, cohesin regulates global gene expression by influencing long-range intrachromosomal interactions between cis regulatory elements.21,23 Cohesin maintains genomic integrity by coordinating interchromosomal interactions during repair of double-stranded DNA breaks by homologous recombination24 and by regulating telomere maintenance by its role in sister telomere cohesion.25-27 Additional functional studies will clarify the relative contribution of different cohesin functions to leukemia biology.
DNA methylation
The notion that aberrant DNA methylation may drive leukemia pathogenesis was reinforced by the discovery of recurrent mutations in the de novo DNA methyltransferase DNMT3A.28 DNMT3A mutations are found in a wide range of myeloid malignancies, including AML,28-31 MDS,32,33 CMML,34 systemic mastocytosis,5 and rarely in the myeloproliferative neoplasms.35,36 DNA methyltransferases (DNMTs) catalyze the conversion of cytosine bases to 5-methylcytosine (5mC), primarily in the context of CpG dinucleotides at the site of gene promoters. The density and distribution of genome methylation identifies regions with distinct transcriptional potential, and aberrant methylation is thought to contribute to oncogenesis by altering transcription of key cancer genes. Methylation profiling in both AML and MDS patient samples defined distinct groups based on global patterns of CpG methylation status that correlated with specific clinical and genetic features.37,38 Notably, aberrant methylation signatures have not yet been associated with DNMT3A mutations, raising the possibility of either alternative mechanisms of action or limitations of current experimental approaches.39
The discovery of recurrent TET2 mutations in a broad range of myeloid malignancies prompted key advances in our understanding of the epigenome. TET2 catalyzes the oxidation of 5mC to 5-hydroxymethylcytosine (5hmC).40-42 Biochemical studies show reduced levels of 5hmC in the bone marrow of patients with TET2-mutated myeloid malignancies, consistent with decreased catalytic activity of mutant TET2 protein.43,44 In AML, TET2 mutations are markedly more frequent in older patients and in those with myelodysplasia-related morphologic changes or secondary disease.44-46
TET2 is among the most frequently mutated genes in MDS (20%-25%),3,47 CMML (40%-50%),48-50 and aggressive systemic mastocytosis (23%-29%).5,51,52 At the same time, somatic TET2 alterations can be found in healthy, aging individuals without hematopoietic malignancies despite association with clonal, myeloid-biased hematopoiesis.53-55 Consistent with this clinical observation, Tet2-deficient mice display increased HSC self-renewal, a competitive growth advantage and a differentiation bias toward the myeloid lineage.56-58
With the discovery of recurrent mutations in the isocitrate dehydrogenase genes IDH1 and IDH2 in AML, glioma, and glioblastoma multiforme, unbiased whole genome surveys uncovered a surprising connection between citrate metabolism and DNA methylation.59-61 Mutations in IDH genes are mutually exclusive, uniformly monoallelic, and occur at 3 conserved arginine residues (IDH1-R132, IDH2-R172, and IDH2-R140). These mutations endow mutant IDH1/2 with neomorphic enzymatic activity that directly converts α-ketoglutarate (2-oxoglutarate [2-OG]) to 2-hydroxyglutarate (2-HG).62 (R)-2HG functions as an “oncometabolite” that inhibits a range of 2-OG–dependent dioxygenases, including the TET DNA hydroxymethylases, JmjC domain-containing histone demethylases, and HIF prolyl hydroxylases.63,64 Primary IDH-mutated AML blasts display global hypermethylation that is consistent with disruption of TET2-mediated 5mC hydroxylation and the observed mutual exclusivity of TET2 and IDH1/2 mutations.1,30,44 Conditional knock-in of the Idh1 R132H allele in murine hematopoietic lineages increases global methylation, focused on promoter and intragenic regions, similar to the methylation profiles of IDH1/2-mutated human leukemias.65
Investigation of IDH1/2 mutations illustrates how the discovery of mutations by genome sequencing can be followed rapidly by dramatic insights into disease biology and entry of novel therapeutic agents into clinical trials. IDH1/2-mutated leukemias are associated with elevated serum 2-HG levels, and serial monitoring has demonstrated correlation between serum 2-HG and disease status.66,67 (R)-2HG alone is sufficient to promote cytokine-independent growth and a differentiation block in cell lines in vitro.68 Importantly, these in vitro effects of (R)-2HG are reversible, suggesting that selective inhibition of mutant IDH1/2 enzymatic activity may represent an effective antileukemic therapy. Indeed, a small-molecule targeting IDH2-R140Q caused reduced 2-HG production and myeloid differentiation in primary IDH2-R140Q mutated human leukemia blasts ex vivo.69
Histone modification
Proper regulation of gene transcription depends on the coordinated function of proteins that add, subtract, and interpret diverse posttranslational modifications at histone H3 (writers, erasers, and readers, respectively). Histone lysine methyltransferases (KMTs) and histone lysine demethylases (KDMs) activate or repress gene expression via dynamic regulation of the methylation status at specific lysine residues. Histone acetyltransferases (HATs) add acetyl groups to lysine residues on the histone H3 tail, causing increased gene expression. Proteins known as histone “readers” recognize specific histone modifications and recruit additional effector complexes.
Dysregulation of histone modification pathways has previously been recognized in the context of recurrent translocations involving HATs (such as the KAT6A-NCOA2/MOZ-TIF2 fusion), KMTs (such as MLL translocations and NUP98-NSD1 fusion), and KDMs (such as the NUP98-JARID1A fusion). Sequencing studies have demonstrated that genes in each of these functional classes are recurrently mutated in myeloid malignancies, with somatic mutations in KMTs (EZH2, MLL family, NSD family, SETD2), KDMs (JARID1A, KDM6A/UTX), and HATs (CREBBP, EP300).
EZH2, among the more commonly mutated chromatin regulators, encodes an H3K27 methyltransferase, which, along with EED, SUZ12, and RBBP4, comprise the polycomb-repressive complex 2 (PRC2). PRC2 regulates gene expression through trimethylation of H3K27 and depending on the cellular context, can function either to promote or suppress tumor development. In myeloid malignancies, EZH2 mutations cause a loss of enzymatic function, resulting from mutations that cause premature chain termination or missense mutations in the C-terminal SET domain or the protein-protein (EED-binding or SUZ12-binding) interaction domains.70,71 By contrast, lymphoid malignancies harbor oncogenic EZH2 mutations, primarily at position Y641, that alter enzymatic function and cause increased H3K27 trimethylation.72,73
Genomic studies identified recurrent alterations of ASXL1 in myeloid malignancies.74-77 ASXL1 contains a plant homeodomain (PHD) finger and several nuclear receptor coregulator binding (NR box) motifs. Proteins that contain PHD fingers are thought to function as epigenetic “readers” via PHD-dependent binding to trimethylated lysine residues on histones. ASXL1 has been shown to interact with EZH2 in human leukemia cells and mutations in ASXL1 cause loss of PRC2-mediated H3K27 trimethylation and activation of a HOXA gene expression signature.78 ASXL1 mutations are largely heterozygous alterations that are predicted to result in a truncated protein lacking the PHD finger and nearly all NR box motifs.
Genome instability
Extensive cancer genome sequencing supports the observation that disruptions in a multitude of heterogeneous pathways converge on a phenotype of chromosomal instability. Genomic analysis permits further inference about the temporality of specific rearrangement events, suggesting that some cancers are characterized by intermittent chromosomal catastrophes while others are subject to more gradual genomic degeneration. A provocative example is the recent description of chromothripsis (Greek, “hripsis” = “shattering”), in which complex chromosomal rearrangements result in a distinctive pattern of 2 to 3 oscillating copy number states limited to 1 or several chromosomes.79 Chromothripsis is thought to reflect an isolated, cataclysmic event capable of initiating a tumor by inducing multiple simultaneous DNA alterations. Mechanistically, the phenomenon may result either from critical telomere shortening or from damage to chromosomes in micronuclei formed during aberrant, asynchronous DNA replication.80 In AML, chromothripsis can result in hemizygous tumor suppressor loss, contrasting with solid tumor malignancies where it is preferentially associated with oncogene amplification.81 Although rare in human cancers as a whole, the phenomenon may be enriched in specific biological contexts, such as tumors with germline or somatic TP53 mutations. One study found that nearly 50% of TP53-mutated AML display evidence of chromothripsis,81 an observation that raises important questions about the dynamics of genome instability among this group of patients.
Myeloid neoplasms are characterized by a variety of recurrent chromosomal rearrangements that are demonstrable by routine cytogenetics and known to influence disease biology. In addition, in the recent TCGA analysis of de novo AML, cryptic gene fusions were identified by RNA sequencing that escaped detection by standard karyotype analysis.1 Most novel fusion events were out-of-frame and few were identified in more than a single sample. As such, their biological significance is largely unclear. At present, the functional relevance of specific fusions may be inferred based on whether the genes involved are otherwise subject to recurrent mutation or involve members of recurrently affected pathways. With more extensive application of RNA sequencing to primary myeloid tumor samples, patterns of recurrence will inevitably become more apparent and attribution of biological significance more straightforward.
With whole-genome sequencing, it is possible to identify global patterns of somatic nucleotide variation, suggesting specific types of mutational processes, such as UV radiation, chemical carcinogens, and chemotherapeutic agents. In AML, mutations affecting coding regions are biased toward C>T/A>G transitions, thought to result from the deamination and mutation of the methylated cytosines.7 After exposure to chemotherapy, however, this profile is relatively enriched in transversions (purine → pyrimidine; pyrimidine → purine), likely reflecting the direct DNA toxicity of treatment and potentially serving as a basis for mutations that confer therapy resistance.18,19
Clonal heterogeneity
Individual cancers are remarkably complex, characterized by cellular, genetic, and phenotypic diversity. By identifying individual somatic variants and enumerating their relative allele frequencies, bulk tumor sequencing can be used to define a cancer’s clonal architecture and infer its clonal evolution (Figure 2). Clustering of mutations by shared variant allele frequency can define genetically distinct subclones from parental clone or sibling subclones. A parent clone and all derivative subclones share a suite of somatic mutations (both passengers and drivers), while unique subclones are defined by additional variants that developed after tumor initiation. Using genome sequencing, it was demonstrated that most de novo AML samples possess a founder clone and at least 1 derivative subclone.1 Whole-genome sequencing of MDS and secondary AML samples demonstrated similar oligoclonality, with identification of a mean of 2.4 clones and 3.1 clones, respectively.19
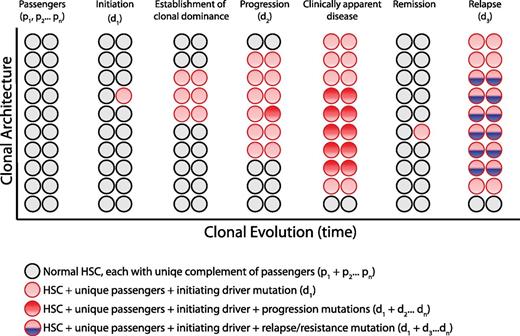
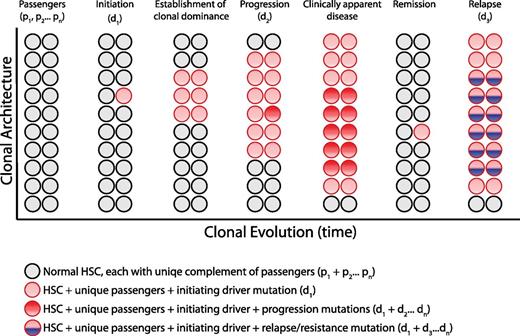
Clonal architecture and genetic heterogeneity. The normal bone marrow contains a heterogeneous mixture of equally represented HSC clones. Each normal HSC clone accumulates a unique set of passenger mutations throughout its lifespan, defining a clonal genetic mosaicism of normal hematopoiesis that is undetectable by bulk sequencing analysis. When 1 clone acquires a driver mutation that causes a selective growth advantage, depicted with a red circle, the progeny of this mutated cell gain clonal dominance and may cause clinically apparent disease. Within this dominant clone, additional mutations can cooperate to drive further clonal advantage, illustrated with darker red circles, and define genetically distinct subclones. Upon treatment, a myeloid malignancy can be driven into remission, although the persistence of any residual disease clones can serve as the substrate for relapse, potentially bearing a new mutation, depicted in blue.
Clonal architecture and genetic heterogeneity. The normal bone marrow contains a heterogeneous mixture of equally represented HSC clones. Each normal HSC clone accumulates a unique set of passenger mutations throughout its lifespan, defining a clonal genetic mosaicism of normal hematopoiesis that is undetectable by bulk sequencing analysis. When 1 clone acquires a driver mutation that causes a selective growth advantage, depicted with a red circle, the progeny of this mutated cell gain clonal dominance and may cause clinically apparent disease. Within this dominant clone, additional mutations can cooperate to drive further clonal advantage, illustrated with darker red circles, and define genetically distinct subclones. Upon treatment, a myeloid malignancy can be driven into remission, although the persistence of any residual disease clones can serve as the substrate for relapse, potentially bearing a new mutation, depicted in blue.
In contrast to bulk tumor sequencing, single-cell techniques can evaluate genetic heterogeneity directly. Using targeted sequencing of single-cell–derived hematopoietic colonies, complex clonal relationships can be visualized with confidence, affording key insights into tumor evolution.4,82,83 However, colony assays are labor intensive, poorly scalable, and susceptible to in vitro selection bias. Recent technologic and computational advances in single-cell genomics, including both RNA and DNA sequencing, raise the hope that large-scale, unbiased characterization of single cells may soon be possible. Whole-exome sequences generated from 58 single cells from a primary human JAK2-negative myeloproliferative neoplasm (MPN) confirmed disease clonality and enabled inferences about genetic events underlying disease initiation and progression.84
Clinical implications
The fundamental objectives of clinical cancer sequencing are to predict the vulnerability of a particular tumor genotype to specific therapeutic interventions and to stratify an individual patient’s risk of disease-associated morbidity and mortality. To that end, we are approaching a complete catalog of all recurrent, disease-causing exonic mutations and beginning to build a robust evidence base of genotype-phenotype associations. Several opportunities for immediate clinical utility and investigation are clear.
Prediction of prognosis
Somatic genetic alterations are already integrated into well-accepted prognostic models for a range of myeloid malignancies, including MDS (Revised International Prognostic Scoring System [IPSS-R]),85 AML (European LeukemiaNet),86 and primary myelofibrosis (Dynamic International Prognostic Scoring System–plus [DIPSS-plus]).87 These risk models combine clinicopathological variables with cytogenetic and molecular genetic findings to stratify patients into distinct risk groups that serve as a rational basis for clinical management decisions. Inclusion of additional genetic data can improve prognostic risk models in both MDS and AML.3,30 A more refined understanding of disease-intrinsic risk will be immediately useful in the clinic, facilitating more confident risk-benefit calculations regarding the timing and modality of treatment.
Prediction of therapeutic response
Somatic genetic alterations are important in driving the clinical and biological heterogeneity of myeloid malignancies, raising the possibility that sensitivity to specific therapies will be enriched in distinct, genetically-defined subsets of patients. One retrospective analysis using a targeted resequencing strategy in young (<60 years of age), mostly de novo AML patients showed that MLL translocations or mutations in DNMT3A or NPM1 predict for improved survival with anthracycline dose intensification (90 vs 45 mg/m2) during remission induction therapy.30 At present, there are few molecular targeted therapies available for patients with myeloid malignancies, but as more novel, genotype-directed agents reach the clinic, such as IDH inhibitors, sequencing will be critical for matching therapies with specific tumor genetic contexts. Comprehensive diagnostic sequencing may also aid in identifying preexisting genetic mechanisms of treatment resistance, such as TP53 mutations in lenalidomide-treated del(5q) MDS.88
Diagnosis
Genetic analyses have the potential to demonstrate clonality and to identify mutations that are universally pathological. Determination of clonality is a key criterion for the diagnosis of myeloid malignancies, particularly those such as MDS and MPNs that lack a single unifying pathological finding and where interobserver agreement can be problematic.89,90 Specific disease-defining genetic abnormalities, such as t(9;22)(q34;q11.2) in chronic myeloid leukemia, or JAK2-V617F in MPNs, are already integrated into World Health Organization (WHO) diagnostic criteria.91 The demonstration of clonality by the presence of a clonal, somatic mutation in the bone marrow or peripheral blood can have clinical utility. For example, in the clinical evaluation of patients with marked platelet elevation, clonality assessment is particularly useful in distinguishing reactive thrombocytosis from essential thrombocythemia. Of note, clonality, by itself, is not diagnostic of disease. Some individuals have clonal hematopoiesis with mutations or deletions of specific genes, such as TET2, without overt evidence of disease.53,54 It remains to be determined whether clonality alone, absent clinically significant hematopoietic functional defects, confers an increased risk of developing a bona fide myeloid malignancy.
Heterogeneity
Tumor heterogeneity has clear implications for understanding sensitivity and resistance to therapy. In some cases, genetic mechanisms of resistance may already be present at the time of diagnosis. Indeed, in patients with low-risk del(5q) MDS, the detection of a minor subclone harboring a TP53 mutation predicts for decreased response to lenalidomide and a more rapid transformation to acute leukemia.88 In a clinical trial setting, serial monitoring of clonal dynamics may prove to be a useful tool in evaluating the effectiveness of investigational agents. For example, the selective disappearance of a genetic subclone would be powerful evidence of on-target therapeutic efficacy, even in the context of a minor hematologic response. Identification of subclonal treatment responses could afford insight into factors that influence drug sensitivity and direct future clinical trial design to specific tumor genetic contexts.
The ability to distinguish initiating mutations (clonal drivers) from progression mutations (subclonal drivers) is conceptually important in considering the application of molecularly targeted treatment strategies in the myeloid malignancies. Achievement of disease cure relies on eradication of the entire complement of tumor-initiating stem cells because these serve as a reservoir of treatment resistance.18 As such, focused treatment of the founder clone would be most likely to render a patient free of disease. At the same time, there may be clinical value in selectively targeting specific subclones. For example, treating abundant or aggressive subclones could be useful in relieving symptoms, restoring functional hematopoiesis, or achieving cytoreduction prior to stem cell transplantation.
Novel therapeutics
Unbiased genome surveys have revealed recurrent mutations in components of multiple core biological pathways whose involvement in myeloid cancer biology may confer susceptibility to novel, targeted therapies. The prospect of rapid clinical genotyping raises the hope that specific drugs may eventually be paired with a particular tumor genomic context. Realization of this goal is complicated by the fact that myeloid driver genes are often subject to loss-of-function alterations, rendering conventional therapeutic strategies somewhat challenging. Alternative approaches, such as targeting a tumor’s obligate compensatory mechanisms, are aimed at inducing synthetic lethality and may be particularly useful in the context of malignancies with spliceosomal dysfunction, defective DNA damage response, or impaired cohesin function. Other approaches target core processes that appear critical for disease biology. In this respect, epigenetic pathways have been attractive targets for the development of novel therapies because of the high frequency of somatic alterations in epigenetic modifiers and prior empiric success with the DNMT inhibitors, 5-azacytidine (azacitidine) and 5-aza-2′-deoxycytidine (decitabine). A number of novel therapies aimed at restoring the epigenome to a normal state are currently in preclinical development or early-phase clinical trials.92
Conclusions
In the early 2000s, an attractive paradigm of leukemia pathogenesis was proposed, in which acquired genetic alterations could be neatly allocated to 2 basic functional classes, those that confer a proliferative or survival advantage (class I) and those that induce a differentiation blockade (class II).93,94 The model was fueled by discovery of highly recurrent mutations in kinase signaling pathways (FLT3 and JAK2) and balanced translocations involving lineage-defining transcription factors. The possibility of collapsing hundreds of distinct genetic alterations into 2 unifying biological groups engendered enthusiasm for a broadly applicable therapeutic rationale aimed at targeting these core pathways. However, a more comprehensive genomic analysis has now revealed a striking complexity that defies this straightforward dichotomy.
Recurrent mutations affecting basic cellular pathways, such as RNA splicing, epigenome regulation, and chromosome dynamics reveal a wide spectrum of pathways capable of driving clonal dominance. Despite apparent biological heterogeneity, the affected pathways are unified by their influence on the cellular transcriptional state. Derangement of the stem cell self-renewal program represents a core biological principle of myeloid oncogenesis that underlies the ability of mutations to promote clonal dominance at the stem cell level. The disparate clinical phenotypes associated with specific mutations may be attributable to the dynamic coregulation of HSC self-renewal and lineage-specific differentiation.
The complete list of common, recurrent exonic mutations in the myeloid malignancies will soon be complete. We have learned from large-scale sequencing efforts that relatively few genes are mutated at a high frequency and that most recurrently affected genes reside in the so-called “long tail” of the mutation distribution, occurring at a frequency of a few percent or less.95 Within this “long tail,” the capacity to resolve robust clinical and biological associations has been limited by insufficient statistical power, a problem only addressed by continued sequencing of well-annotated primary patients samples. In parallel, ongoing efforts in genetic discovery will help determine whether somatic mutations in the nonprotein coding genome have important biological and clinical roles in the pathobiology of myeloid malignancies.
The genes identified by genome sequencing have transformed our understanding of the key biological pathways that drive myeloid malignancies. These discoveries are being rapidly investigated, revealing mechanistic insights into myeloid transformation. New mouse models are being created using disease-causing alleles, and new therapeutic strategies are being developed. Most immediately, the commonly mutated genes are being studied in a clinical setting with the aim of improving the diagnosis and classification of myeloid malignancies and enhance the prediction of prognosis and response to therapy.
Authorship
Contribution: R.C.L. and B.L.E. reviewed the literature, cowrote the manuscript, and made the figures.
Conflict-of-interest disclosure: B.L.E. has received consulting fees from Genoptix, Celgene, and H3 Biomedicine. R.C.L. declares no competing financial interests.
Correspondence: Benjamin L. Ebert, Brigham and Women’s Hospital, Karp Research Building, CHRB 05.210, 1 Blackfan Circle, Boston, MA 02115; e-mail: bebert@partners.org.